Abstract
DNA topoisomerase II (topo II) is an essential determinant of chromosome structure and function, acting to resolve topological problems inherent in recombining, transcribing, replicating and segregating DNA. In particular, the unique decatenating activity of topo II is required for sister chromatids to disjoin and separate in mitosis. Topo II exhibits a dynamic localization pattern on mitotic chromosomes, accumulating at centromeres and axial chromosome cores prior to anaphase. In organisms ranging from yeast to humans, a fraction of topo II is targeted for SUMO conjugation in mitotic cells, and here we review our current understanding of the significance of this modification. As we shall see, an emerging consensus is that in metazoans SUMO modification is required for topo II to accumulate at centromeres, and that in the absence of this regulation there is an elevated frequency of chromosome non-disjunction, segregation errors, and aneuploidy. The underlying molecular mechanisms for how SUMO controls topo II are as yet unclear. In closing, however, we will evaluate two possible interpretations: one in which SUMO promotes enzyme turnover, and a second in which SUMO acts as a localization tag for topo II chromosome trafficking.
Keywords: DNA topoisomerase II, SUMO, sumoylation, mitosis, centromere, kinetochore, spindle assembly checkpoint, Aurora B/Ipl1, chromosome segregation, chromosome dynamics
1. INTRODUCTION
Post-translational modification through the SUMO pathway has emerged as an important determinant of genome stability. While this requirement is clearly multi-faceted, increasing evidence points towards an especially prominent role for sumoylation in mitosis [1]. Studies in yeast and vertebrates have shown that a distinct set of SUMO conjugates accumulates in mitotic cells [2,3], and vertebrate SUMO isoforms display dynamic localization patterns on mitotic chromosomes [4]. More directly, blocking SUMO conjugation by interfering with SUMO E2 and E3 activities delays progression through mitosis and is associated with both severe chromosome segregation defects [2,5–11] and early embryonic lethality [12]. Recent proteomic approaches have identified a wide range of SUMO substrates with mitotic functions [13–16]. Insight into how SUMO conjugation and removal regulates individual substrates, however, will necessitate a considerably more detailed analysis.
An illustrative “case study” that highlights some of the complexities in dissecting functional roles for SUMO concerns DNA topoisomerase II (topo II or Top2 in yeast). Topo II was one of the first mitotic SUMO substrates to be described [7,17], and in this review we provide an overview of how sumoylation impacts topo II function. As a starting point, we first introduce some aspects of topo II catalysis, pharmacology and chromosome dynamics. Second, research on topo II sumoylation currently points towards a connection between this modification and topo II function at centromeres, leading us to discuss possible roles for topo II within centromeric chromatin. Finally, we examine topo II SUMO modification in two contexts: as an induced modification that occurs in response to topo II enzyme stress, and as a component of topo II regulation in mitosis.
2. OVERVIEW OF TOPO II
2.1. Reaction mechanism and pharmacology
Eucaryotic topo IIs comprise a highly conserved family of homodimeric enzymes [18,19]. Each subunit contains an N-terminal ATPase domain, a central DNA cleavage and ligation domain, and a variable C-terminal domain (CTD; Figure 1A). The CTD is not required for enzyme activity [20–23] but has interesting regulatory properties. For one thing, the CTD has been proposed to allow the enzyme to distinguish the handedness of DNA supercoils [24,25]. The CTD is also targeted by a variety of kinases, including casein kinase II [26–29], Polo kinases [30,31], and Aurora B [32]; the significance of these phosphorylation events is largely unknown. In addition, in budding yeast the CTD is a prominent SUMO modification site ([17,33]; Section 4.2).
Figure 1. SUMO consensus sites in topo II CTDs.
