Abstract
Nitric oxide synthase (NOS) inhibitors are potential drug candidates because it has been well demonstrated that excessive production of NO critically contributes to a range of diseases. Most inhibitors have been screened in vitro using recombinant enzymes, leading to the discovery of a variety of potent compounds. To make inhibition studies more physiologically relevant and bridge the gap between the in vitro assay and in vivo studies, we report here a cellular model for screening NOS inhibitors. Stable transformants were generated by overexpressing rat neuronal NOS in HEK 293T cells. The enzyme was activated by introducing calcium into cells, and its activity was assayed by determining the amount of nitrite that was formed in culture media using the Griess reagent. We tested a few NOS inhibitors with this assay and found that the method is sensitive, versatile, and easy to use. The cell-based assay provides more information than in vitro assays regarding the bioavailability of NOS inhibitors, and it is suitable for high-throughput screening.
Introduction
Nitric oxide (NO) is endogenously produced from L-arginine, catalyzed by nitric oxide synthases (NOS) [1]. This pleiotropic signaling molecule has several biological functions, including neurotransmission, regulation of blood-vessel tone, and the immune response [2], [3], [4], and [5]. Despite the pivotal role of NO under physiological conditions, recent studies have also unambiguously demonstrated that excess production of NO critically contributes to a range of diseases [2], [3], [5], and [6]. Hence, inhibition of NOS to decrease NO biosynthesis has been an attractive approach for the design of potential new drugs for diseases caused by NO overproduction [7], [8], [9], and [10]. Many NOS inhibitors have been developed and tested based on an in vitro assay using recombinant enzymes [8], [10], [11], [12], [13], and [14]. An in vitro assay is important for initial inhibitor screening and for enzyme mechanism studies. However, it is only the first step in drug development because in vitro results do not provide adequate information regarding bioavailability of the compounds. To bridge the gap between the in vitro assay and in vivo studies, we developed a cell-based neuronal NOS (nNOS) inhibition assay. A cell-based assay for iNOS is well documented [15], [16], [17], [18], and [19], because iNOS is easily induced in a variety of cells by different stimulants. Cell-based eNOS and nNOS inhibition methods have also been reported in recent years using radiolabeled materials or a rhodamine-based fluorescent probe [20] and [21]. The inhibition of eNOS was initially assayed in columns of vascular endothelial cells, using the relaxation of smooth muscle strips as a read-out [22]. More recently, NO production by eNOS was indirectly monitored in living cells via soluble guanylate cyclase activation and calcium ion influx [23]. Both of these methods, however, are very inconvenient to implement. We report here an alternative colorimetric assay, which is a convenient and easy-to-use method to study nNOS inhibition in human cells. Stable transformants were generated by overexpressing nNOS in HEK 293T cells (293T/nNOS), and the enzyme was activated by introducing calcium to the cells. The formation of nitrites, a stable metabolite of NO, was detected in the culture medium by the Griess reagent, which correlates with the enzyme activity.
Materials and Methods
Materials
Nω-Nitro-L-arginine (NNA) was purchased from Sigma (St. Louis, MO), 1400W was bought from Alexis (San Diego, CA), and N-[4-(6-amino-4-methylpyridin-2-ylmethyl)pyrrolidin-3-yl]-N′-(3-fluorophenylethyl) ethane-1,2-diamine (3) was synthesized in our lab [24].
Establishment of a stable cell line overexpressing rat nNOS
HEK 293T cells were grown in DMEM (Invitrogen, Carlsbad, CA) medium containing 10% FBS (Hyclone, Logan, UT)), 100 U/mL penicillin, and 0.1 mg/mL streptomycin in a 5% CO2 atmosphere. The cells were transfected with the rat nNOS-cDNA construct by the Lipofectamine method (Invitrogen) [25]. Transfectants were selected by Geneticin (0.8 mg/ml, Invitrogen) and isolated by single-cell cloning. nNOS expression was analyzed by immunoblotting, and the clone with the highest expression rate (293T/nNOS) was chosen for further experiments. The 293T/nNOS cells were cultured by the same procedure as WT 293T cells but with the addition of 0.4 mg/ml Geneticin in the DMEM medium.
