RIG-I is important in recognizing viral RNA and stimulating the immune system. Human RIG-I protein was crystallized in complex with 18-mer double-stranded RNA to study the recognition mechanism.
Keywords: retinoic acid inducible gene-I
Abstract
Retinoic acid inducible gene-I (RIG-I) is an essential component of the innate immune system that is responsible for the detection and elimination of invading viruses. RIG-I recognizes viral RNAs inside the cell and then initiates downstream signalling to activate the IRF-3 and NF-κB genes, which results in the production of type I interferons. RIG-I is composed of an N-terminal CARD domain for signalling and C-terminal helicase and repressor domains for RNA recognition. A RIG-I–RNA binding assay was performed to investigate the in vitro RIG-I–RNA binding properties. Selenomethionine-incorporated RIG-I was expressed using Escherichia coli and purified for crystallization. X-ray data were collected from RIG-I–dsRNA complex crystals to 2.8 Å resolution using synchrotron radiation.
1. Introduction
When we are infected with pathogens, it is important to detect the presence of the pathogenic molecules immediately through the innate immune system in order to prevent the progression of infectious diseases. RIG-I (retinoic acid inducible gene-I) is an element of the innate immune system that is responsible for the detection of the RNA molecules of invading viruses. RIG-I is located in the cytoplasm and the accumulation of viral RNA inside the cell activates a RIG-I-mediated signalling cascade that produces type I interferons (IFNs) (Yoneyama et al., 2004 ▶; Meylan & Tschopp, 2006 ▶). In addition to RIG-I, there are two other proteins in the cytoplasm that can recognize viral RNA: melanoma differentiation-associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2) (Yoneyama et al., 2005 ▶). Conversely, toll-like receptor 3 (TLR-3), which also detects viral RNA, is primarily located in the endosome, where an acidic pH is required for the recognition of RNA (Matsumoto et al., 2003 ▶).
RIG-I is composed of an N-terminal caspase activation and recruitment domain (CARD), which is involved in signalling via the adaptor molecule IPS-1, and C-terminal helicase and repressor domains (RD) for viral RNA recognition (Saito et al., 2007 ▶; Yoneyama et al., 2005 ▶). The RD recognize viral RNAs and are believed to undergo viral RNA-induced conformational changes that enable the CARD domain to interact with the CARD domain of the downstream IPS-1 protein (Kawai et al., 2005 ▶; Xu et al., 2005 ▶; Seth et al., 2005 ▶; Meylan et al., 2005 ▶). This interaction initiates the signalling pathway that leads to IRF-3 and NF-κB activation and subsequent induction of IFNs (Yoneyama & Fujita, 2004 ▶; Heim, 2005 ▶; Cui et al., 2008 ▶)
RIG-I is known to recognize various types of RNA molecules including short double-stranded RNA (dsRNA) with or without overhangs and single-stranded RNA with a 5′-triphosphate group (Hornung et al., 2006 ▶; Pichlmair et al., 2006 ▶; Takahasi et al., 2008 ▶; Kato et al., 2008 ▶). RIG-I–RNA binding is known to be sequence-nonspecific, although some sequence preferences have been reported (Saito et al., 2008 ▶; Uzri & Gehrke, 2009 ▶). In order to understand the mechanism by which RIG-I recognizes viral RNAs, we conducted RIG-I–RNA binding assays and carried out crystallization and preliminary X-ray crystallographic studies of RIG-I (C-terminal domain) with bound double-stranded RNA.