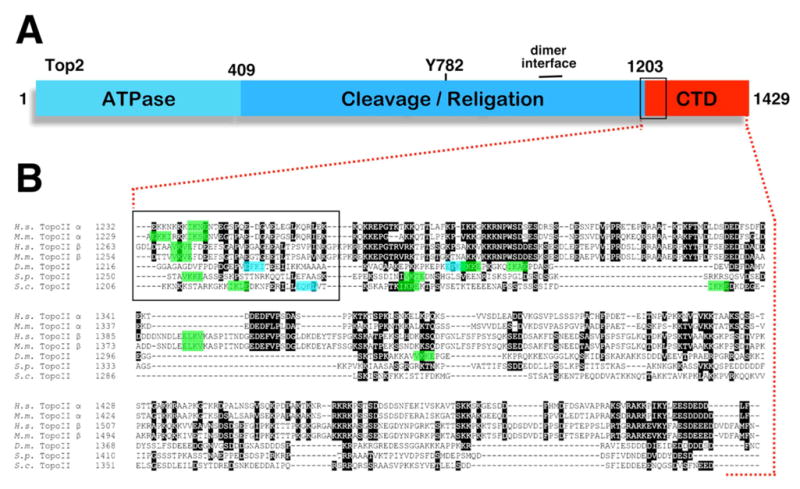
(A)Topo II domain structure. Numbers refer to amino acid coordinates for S. cerevisiae Top2. Y782 is the active site tyrosine. (B) Alignment of CTD sequences from human, mouse, Drosophila and yeast topo IIs. In these organisms, the boundary between the CTD and the cleavage/ligation domain has been mapped through limited proteolytic cleavage [23,180–183]. With the exception of human topo IIb (which starts right at the CTD boundary), this alignment is based on sequences starting 10 amino acids prior to the protease sensitive site. Highlighted green sequences correspond to the SUMO modification/Ubc9 recognition consensus sequence y-K-X-E/D, where y is a hydrophobic residue [184,185]. Blue sequences are inverted consensus sites, as there is evidence that this motif can also be targeted in the reverse orientation [186]. The box indicates a region close to the CTD boundary where most topo IIs we examined have at least one sumoylation consensus site. The exception was Drosophila, which has an inverted consensus site within this region and two forward consensus sequences that are just beyond the boxed area (VK1269KE, IK1278AE). Although their sequences are not included in this alignment, we also found SUMO modification consensus sites in what would equate to the boxed region in chicken topo II (IK1241SE), C. elegans topo II (VK1299KE, VK1312KE), and the probable Xenopus topo II (hypothetical protein LOC398512; IK1235GE, IK1276KE, VK1298KE).
Vertebrates express two isoforms of topo II, a and b [34]. Both forms are catalytically active and differ mainly in their CTD regions (Figure 1B). Although there is some functional overlap [35], topo IIa is largely responsible for the essential functions of the enzyme in chromosome structure and cell division [36]. Topo IIb, on the other hand, plays a critical role in neural development [37], possibly corresponding with roles for topo IIb in regulating gene expression [38,39].
Topo IIs modulate DNA topology through an ATP-dependent “cross inversion” mechanism [18,40,41]. During catalysis, each subunit cleaves opposing strands of a DNA helix (the G segment), forming a covalent linkage with the enzyme. This topo II-DNA adduct – the cleavable complex – is normally a short-lived reaction intermediate. When bound to ATP, the two ATPase domains form a clamp that quickly transports a second DNA segment (the T segment) through the DNA-protein gate, followed by re-ligation, ATP hydrolysis, and release of the enzyme. The T and G segments can be on the same or different DNA molecules, endowing topo II with the unique ability to catenate/decatenate and knot/unknot DNA. Topo II also shares functional overlap with topo I in relaxing (+) or (−) superhelical winding. Interestingly, topo II is actually more proficient than topo I at relieving helical tension on chromatin substrates, possibly because nucleosomes constrain the strand rotation required for topo I-mediated twist-diffusion [42]. Thus, there may be chromatin environments where topo II is specifically required to relax torsional strain.
Topo II is targeted by a variety of compounds classified as either poisons or catalytic inhibitors. Topo II poisons (e.g. etoposide/VP-16, doxorubicin/adriamycin, and amsacrine) interfere with the enzyme in ways that stabilize the cleavable complex and generate DNA breaks [43,44]. These clastogenic properties place topo II poisons among the most effective chemotherapeutic compounds in clinical use today [45], although treatment is associated with toxicity [46,47] and a predisposition to certain secondary tumors [48,49]. Catalytic inhibitors, on the other hand, block topo II at steps other than the cleavable complex [50,51]. In particular, bis-dioxopiperazine derivatives such as ICRF-187 and ICRF-193 act by stabilizing topo II in a closed clamp conformation, potentially trapping the enzyme on DNA [52,53]. The effects of these compounds may therefore not be equivalent to the absence of topo II function (for example, see [54]). To some extent, the same criticism can be levied against temperature sensitive top2 alleles in yeast, most of which have poorly defined effects on catalysis. Because of this, RNAi knockdown and cell lines that allow conditional depletion of topo II [55,56] are increasingly being used as experimental tools to assess topo II function.
2.2. Mitotic chromosome dynamics
During DNA replication, a fraction of the torsional strain associated with separating parental DNA strands is relieved by winding daughter chromosomes around each other, leading to the formation of catenates [57–59]. Topo II is the only enzyme that can resolve these linkages, and, consistent with this, early genetic and inhibitor studies revealed that the most prominent phenotype associated with perturbations to topo II was anaphase chromosome bridging, indicative of a partial defect in sister chromatid disjunction [60–69]. One process that may be particularly important in helping topo II drive out catenates is mitotic chromosome condensation, with the twisting and coiling of chromosomes into smaller structures imparting a direction to the topo II reaction that favors decatenation [70,71]. During mitotic chromosome assembly, there is a dynamic series of changes to topo II localization on chromosomes, suggesting topo II may be channeled to functionally relevant sites [72]. A brief description of this localization pattern is necessary for a consideration of how topo II is acting in mitosis.