Detection of protein expression and cell proliferation
The nNOS protein levels in WT 293T cells and 293T/nNOS cells were detected by Western blotting. For the cell proliferation assay, WT 293T cells and 293T/nNOS cells were plated in 96-well culture plates (5,000cells/well). Cell proliferation was detected after 24 h and 48 h by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assay [26].
Enzyme activation and inhibition
The 293T/nNOS cells were cultured in 6-well plates. When cell density reached ~80% confluence, the cells were treated with 5 μM A23187, and the amount of nitrite in media was quantified at different times with the Griess reagent [16] and [18]. For the inhibition assay, cells were treated with 5 μM A23187 (Calbiochem, San Diego, CA) and different concentrations of inhibitors for 8 h. The formation of nitrite in media was determined with the Griess reagent. The enzyme activity was assumed to be 100% without inhibitors (control). The enzyme activity in the presence of inhibitors was calculated compared to the control and was expressed as relative values.
Inhibition of nNOS in vitro
Recombinant rat nNOS was prepared as described before [27] and [28]. The method used for the in vitro inhibition assay was used [8] and [10].
Results
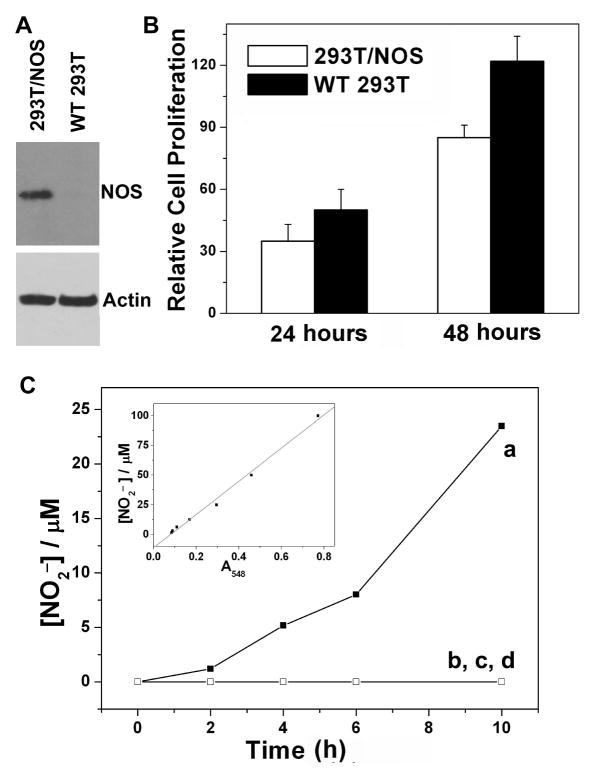
Stable 293T/nNOS transformants were generated; to verify the protein expression level, an immunoblot was performed. As shown in Figure 1A, a dramatic increase of nNOS in 293T/nNOS cells was detected, while almost no detectable protein was detected in the wild type (WT) HEK 293T cells. Actin was used as the loading control to ensure equal amounts of total proteins were loaded. To determine if overexpression of nNOS was toxic to the cells, the MTT assay was used. We found 293T/nNOS cells grew a little slower than did the WT cells (Fig. 1B), but no significant cell death was observed. Since nNOS was overexpressed, and this overexpression had no apparent toxicity to the cells, we tried to activate the nNOS in the cells. nNOS activity is tightly regulated by calcium [29]; however, under normal conditions the intracellular calcium concentration is extremely low (nM level compared to mM level in culture media). A previous report showed that calcium ionophore A23187 could induce an increase in intracellular calcium levels [30] and [25]. The 293T/nNOS cells or WT 293T cells were treated with or without A23187 for an indicated period of time, and it was found that only the 293T/nNOS cells produced nitrite, a metabolite of NO, under A23187 stimulation (Fig. 1C). This result indicated that the formation of nitrite predominated from the overexpressed nNOS, and the amount of nitrite production reflected nNOS activity in 293T/nNOS cells. After 2 hours of A23187 stimulation, a significant increase of nitrite was detected, and the nitrite production was time-dependent. There was no obvious cell death after 10 h with A23187 stimulation.