2. Materials and methods
2.1. Cloning, expression and purification
Human RIG-I (residues 218–925) was cloned into the pET30b (Novagen) vector and contained a linker and six successive histidine residues (–AAALEHHHHHH) at the C-terminus that were not cleaved prior to crystallization. The expression construct was transformed into Escherichia coli BL21 (DE3) strain (Novagen) and cultured using M9 minimal media containing selenomethionine, six other amino acids (Lys, Thr, Phe, Leu, Ile and Val) and 50 µg ml−1 kanamycin at 310 K until the OD at 600 nm reached approximately 0.7. The temperature was lowered to 297 K and isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM. After overnight incubation, the cells were harvested by centrifugation and disrupted by sonication in ice-cold buffer A (50 mM sodium phosphate pH 7.0 and 500 mM NaCl). The lysate was then clarified by centrifugation at 12 000g for 60 min. The protein was first purified using an Ni Sepharose Fast Flow column (GE Healthcare) with gradient elution from 50 to 400 mM imidazole in buffer A. The fractions containing RIG-I were concentrated and applied onto a Superdex 200 column (GE Healthcare) in 10 mM Tris–HCl pH 7.5, 250 mM NaCl, 1 mM EDTA and 1 mM DTT. The purified protein was concentrated to a final concentration of 6–10 mg ml−1 using centrifugal filter devices (Microcon Centrifugal Filters, nominal molecular-weight limit 10 000; Millipore). The purity of the protein was analyzed by Coomassie-blue-stained SDS–PAGE.
2.2. Crystallization
All crystallization trials were conducted using the hanging-drop vapour-diffusion method by mixing equal volumes (0.5 µl) of the protein and reservoir solutions and equilibrating the mixed solutions against 500 µl reservoir solution in a 24-well plate at 293 K. The protein solution was made by mixing 6 µl 5.7 mg ml−1 (70 µM) purified human RIG-I (218–925) in 10 mM Tris–HCl pH 7.5, 250 mM NaCl, 1 mM EDTA, 1 mM DTT and 0.6 µl 1.5 mM 18-mer dsRNA (see §2.3 for RNA composition) in 10 mM Tris–HCl pH 7.5 and 100 mM NaCl to give a 1:2.2 molar ratio with excess dsRNA. The conditions under which crystalline precipitates appeared were optimized by changing the concentration of the precipitant and the pH of the buffer solutions. The optimized reservoir solution consisted of 1.46 M sodium citrate and 0.1 M HEPES pH 7.0. Crystals with dimensions of 0.05 × 0.01 × 0.01 mm appeared in approximately four weeks.
2.3. Binding assay
The 18-mer single-stranded RNAs (5′-GGGUUUUGGGGUUUUGGG and 5′-CCCAAAACCCCAAAACCC) used for the crystallization and binding assays were purchased from Dharmacon Inc. (USA). The two complementary single-stranded RNAs were mixed in equal amounts in 10 mM Tris–HCl pH 7.5 and 100 mM NaCl, heated to 363 K and slowly cooled to room temperature to form a blunt-ended 18-mer double-stranded RNA without a 5′-triphosphate group. Human RIG-I protein was mixed with 18-mer dsRNA in 100 mM HEPES pH 7.0 and incubated at room temperature for 20 min prior to analysis on a 15% TBE–polyacrylamide gel. The gel was run at 100 V for 60 min on ice and the RNA was then stained by incubation with 0.005% ethidium bromide solution and the stained bands were recorded using an Image Quant 300 imaging system (GE Healthcare).