As part of condensation, vertebrate chromosomes undergo a process called resolution, in which arm regions of chromosome pairs split and form separately coiled sister chromatids. Immuno-staining and live cell analyses have indicated that topo II localizes within the axial cores of chromatid arms, forming paired stripes running down the middle of the chromosomes [73–80] (this has not been universally observed, however [60,81–83], and has been suggested to potentially represent a specimen preparation artifact [83]). Chromatid resolution has been reported to require topo II activity, suggesting this may represent a step where a significant fraction of the catenate load is removed [84]. A second place where topo II localizes during mitotic chromosome assembly in vertebrates is at centromeres. In a variety of studies, topo II has been shown to accumulate prominently at the centromere/primary constriction during prophase or prometaphase, to persist there while chromosomes align upon the spindle, and to rapidly disperse following anaphase entry ([10,11,66,75,77,79,80,83,85–88]; Figure 2).
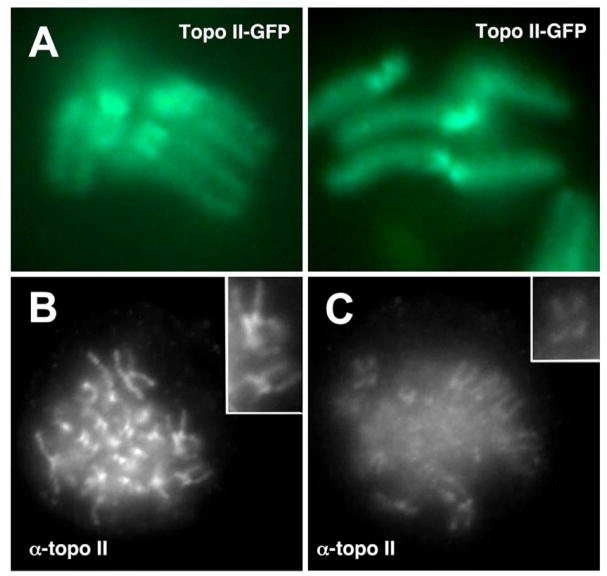
Figure 2. Topo II at vertebrate centromeres.
(A) Distribution of topo IIa-GFP on Indian muntjac chromosomes. Cell lines stably expressing human topo IIa-GFP were arrested with nocodazole and chromosome pairs were visualized in live cells. Images are courtesy of Duncan Clarke, Univ. of Minnesota Medical School. (B and C) Requirement for PIASg in topo II accumulation on mitotic chromosomes. HeLa cells were either left untreated (B) or depleted of PIASy using RNAi (C). Metaphase-arrested cells grown on glass coverslips were fixed with paraformaldehyde and immuno-stained to visualize topo II. Insets show slight enlargements of selected chromosomes. Data in B and C is reproduced here from [10] under the Creative Commons Attribution License and with the permission of D. Clarke.
For some time, the chromosomal population of topo II, especially topo II within axial cores, was considered to be relatively static, acting as a scaffold for anchoring chromosome loops [89–91]. Over the past several years, however, it has emerged that topo II exhibits robust catalytic activity on mitotic chromosomes [92,93], corresponding with specific regions of topo II cleavage within centromeric heterochromatin [94–98]. Moreover, as evaluated by fluorescence recovery after photobleaching, the vast majority of topo II at centromeres and chromatid arms is actually remarkably dynamic, exchanging between chromosomal and non-chromosomal pools with a half-life on the order of several seconds [79,83]. As discussed below, these observations have greatly changed how we think about topo II dynamics on mitotic chromosomes, which in turn has a bearing on our conceptions of topo II function.
3. TOPO II AT CENTROMERES
Topo II enrichment at centromeres is arguably the most prominent aspect of topo II localization on mitotic chromosomes (Figure 2A). This behavior has primarily been described for vertebrate cells in culture, but may well be a general feature of metazoan cells. Whether an equivalent enrichment occurs at centromeres in yeast is not yet clear (we will refer to yeast centromeres as CENs, using the term to encompass both the genetically defined CEN locus and the ~50 kB of flanking chromatin in which cohesin [99–101] and potentially other factors are enriched). A recent report examining Top2 distribution on budding yeast chromosomes III and VI by chromatin immunoprecipitation (ChIP) did not specifically describe a CEN population, although the primary focus of this study was not on mitotic cells [102]. In our own ChIP analysis, Top2 appears to be broadly distributed at CEN regions during mitosis, but there is no obvious preferential association with CENs compared to several sites along chromosome arms (unpub. obs.). The literature on topo II localization and function at centromeres was comprehensively reviewed in 2004 [103]. Here we highlight some more recent observations, structured around three possible roles for topo II at centromeres.