Figure 1.
A. Detection of nNOS expression in 293T/nNOS cells and WT 293T cells. B. Comparison of the cell proliferation rate of 293T/nNOS cells and WT 293T cells. The results are from three independent experiments and are expressed as mean ± S.D. C. Activation of nNOS in 293T/nNOS cells by A23187 (5 μM). Cells were treated with or without A23187 for indicated times, and the amount of nitrite in the culture media was quantified with the Griess reagent. 293T/nNOS cells were treated with (a) or without (b) A23187 and WT 293T cells were treated with (c) or without (d) A23187.
To determine if nNOS activation could be inhibited, we tested a variety of nNOS inhibitors on nNOS activity in 293T/nNOS cells. The potent, but non-selective, NOS inhibitor, Nω-nitro-L-arginine (NNA) was found to inhibit nitrite production in a dose-dependent fashion with an IC50 value of 9.9 μM (Fig. 2A). No cell death was observed under the experimental conditions. AR-R17477 (1), a potent nNOS inhibitor [31] and [32], and 1400W, (2), a selective inducible NOS (iNOS) inhibitor [33] and [34], also were tested (Fig. 2B). AR-R17477 greatly inhibited the cellular nNOS activity, while 1400W showed only a little inhibition under the experimental conditions. The cells remained healthy after treatment by NNA, AR-R17477, or 1400W during the period of A23187 stimulation.
Figure 2.
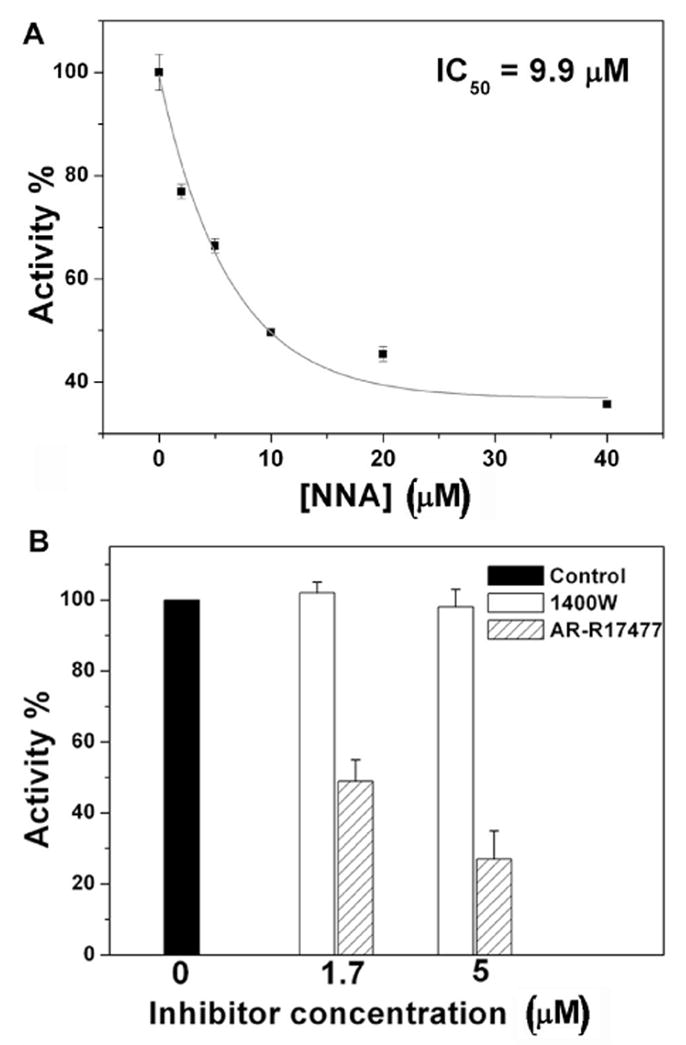
Inhibition of nNOS in 293T/nNOS cells. 293T/nNOS cells were treated with A23187 (5 μM) and different inhibitors. After 8 h, the amount of nitrite in the culture media was quantified by the Griess reagent. A. Dose-dependent inhibition of the enzyme by NNA. The results are from two independent experiments and are expressed as mean ± S.D. B. Inhibition of the enzyme by AR-R17477 and 1400W. The results are from two independent experiments and are expressed as mean ± S.D.