2.4. Data collection and processing
Crystals were transferred into a cryoprotectant solution composed of reservoir solution with 5–10%(v/v) glycerol and then flash-frozen in liquid N2. The crystals were placed in the 100 K nitrogen-gas stream of an Oxford Cryojet system on the MAXII6C beamline equipped with an ADSC Q210 CCD detector at Pohang Accelerator Laboratory (PAL, Korea). The data were collected using a 1° oscillation per image with a crystal-to-detector distance of 200 mm. The crystal was exposed to X-rays for 60 s per image and a total of 360 frames were recorded. A data set was collected to 2.80 Å resolution from a single crystal and was processed using the HKL-2000 software package (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
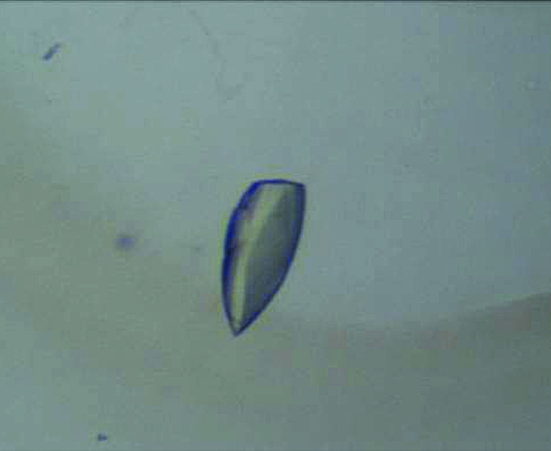
Human RIG-I (218–925) was overexpressed in E. coli using M9 media to replace methionine by selenomethionine in order to enable structure determination by MAD analysis. The human RIG-I (218–925) protein was purified for crystallization experiments using Ni-affinity and Superdex 200 size-exclusion columns. RIG-I–double-stranded RNA complex crystals were obtained in 1.32–1.48 M sodium citrate and 0.1 M HEPES pH 7.0 (Fig. 1 ▶). The data-collection statistics for the RIG-I (218–925)–18-mer dsRNA complex crystal are summarized in Table 1 ▶. The crystal diffracted to 2.80 Å resolution and belonged to the monoclinic space group C2, with unit-cell parameters a = 78.45, b = 45.29, c = 143.24 Å, β = 100.55°. Assuming the presence of one molecule of RIG-I–18-mer dsRNA in the asymmetric unit, the Matthews coefficient was 1.70 Å3 Da−1 (Matthews, 1968 ▶).
Figure 1.
Crystals of human RIG-I (218–925)–18-mer dsRNA complex.
Table 1. Data-collection statistics of the human RIG-I (218–925)–18-mer dsRNA complex crystal.
Values in parentheses are for the last resolution shell.
| Wavelength (Å) | 0.979 |
| Temperature (K) | 100 |
| Oscillation range (°) | 1 |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 78.45, b = 45.29, c = 143.24, β = 100.55 |
| Resolution (Å) | 20–2.8 (2.9–2.8) |
| Reflections (total/unique) | 73657/11788 |
| Completeness (%) | 94.8 (95.6) |
| Redundancy | 6.2 (6.3) |
| 〈I/σ(I)〉 | 16.3 (1.4) |
| Rmerge† | 0.055 (0.519) |
R
merge = 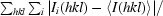
 .
.
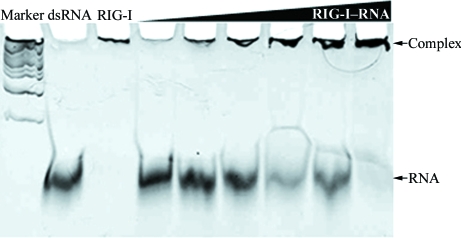
The RIG-I–RNA binding assay conducted at pH 7.0 shows that the intensity of the RNA band decreased as the concentration of the RIG-I protein increased (Fig. 2 ▶). The shift of the RNA band from the original position to the top of the gel near the the sample-loading well indicates an increase in the mass-to-charge ratio of the RNA. This shift was interpreted as the formation of a dsRNA–RIG-I protein complex because the migration of the highly negatively charged RNA will be reduced when bound to RIG-I. When the gel was stained with Coomassie blue to visualize protein, the bands at the top of the gel were stained, confirming the presence of protein in these bands (data not shown). Furthermore, our data showed that RIG-I (218–925) bound to the 18-mer dsRNA that was used in the crystallization experiments in a concentration-dependent manner.
Figure 2.
Human RIG-I (218–925)–18-mer dsRNA binding assay at pH 7.0 using an electrophoretic mobility shift assay. Human RIG-I proteins were mixed with 18-mer dsRNA in 100 mM HEPES pH 7.0. After incubation at room teperature for 20 min, the mixture was applied onto a 15% TBE–polyacrylamide gel and stained with ethidium bromide. The lanes are 1 kb DNA marker, 18-mer dsRNA, RIG-I protein and 18-mer dsRNA and RIG-I mixtures with RIG-I:dsRNA molar ratios of 0.12, 0.25, 0.5, 1, 2 and 4, respectively.