3.1. A role for topo II in decatenating centromeres
In those cells where topo II enrichment at centromeres has been described, the centromere/primary constriction represents one of the last major sites of attachment between sister chromatids. It seems likely that a main functional requirement for topo II at centromeres is to resolve a final population of catenates linking daughter chromosomes. Interestingly, the physical act of segregating chromosomes may be necessary to actually complete this task. Highly extended chromatin filaments have recently been observed to stretch between vertebrate kinetochores during anaphase [104,105]. The incidence of these filaments increases following topo II inhibition or depletion, suggesting they are likely to represent residual topological linkages [105–107]. Another argument for the continued presence of catenates late into mitosis is that the addition of topo II inhibitors immediately prior to anaphase can block or severely restrict chromatid separation [10,66–68,108–111] (although it is difficult to exclude that topo II inhibition generates an artificial link between the chromatids). Early experiments in budding yeast mapped the Top2 execution point to the metaphase to anaphase transition, suggesting Top2 must act during anaphase to resolve persistent catenation [112]. Cell synchrony experiments have shown that chromosome regions adjacent to CENs separate with normal timing in budding yeast top2–4 mutants [113]. Telomeric regions, however, fail to disjoin, producing the characteristic Cut- phenotype associated with most top2 alleles. Based on this, one presumes that CENs must not be extensively linked by catenates in budding yeast, or that the path of CEN DNA in this organism does not trap catenates between points of microtubule attachment.
3.2. A role for topo II in cohering centromeres
As the converse of decatenation, a second possible role for topo II at centromeres is to modulate chromatid cohesion [114]. One way in which this might occur would be if topo II were held in check to allow catenation to work in parallel with the cohesin complex to reinforce chromatid attachment [115]. In possible support for such a strategy, inhibiting or down-regulating topo II has been observed to block premature sister chromatid separation in yeast and vertebrate cohesin mutants [10,108,116,117]. Following the loss of cohesin function, the Ipl1/Aurora B kinase becomes activated to detach kinetochores from the spindle and induces spindle assembly checkpoint (SAC) cell cycle arrest, possibly because centromeres can no longer be placed under mechanical tension [118,119]. Under these conditions, inactivating topo II can restore chromosome-spindle attachment and silence the SAC, suggesting unresolved catenates provide a sufficiently robust linkage to generate tension [108,116,117]. The question, however, is to what extent catenation normally contributes to cohesion, and, if it does, whether topo II is specifically regulated to stabilize chromatid entanglements. At face value, the fact that chromatids do indeed separate in cohesin mutants (although typically with incomplete penetrance, summarized in [115]) would seem to challenge the idea that catenates are stabilized to produce cohesion. This is in keeping with robust topo II activity at vertebrate centromeres [92,93] and in Xenopus extracts [67] prior to anaphase (for a different view, see [10,120,121]).
One recent report suggests that roles for topo II in centromeric cohesion may not be restricted to chromatid inter-twining. In a screen for C. elegans chromosome instability mutants, an essential locus was identified that was named CIN-4 [122]. Analysis of cin-4 mutants revealed they were unable to resolve sister centromeres into paired CENP-A foci, leading to lethal chromosome segregation errors. The defect in centromere resolution corresponded with a failure to dissociate cohesin from centromeres, and loss of cohesin function in cin-4 mutants restored centromere resolution. Remarkably, the wild type CIN-4 locus was found to encode a truncated form of topo II, consisting mostly of the DNA cleavage/ligation domain but lacking the amino-terminal ATPase region and most of the CTD. The presumptive active site tyrosine was also changed to serine, indicating the CIN-4 protein is unlikely to possess even vestigial catalytic activity. Thus, CIN-4 appears to be a partial topo II duplication that has acquired (or retained) a catalysis-independent function in releasing cohesin at centromeres.