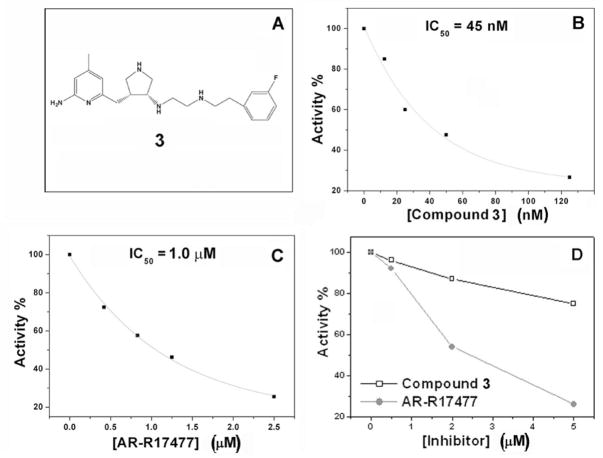
Two inhibitors were tested to compare inhibition of nNOS in vitro and in cells. As shown in Fig. 3, compound 3 was a much more potent inhibitor of nNOS than was AR-R17477 (1) in vitro (Fig. 3B,C). However, in the cell-based assay, AR-R17477 was more potent than 3 (Fig. 3D). Neither compound 3 nor AR-R17477 inhibited cell growth, and there was no obvious cytotoxicity under our experimental conditions (by visual inspection under a microscope), which excludes the possibility that the decreased formation of nitrite is caused by inhibition of cell growth or cell death.
Figure 3.
Inhibition of nNOS by 3 and AR-R17477 in vitro and in 293T/nNOS cells. A. Chemical structure of 3. B. Dose-dependent inhibition of the recombinant enzyme by 3 and C. by AR-R17477. D. Inhibition of the enzyme in 293T/nNOS cells by AR-R17477 and 3.
Discussion
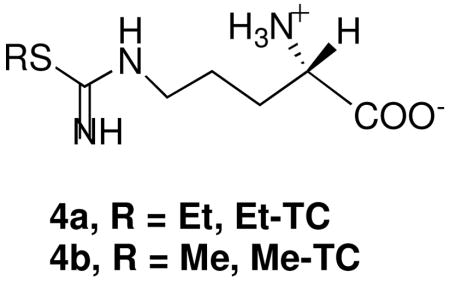
NO is involved in a variety of biological functions in vivo, although its overproduction is closely linked to several diseases [2], [3], [5] and [6]. Thus, regulation of NO production by inhibition of NOS is a potential approach for new drug development. At present, most NOS inhibitors are tested in vitro using recombinant enzymes. However, data from an in vitro assay does not necessarily reflect the in vivo effect. For example, S-ethyl-L-thiocitrulline (Et-TC) (4a) is more selective in vitro for nNOS inhibition over eNOS than is S-methyl-L-thiocitrulline (Me-TC) (4b); however, when both inhibitors were tested in rat tissue, Me-TC was better than Et-TC for inhibition of nNOS over eNOS [35].