The crystals suffered from significant radiation damage during data collection; therefore, attempts to obtain improved crystals for structure determination are currently under way.
Acknowledgments
We thank the staff members of the Pohang Accelerator Laboratory MAXII6C beamline. This work was supported by the University of Seoul 2007 research fund.
References
- Cui, S., Eisenacher, K., Kirchhofer, A., Brzozka, K., Lammens, A., Lammens, K., Fujita, T., Conzelmann, K. K., Krug, A. & Hopfner, K. P. (2008). Mol. Cell, 29, 169–179. [DOI] [PubMed]
- Heim, M. H. (2005). J. Hepatol.42, 431–433. [DOI] [PubMed]
- Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., Poeck, H., Akira, S., Conzelmann, K. K., Schlee, M., Endres, S. & Hartmann, G. (2006). Science, 314, 994–997. [DOI] [PubMed]
- Kato, H., Takeuchi, O., Mikamo-Satoh, E., Hirai, R., Kawai, T., Matsushita, K., Hiiragi, A., Dermody, T. S., Fujita, T. & Akira, S. (2008). J. Exp. Med.205, 1601–1610. [DOI] [PMC free article] [PubMed]
- Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., Ishii, K. J., Takeuchi, O. & Akira, S. (2005). Nature Immunol.6, 981–988. [DOI] [PubMed]
- Matsumoto, M., Funami, K., Tanabe, M., Oshiumi, H., Shingai, M., Seto, Y., Yamamoto, A. & Seya, T. (2003). J. Immunol.171, 3154–3162. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Meylan, E., Curran, J., Hofmann, K., Moradpour, D., Binder, M., Bartenschlager, R. & Tschopp, J. (2005). Nature (London), 437, 1167–1172. [DOI] [PubMed]
- Meylan, E. & Tschopp, J. (2006). Mol. Cell, 22, 561–569. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pichlmair, A., Schulz, O., Tan, C. P., Naslund, T. I., Liljestrom, P., Weber, F. & Reis e Sousa, C. (2006). Science, 314, 997–1001. [DOI] [PubMed]
- Saito, T., Hirai, R., Loo, Y. M., Owen, D., Johnson, C. L., Sinha, S. C., Akira, S., Fujita, T. & Gale, M. Jr (2007). Proc. Natl Acad. Sci. USA, 104, 582–587. [DOI] [PMC free article] [PubMed]
- Saito, T., Owen, D. M., Jiang, F., Marcotrigiano, J. & Gale, M. Jr (2008). Nature (London), 454, 523–527. [DOI] [PMC free article] [PubMed]
- Seth, R. B., Sun, L., Ea, C. K. & Chen, Z. J. (2005). Cell, 122, 669–682. [DOI] [PubMed]
- Takahasi, K., Yoneyama, M., Nishihori, T., Hirai, R., Kumeta, H., Narita, R., Gale, M. Jr, Inagaki, F. & Fujita, T. (2008). Mol. Cell, 29, 428–440. [DOI] [PubMed]
- Uzri, D. & Gehrke, L. (2009). J. Virol.83, 4174–4184. [DOI] [PMC free article] [PubMed]
- Xu, L. G., Wang, Y. Y., Han, K. J., Li, L. Y., Zhai, Z. & Shu, H. B. (2005). Mol. Cell, 19, 727–740. [DOI] [PubMed]
- Yoneyama, M. & Fujita, T. (2004). Tanpakushitsu Kakusan Koso, 49, 2571–2578. [PubMed]
- Yoneyama, M., Kikuchi, M., Matsumoto, K., Imaizumi, T., Miyagishi, M., Taira, K., Foy, E., Loo, Y. M., Gale, M. Jr, Akira, S., Yonehara, S., Kato, A. & Fujita, T. (2005). J. Immunol.175, 2851–2858. [DOI] [PubMed]
- Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S. & Fujita, T. (2004). Nature Immunol.5, 730–737. [DOI] [PubMed]