There is also evidence for a relationship between Top2 and cohesion in budding yeast. Mutations affecting the SUMO isopeptidase Smt4/Ulp2 fail to maintain cohesion at CEN chromosomal regions [17]. We identified TOP2 as a dosage suppressor of this cohesion defect, and found that a top2 mutant that was resistant to SUMO (see section 4.2) was also able to partially restore cohesion. Independently, TOP2 was recovered as a suppressor of temperature sensitive lethality in pds5 strains [123], which are also defective in maintaining cohesion [124]. In this case, however, the ability of Top2 to act as a pds5 suppressor did not require catalytic activity and did not correspond with restoration of chromatid attachment [123]. To better understand this complex set of observations, we have been examining whether the ability of top2 mutations to rescue cohesion is indeed attributable to unresolved catenates. Using circular minichromosomes where catenation can be monitored directly, we find that strains that are deficient for both cohesin and Top2 retain minichromatid attachment even in the absence of catenation (unpub. obs). One hypothesis to explain this cohesive force is that defective Top2 catalysis or overproducing Top2 could increase the persistence or relative abundance of Top2-mediated linkages between sister chromatids. In this scenario, the ability of topo II to hold two DNA strands within the enzyme is potentially analogous to topological entrapment of chromatids within cohesin rings (at a minimum, related ideas have been discussed in [7,33,103,123,125]).
3.3. A role for topo II in centromere dynamics
There has also been recent interest in whether topo II activity at the centromere could impact kinetochore-spindle interactions. In their seminal 1996 paper, Rattner et al [87] observed that treating vertebrate cells with high concentrations of ICRF-193 prevented topo II from accumulating at centromeres. As evaluated by electron microscopy, this was accompanied by a disorganization of centromere structure, with the chromatin underlying the kinetochore tending to exhibit a less compact morphology. More recently, simultaneously depleting topo IIa and b in HeLa cells or treating cells with ICRF-193 was found to disrupt the organization of the metaphase plate, making it difficult to distinguish individual centromere pairs [108]. In those cases where it was possible to make this assignment, however, the distance across bioriented kinetochores was shortened by ~20–30% compared to topo II-proficient controls. A second study also reported an ~10–25% reduction in inter-kinetochore distance in a human topo IIa conditional null cell line [106]. One way of looking at this is that topo II activity has a bearing on how centromeric chromatin deforms under tension. For example, increased tangling could impede centromere stretching [106].
Work from our lab is in keeping with a requirement for topo II in the mechano-chemical properties of centromeres [126]. In budding yeast, sister CENs are pulled apart during biorientation, accompanied by distension of separated chromatid fibers [127,128]. Once stable bipolar connections are achieved, a localized recoil of the stretched chromatin occurs, forming two compact CEN foci (Figure 3A). We observed that top2–4 mutants have difficulty maintaining a compact CEN morphology under tension, resulting in an apparent unraveling of CEN chromatin and increased sister kinetochore displacement (Figure 3B; [126]). Using a Cre/lox excision assay, CEN stretching in top2–4 strains corresponded with an apparent overwinding of CEN chromatin in response to spindle tension. Unexpectedly, increased CEN stretching in top2–4 mutants also corresponded with genetically crippled kinetochores (harboring mutations in the Mtw1 protein) becoming resistant to Ipl1/Aurora B-mediated detachment from the spindle. top2–4 mutants remained proficient for Ipl1/Aurora B activity towards monopolar and syntelic attachments, suggesting stabilization of defective kinetochore-spindle interactions in top2–4 strains was specific for a partial reduction, rather than the absence, of tension. We interpreted these observations to suggest Top2 is required to maintain a compact CEN organization that can resist the pulling force of the spindle. Perturbations to this function correspond with an attenuation of a sub-set of Ipl1/Aurora-B activities in the tension checkpoint.
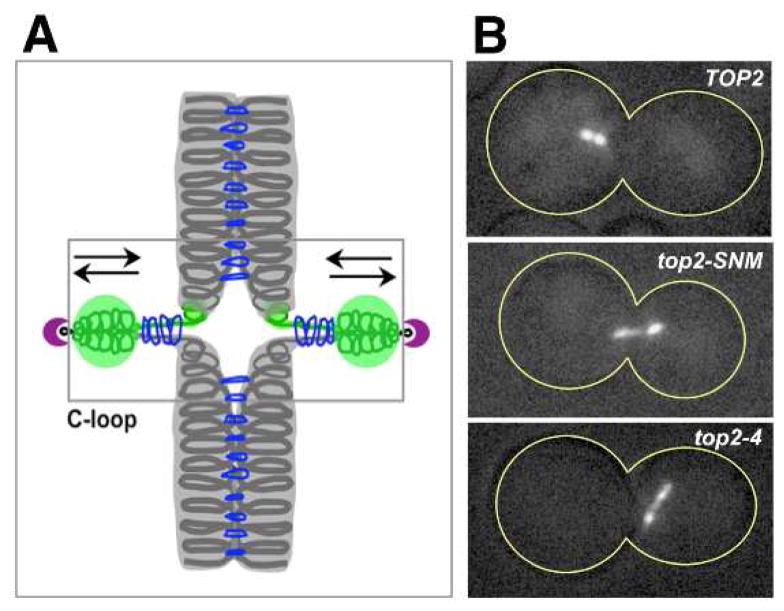
Figure 3. Influence of Top2 on budding yeast centromere dynamics.