We have generated stable 293T/nNOS transformants and have demonstrated expression of NOS relative to wild-type HEK 293T cells (Figure 1A). The overexpression of nNOS was not toxic to the cells (Figure 1B). Only the 293T/nNOS cells were activated by the calcium ionophore A23187 (Figure 1C), suggesting that an increase in calcium ion concentration is important to activation. The nNOS activity in 293T/nNOS cells was inhibited by NNA, a nonselective NOS inhibitor as well as by AR-R17477, a potent nNOS-selective inhibitor, but not by 1400W, an iNOS-selective inhibitor (Figure 2). In all cases the cells remained healthy during A23187 stimulation. When 3, a potent and selective nNOS inhibitor and AR-R17477, a potent but less selective nNOS inhibitor, were compared in 293T/nNOS cells, only weak inhibition was observed for both compounds (Figure 3). The relatively weak inhibition of 3 and AR-R17477 in the cell-based assay indicates that these compounds do not penetrate the cellular membrane well, probably because of their polarity. Compound 3 is a much better inhibitor of nNOS than AR-R17477 in the in vitro assay; however, in the cell-based assay, we found AR-R17477 is more potent. The much higher polarity of 3 relative to AR-R17477, which is detrimental to membrane permeability, is apparent because of the greater inhibitory activity of AR-R17477 in cells, despite its lower potency in vitro.
Cell-based assays are good models to mimic in vivo activity and can provide useful feedback for lead modification. Several studies have used cellular models to test iNOS inhibition [16], [17], [18] and [19]. Unlike nNOS and eNOS, iNOS is inducible and calcium-independent, which makes a cell-based iNOS assay more straightforward; because it is inducible, DNA does not need to be transfected into the cells, and calcium is not needed to activate the enzyme. A cell-based eNOS assay is currently being developed. While this project was underway, Surup et al. reported the assay of some NOS inhibitors using Chinese hamster ovary (CHO) cells overexpressing nNOS and eNOS [20]. The authors also activated the enzymes by stimulation of the cells with A23187. However, they assayed the enzyme activities by measuring the formation of radioactive L-citrulline from radiolabelled L-arginine. In this assay, radioactive compounds had to be handled, and the cells had to be broken prior to extraction of the L-citrulline. Our method is rather simple; the production of nitrite is detected colorimetrically, which is especially suitable for high throughput screening. Another advantage of our assay is that we treat cells with inhibitors for a much longer time (8 hours) than the previous method (15 minutes) [20]. This longer incubation time gives more physiologically relevant information of inhibition and helps determine if the inhibitors are toxic to cells. The cells were treated with A23187 and inhibitors at the same time point. The longer incubation time should overcome the problem caused by inhibitor “lag time” (the time that an inhibitor takes to penetrate the cell membrane and reach a steady intracellular concentration). We can deduce from Fig. 1C that the amount of nitrite formation is extremely low (~ 0.5 μM) within the initial 30 min, which is negligible compared to the nitrite concentration after the 8 h-incubation (~ 18 μM). Wood et al. investigated nNOS inhibition in cells using a rhodamine-based fluorescent probe DAR-4M [21]. It is well known that many rhodamine-based fluorescent probes are versatile reagents for detecting reactive oxygen species (ROS) or reactive nitrogen species (RNS) [36]. Furthermore, recent reports have shown that DAR-4M also readily reacts with dehydroascobic acid (DHA) and gives the same fluorescence spectra as does NO [37] and [38]. Therefore, the specificity of this fluorescence-based method is not apparent. Under appropriate conditions, NO undergoes a series of reactions to form the stable final products, nitrite and nitrate. The measurement of nitrite and nitrate is commonly used as a rapid and simple method to detect NO production [18], [39], and [40]. Since the ratio of nitrite to nitrate is frequently the same (usually 3:2 in cultured cells) [18], the measurement of nitrite alone is sufficient to serve as an indicator of NOS activity.
In conclusion, in vitro assays do not mimic the in vivo environment, and, therefore, may not give an accurate assessment of drug effects in vivo. We have demonstrated a convenient method to detect NOS activity in cells. The cell-based colorimetric assay described here is easy-to-use and is suitable for high throughput screening. It should provide more relevant data for NOS inhibitor-based drug development than isolated enzyme assays.
Acknowledgments
We are grateful for financial support from the National Institutes of Health (GM049725). We thank Dr. Haitao Ji for compound 3.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Griffith OW, Stuehr DJ. Nitric oxide synthases: properties and catalytic mechanism. Annu Rev Physiol. 1995;57:707–736. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- 2.Calabrese V, Mancuso C, Calvani M, Rizzarelli E, Butterfield DA, Stella AM. Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat Rev Neurosci. 2007;8:766–775. doi: 10.1038/nrn2214. [DOI] [PubMed] [Google Scholar]
- 3.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rastaldo R, Pagliaro P, Cappello S, Penna C, Mancardi D, Westerhof N, Losano G. Nitric oxide and cardiac function. Life Sci. 2007;81:779–793. doi: 10.1016/j.lfs.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 5.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 6.Moncada S, Bolanos JP. Nitric oxide, cell bioenergetics and neurodegeneration. J Neurochem. 2006;97:1676–1689. doi: 10.1111/j.1471-4159.2006.03988.x. [DOI] [PubMed] [Google Scholar]
- 7.Salerno L, Sorrenti V, Di Giacomo C, Romeo G, Siracusa MA. Progress in the development of selective nitric oxide synthase (NOS) inhibitors. Curr Pharm Des. 2002;8:177–200. doi: 10.2174/1381612023396375. [DOI] [PubMed] [Google Scholar]
- 8.Ji H, Gomez-Vidal JA, Martasek P, Roman LJ, Silverman RB. Conformationally restricted dipeptide amides as potent and selective neuronal nitric oxide synthase inhibitors. J Med Chem. 2006;49:6254–6263. doi: 10.1021/jm0604124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nason DM, Heck SD, Bodenstein MS, Lowe JA, 3rd, Nelson RB, Liston DR, Nolan CE, Lanyon LF, Ward KM, Volkmann RA. Substituted 6-phenyl-pyridin-2-ylamines: selective and potent inhibitors of neuronal nitric oxide synthase. Bioorg Med Chem Lett. 2004;14:4511–4514. doi: 10.1016/j.bmcl.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 10.Hah JM, Roman LJ, Martasek P, Silverman RB. Reduced amide bond peptidomimetics. (4S)-N-(4-amino-5-[aminoakyl]aminopentyl)-N′-nitroguanidines, potent and highly selective inhibitors of neuronal nitric oxide synthase. J Med Chem. 2001;44:2667–2670. doi: 10.1021/jm0101491. [DOI] [PubMed] [Google Scholar]
- 11.Hevel JM, Marletta MA. Nitric-oxide synthase assays. Methods Enzymol. 1994;233:250–258. doi: 10.1016/s0076-6879(94)33028-x. [DOI] [PubMed] [Google Scholar]
- 12.Ijuin R, Umezawa N, Higuchi T. Design, synthesis, and evaluation of new type of L-amino acids containing pyridine moiety as nitric oxide synthase inhibitor. Bioorg Med Chem. 2006;14:3563–3570. doi: 10.1016/j.bmc.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 13.Seo J, Igarashi J, Li H, Martasek P, Roman LJ, Poulos TL, Silverman RB. Structure-based design and synthesis of N(omega)-nitro-L-arginine-containing peptidomimetics as selective inhibitors of neuronal nitric oxide synthase. Displacement of the heme structural water. J Med Chem. 2007;50:2089–2099. doi: 10.1021/jm061305c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seo J, Martasek P, Roman LJ, Silverman RB. Selective L-nitroargininylaminopyrrolidine and L-nitroargininylaminopiperidine neuronal nitric oxide synthase inhibitors. Bioorg Med Chem. 2007;15:1928–1938. doi: 10.1016/j.bmc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naureckiene S, Edris W, Ajit SK, Katz AH, Sreekumar K, Rogers KE, Kennedy JD, Jones PG. Use of a murine cell line for identification of human nitric oxide synthase inhibitors. J Pharmacol Toxicol Methods. 