(A) Diagram of proposed C-loop structure for mitotic CEN chromatin [187]. In this model, the separation of CEN chromatid fibers that occurs in response to bipolar spindle traction is stabilized by a localized reorganization of cohesin (blue rings) to form intra-stand cohesive linkages. GFP labeled chromatin (green; [188]) and kinetochores (purple) are also depicted. (B) Wild type (TOP2), top2–4 and SUMO conjugation resistant top2-SNM (see section 4.2.) strains were blocked in mitosis and the stretching of CEN chromatin under tension was evaluated using a CEN-proximal GFP tag on chromosome IV (CEN4-GFP). These images are reproduced here from [126] and from J. Bachant, InCytes, Mol. Biol. Cell 19 (2008) 4019 with permission from the publisher.
While this review was under preparation, a second study emerged supporting a positive-acting role for topo II in Ipl1/Aurora B function [129]. Based on time-lapse imaging of Drosophila S2 cells, these investigators observed that centromeres did not align properly on the metaphase plate after topo II RNAi, leading to co-segregation of sister chromatids to the same spindle pole. Remarkably, this segregation defect was found to be associated with a high frequency of unresolved syntelic kinetochore attachments, and similar alignment errors were observed in ICRF-187 treated HeLa cells. To account for this, the authors suggested decatenation might be necessary to produce a plastic chromatin environment in which kinetochores could biorient to opposing sides of the centromere (see also [130]). Further analysis of why these defective attachments were resistant to Ipl1/Aurora B suggested topo II was required for Ipl1/Aurora B to operate efficiently at centromeres. In brief, Ipl1/Aurora B exhibited reduced kinase activity in topo II depleted cells, possibly due to a failure of Ipl1/Aurora B to localize to a position within centromeric chromatin where it could escape inhibition by BubR1.
In evaluating this study, it is necessary to point out that biorientation defects were not reported in a previous RNAi study of topo II function in Drosophila [131], nor in recent reports examining centromere alignment in topo II-depleted human cells [106,108]. If substantiated, however, the remarkable results reported by Coelho et al [129] will necessitate a considerable re-evaluation of topo II function at centromeres.
3.4. Topo II-responsive checkpoints
A discussion of topo II at centromeres also requires a consideration of topo II-responsive checkpoints. In vertebrates, perturbing topo II in mitosis (using poisons, inhibitors, or RNAi) can activate a checkpoint that delays the metaphase to anaphase transition [10,108-111,132,133]. One of the first studies reporting this phenomenon suggested topo II inhibition might lead to DNA breaks in the vicinity of centromeres, causing kinetochore defects that activated the SAC [110]. In support of this, at least one kinetochore per cell showed persistent Mad2 staining, and metaphase delay induced by ICRF-193 was found to be Mad2-dependent. A second study, however, failed to find evidence for ICRF-193 induced DNA damage in mitosis (although see [134–137]) and did not detect Mad2 or Bub1 on kinetochores in ICRF-193 arrested cells [109]. Based on this, these investigators posited the existence of a distinct Mad2-dependent surveillance mechanism that monitored topo II activity or unresolved catenates. Conceptually this is similar to the ATM/ATR-dependent “catenation checkpoint” that has been proposed to operate at the G2/M transition [136,138–143]. To complicate the situation further, yet a third study found that Bub1, but not Mad2, showed intense kinetochore staining in ICRF-193 treated cells [108]; in the SAC lexicon Bub1 staining is considered to reflect a deficit of inter-kinetochore tension [144–146].
In what may prove to be a unifying set of observations, metaphase delay induced by ICRF-193 was recently found to not only require Mad2 but to also be dependent on Ipl1/Aurora B [10]. Similarly, although most yeast top2 mutants do not delay progression through mitosis [62,64], a new group of top2 alleles (e.g. top2-B44) has been identified that exhibit an ~30 minute delay in anaphase onset at the non-permissive temperature [147]. This delay required all SAC proteins that were examined, including Mad2 and Ipl1/Aurora B. Thus, topo II-responsive mitotic checkpoints appear to be conserved, at least in genetic terms, and bear a strong resemblance to the tension-monitoring branch of the SAC. The idea that topo II depletion might either cause or be perceived as a reduction in inter-kinetochore tension could explain a number of observations. But several questions remain. First, in both yeast and vertebrates, cell cycle arrest at the topo II-responsive checkpoint has been shown to be independent of the anaphase inhibitor Pds1/securin, which is the only known target of Mad2 [133,147]. It will be important to define how Mad2 enforces cell cycle arrest under these conditions (for a discussion see [148]). Second, why do some yeast top2 mutants activate a topo II-responsive checkpoint while others do not? Finally, how can perturbations to topo II variously activate Ipl1/Aurora B, interfere with Ipl1/Aurora B, or silence the SAC under different experimental conditions? Resolution of these issues will probably tell us something important about what topo II is doing at centromeres.