2007;55:303–313. doi: 10.1016/j.vascn.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Davey DD, Adler M, Arnaiz D, Eagen K, Erickson S, Guilford W, Kenrick M, Morrissey MM, Ohlmeyer M, Pan G, Paradkar VM, Parkinson J, Polokoff M, Saionz K, Santos C, Subramanyam B, Vergona R, Wei RG, Whitlow M, Ye B, Zhao ZS, Devlin JJ, Phillips G. Design, synthesis, and activity of 2-imidazol-1-ylpyrimidine derived inducible nitric oxide synthase dimerization inhibitors. J Med Chem. 2007;50:1146–1157. doi: 10.1021/jm061319i. [DOI] [PubMed] [Google Scholar]
- 17.Tseng HY, Wu SH, Huang WH, Wang SF, Yang YN, Mahindroo N, Hsu T, Jiaang WT, Lee SJ. Benzothiazolium compounds: novel classes of inhibitors that suppress the nitric oxide production in RAW264.7 cells stimulated by LPS/IFNgamma. Bioorg Med Chem Lett. 2005;15:2027–2032. doi: 10.1016/j.bmcl.2005.02.063. [DOI] [PubMed] [Google Scholar]
- 18.Xie QW. Inducible Nitric Oxide Synthase Expression. Current Protocols in Toxicology. 2000:10.9.1–10.9.16. doi: 10.1002/0471140856.tx1009s04. [DOI] [PubMed] [Google Scholar]
- 19.Murakami A, Gao G, Omura M, Yano M, Ito C, Furukawa H, Takahashi D, Koshimizu K, Ohigashi H. 1,1-Dimethylallylcoumarins potently suppress both lipopolysaccharide- and interferon-gamma-induced nitric oxide generation in mouse macrophage RAW 264.7 cells. Bioorg Med Chem Lett. 2000;10:59–62. doi: 10.1016/s0960-894x(99)00578-8. [DOI] [PubMed] [Google Scholar]
- 20.Surup F, Wagner O, von Frieling J, Schleicher M, Oess S, Muller P, Grond S. The iromycins, a new family of pyridone metabolites from Streptomyces sp. I. Structure, NOS inhibitory activity, and biosynthesis. J Org Chem. 2007;72:5085–5090. doi: 10.1021/jo0703303. [DOI] [PubMed] [Google Scholar]
- 21.Wood MW, Hastings RC, Sygowski LA. A homogeneous fluorescent cell-based assay for detection of heterologously expressed nitric oxide synthase activity. J Biomol Screen. 2005;10:849–855. doi: 10.1177/1087057105280640. [DOI] [PubMed] [Google Scholar]
- 22.Moncada S, Palmer RM, Gryglewski RJ. Mechanism of action of some inhibitors of endothelium-derived relaxing factor. Proc Natl Acad Sci U S A. 1986;83:9164–8. doi: 10.1073/pnas.83.23.9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wunder F, Buehler G, Hüser J, Mundt S, Bechem M, Kalthof B. A cell-based nitric oxide reporter assay useful for the identification and characterization of modulators of the nitric oxide/guanosine 3′,5′-cyclic monophosphate pathway. Anal Biochem. 2007;363:219–227. doi: 10.1016/j.ab.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Ji H, Li H, Martásek P, Roman LJ, Poulos TL, Silverman RB. Discovery of highly potent and selective inhibitors of neuronal nitric oxide synthase by fragment hopping. J Med Chem. 2009;52:779–797. doi: 10.1021/jm801220a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci U S A. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee L, Davis R, Vanderham J, Hills P, Mackay H, Brown T, Mooberry SL, Lee M. 1,2,3,4-Tetrahydro-2-thioxopyrimidine analogs of combretastatin-A4. Eur J Med Chem. 2008;43:2011–2015. doi: 10.1016/j.ejmech.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 27.Gerber NC, Ortiz de Montellano PR. Neuronal nitric oxide synthase. Expression in Escherichia coli, irreversible inhibition by phenyldiazene, and active site topology. J Biol Chem. 1995;270:17791–17796. doi: 10.1074/jbc.270.30.17791. [DOI] [PubMed] [Google Scholar]