4. SUMO MODIFICATION OF TOPO I AND II
4.1. Sumoylation as a topo stress response
Topo II sumoylation was first reported in 2000 [149], shortly following the discovery of this modification on topo I [150]. It had been observed that treating human cells with the topo I poison camptothecin (CPT) caused a fraction of topo I to be degraded through the ubiquitin/26S proteasome pathway, leading to the proposal that proteolysis might be involved in repairing topo I cleavable complexes [151]. Consistent with ubiquitin chain formation, a ladder of higher molecular weight topo I species rapidly accumulated (5 min; [152,153]) after CPT treatment [150]. These species, however, reacted with antibodies against SUMO. This prompted a related study with topo II, revealing that SUMO modification of both topo IIa and b could be induced by the topo II poison VM-26 [149]. Unexpectedly, ICRF-193 also stimulated topo II sumoylation, suggesting this modification was a response to altered topo II conformation rather than an accumulation of cleavable complexes or DNA lesions. In keeping with this, treatments that induced protein conformational stress, but not DNA damaging treatments, were found to generally stimulate sumoylation of nuclear proteins [149,154]. The behavior of topo II appeared to further parallel topo I in that a fraction of topo IIb, and, to a much lesser extent, topo IIa, was degraded in a proteasome-dependent manner in response to inhibition or poisoning [155]. Further analysis of this “inhibitor induced” component of Topo SUMO modification has tended to focus on the nature of the relationship between sumoylation and enzyme proteolysis.
Sumoylation of human topo I occurs predominantly at three N-terminal sites, K103, K117 and K153, with K117 appearing to be the most important acceptor residue [156–158]. The p53- and topo I-binding protein Topors has been implicated as an E3 ligase for topo I sumoylation [159,160]. Sumoylation of budding yeast Top1 also occurs at three N-terminal lysines (K65, K91 and K92), with PIAS family members Siz1 and Siz2 acting as E3s for this modification [161]. For both human and yeast topo I, catalytically inactive proteins bearing mutations at the active site tyrosines are extensively SUMO modified, even in the absence of topo I poisons [158,161]. Thus, as suggested for topo II [149], topo I sumoylation may be primarily triggered by aberrant topo I conformation rather than topo I-DNA adducts. Whether CPT induces sumoylation and down-regulation of yeast Top1 appears to be untested. However, in the absence of Siz1 and Siz2, DNA repair processes become essential for yeast cell viability, suggesting these E3s act to prevent genome damage [161]. Top1 appears to be a key determinant of this lethality, as deletion of Top1 restores viability to siz1siz2 double mutants that are deficient for DNA repair. Furthermore, yeast and human cells that are deficient for SUMO conjugation are sensitive to topo I poisons [150,162,163]. Thus, part of the role of the SUMO pathway in genome stability may be mediated through counter-acting deleterious effects of Top1 or facilitating repair of Top1-induced DNA damage.
The availability of topo I mutants that are resistant to SUMO modification has allowed the relationship between sumoylation and proteolytic degradation to be examined directly. In three studies, the apparent absence of topo I SUMO modification did not obviously interfere with topo I ubiquitin conjugation or down-regulation after CPT treatment, revealing sumoylation is unlikely to be causally linked to topo I proteolysis [156,158,164]. As an additional aspect of the response to CPT, however, Topo I redistributes from the nucleolus, where it is normally enriched, to the nucleoplasm. Sumoylation may be part of a transport mechanism to direct this re-localization, as either dominant negative Ubc9 [152] or specifically blocking topo I SUMO conjugation [156] was reported to prevent topo I from leaving the nucleolus after CPT treatment. These findings, however, were contradicted by a second study, which, using the exact same SUMO consensus site mutations, found no evidence that sumoylation was required for topo I nucleolar clearance [157].
Topo II sumoylation in response to poisoning/inhibition has received somewhat less attention. However, in one study it was shown that human topo IIb becomes extensively modified with both SUMO-1 and SUMO-2/3, as well as polyubiquitin, following VM-16 or ICRF-193 treatment [164]. This same analysis showed that chicken DT40 cells producing dominant negative Ubc9 were largely defective for topo IIb down-regulation in response to ICRF-193. Thus, it is possible that sumoylation is coupled to topo IIb proteolysis, but whether this is directed through sumoylation of topo II or other substrates is unclear. Genetic studies in yeast have shown that SUMO conjugation mutants show increased resistance to the topo II poison doxorubicin [165]. Interestingly, however, specifically abolishing sumoylation on Top2 by itself had no effect on doxorubicin toxicity.