- 28.Roman LJ, Sheta EA, Martasek P, Gross SS, Liu Q, Masters BS. High-level expression of functional rat neuronal nitric oxide synthase in Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:8428–8432. doi: 10.1073/pnas.92.18.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorren AC, Mayer B. Nitric-oxide synthase: a cytochrome P450 family foster child. Biochim Biophys Acta. 2007;1770:432–445. doi: 10.1016/j.bbagen.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 31.Fedorov R, Vasan R, Ghosh DK, Schlichting I. Structures of nitric oxide synthase isoforms complexed with the inhibitor AR- R17477 suggest a rational basis for specificity and inhibitor design. Proc Natl Acad Sci U S A. 2004;101:5892–5897. doi: 10.1073/pnas.0306588101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reif DW, McCarthy DJ, Cregan E, Macdonald JE. Discovery and development of neuronal nitric oxide synthase inhibitors. Free Radic Biol Med. 2000;28:1470–1477. doi: 10.1016/s0891-5849(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, Nikolic D, Van Breemen RB, Silverman RB. Mechanism of inactivation of inducible nitric oxide synthase by amidines. Irreversible enzyme inactivation without inactivator modification. J Am Chem Soc. 2005;127:858–68. doi: 10.1021/ja0445645. [DOI] [PubMed] [Google Scholar]
- 34.Garvey EP, Oplinger JA, Furfine ES, Kiff RJ, Laszlo F, Whittle BJ, Knowles RG. 1400W is a slow, tight binding, and highly selective inhibitor of inducible nitric-oxide synthase in vitro and in vivo. J Biol Chem. 1997;272:4959–4963. doi: 10.1074/jbc.272.8.4959. [DOI] [PubMed] [Google Scholar]
- 35.Furfine ES, Harmon MF, Paith JE, Knowles RG, Salter M, Kiff RJ, Duffy C, Hazelwood R, Oplinger JA, Garvey EP. Potent and selective inhibition of human nitric oxide synthases. Selective inhibition of neuronal nitric oxide synthase by S-methyl-L-thiocitrulline and S-ethyl-L-thiocitrulline. J Biol Chem. 1994;269:26677–26683. [PubMed] [Google Scholar]
- 36.Wardman P. Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: progress, pitfalls, and prospects. Free Radic Biol Med. 2007;43:995–1022. doi: 10.1016/j.freeradbiomed.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 37.Ye X, Rubakhin SS, Sweedler JV. Simultaneous nitric oxide and dehydroascorbic acid imaging by combining diaminofluoresceins and diaminorhodamines. J Neurosci Methods. 2008;168:373–382. doi: 10.1016/j.jneumeth.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim WS, Ye X, Rubakhin SS, Sweedler JV. Measuring nitric oxide in single neurons by capillary electrophoresis with laser-induced fluorescence: use of ascorbate oxidase in diaminofluorescein measurements. Anal Chem. 2006;78:1859–1865. doi: 10.1021/ac051877p. [DOI] [PubMed] [Google Scholar]
- 39.Giustarini D, Rossi R, Milzani A, Dalle-Donne I. Nitrite and nitrate measurement by Griess reagent in human plasma: evaluation of interferences and standardization. Methods Enzymol. 2008;440:361–380. doi: 10.1016/S0076-6879(07)00823-3. [DOI] [PubMed] [Google Scholar]
- 40.Giustarini D, Dalle-Donne I, Colombo R, Milzani A, Rossi R. Adaptation of the Griess reaction for detection of nitrite in human plasma. Free Radic Res. 2004;38:1235–1240. doi: 10.1080/10715760400017327. [DOI] [PubMed] [Google Scholar]