Ultimately, at a current level of resolution, what emerges from these studies seems to be that sumoylation of topo I and topo II is greatly stimulated by catalytic inhibition, potentially in response to an altered conformation of the enzyme on DNA. The adaptive value of this modification with respect to tolerating Topo inhibition, however, remains to be clearly defined. It is worth pointing out that the hypothesis that topo I and topo IIb are targeted for degradation as a stress response to clear cleavable complexes has largely been validated. Based on a series of articles [47,153,166–170], DNA or RNA polymerase collisions with topoisomerase/DNA complexes appear to target the stalled Topos for ubiquitin-mediated proteolysis. In the case of cleavable complexes, this degradation step appears to uncover the underlying strand lesion for repair.
4.2. Topo II sumoylation in mitosis
Sumoylation has also been shown to occur as a normal aspect of topo II regulation in mitosis. In budding yeast, at least three Top2 SUMO conjugates accumulate in mitotic cells, representing ~10% of the enzyme pool [17]. Both in vivo and in vitro, Top2 SUMO modification is largely dependent on Siz1 or Siz2, suggesting these E3s act redundantly to facilitate Top2 SUMO conjugation [33]. There is also a tendency for Top2 SUMO species to accumulate in strains defective for the SUMO de-conjugating enzyme Smt4/Ulp2, indicating this isopeptidase is likely to catalyze the removal of SUMO from Top2 [17].
Top2 contains three consensus sites for SUMO modification within the CTD portion of the protein (Figure 1B). Mutation of all five lysines within these sequences to arginine (the top2-SNM allele) largely blocked Top2 sumoylation, indicating these are likely to be physiologically relevant SUMO acceptor sites [17]. This was confirmed by a thorough in vivo and in vitro analysis of Top2 SUMO conjugation, which showed that top2-SNM, top2 mutants in which only the three predicted SUMO acceptor lysines were altered (top2-3xKR; top2-201), or a Top2 CTD deletion were all equivalently defective for SUMO conjugation [33]. However, prolonged incubation of the Top2-SNM protein in in vitro sumoylation reactions [33] or over-exposure of immunoblots from top2-SNM cells (unpub. obs.) reveals weaker bands that are likely to represent SUMO conjugation at lower affinity sites.
SUMO conjugation on a relatively small population of topo II has also been observed in mitotic Xenopus egg extracts [7,8], as well as in mitotic extracts from human [171] and murine [11] cells. SUMO acceptor residues on any of these topo IIs have yet to be reported, and their identification will be an important next step in evaluating the significance of topo II regulation by SUMO. Interestingly, sequence analysis suggests that although the budding yeast CTD SUMO consensus sites are not strictly conserved, all the topo IIs we examined have at least one SUMO consensus site within the CTD, often closely juxtaposed to the boundary between the CTD and the catalytic portion of the molecule (Figure 1B).
In Xenopus, topo II SUMO conjugation has been found to be mediated exclusively by SUMO-2/3, although SUMO-1 could also be used when present at high concentration [7]. SUMO modification of topo II, as well as virtually all other chromatin-associated SUMO-2/3 conjugates, was shown to be dependent on the PIASy SUMO E3 ligase [8]. PIASy itself was also tightly associated with chromatin, potentially through a cell cycle-regulated modification, leading to the suggestion that PIASy recruited Ubc9 to chromosomal substrates. In nocodazole treated extracts, the majority of the SUMO-2/3 signal, presumably including sumoylated forms of topo II, co-localized with Aurora B at the inner centromere. This accumulation failed to occur following PIASy-immunodepletion, suggesting a probable defect in topo II enrichment at centromeres [8]. One particularly interesting finding was that blocking SUMO conjugation with dominant negative Ubc9 increased the resistance of a fraction of topo II to salt extraction off mitotic chromatin [7]. The presence of this higher affinity fraction corresponded with increased topo II retention on extracted chromosomes.
In human cells, PIASy has not been shown to be required for topo II SUMO modification, at least directly. However, a recent study found both PIASy and bulk SUMO-2/3 conjugates were detectable at low levels on both the axial cores and centromeres of mitotic HeLa chromosomes [171]. These chromosomal SUMO-2/3 conjugates, as well as topo IIa SUMO-2/3 modification, were greatly stimulated by even a brief (15 min) exposure to ICRF-187 or topo II poisons. The ICRF-187-induced accumulation of SUMO conjugates on chromosomes was largely abolished after RNAi depletion of either topo IIa or PIASy, strongly implying that the vast majority were PIASy-dependent topo II SUMO species. Using a variety of compounds that block topo II, it was found that stimulation of SUMO modification required that topo II be able to at least initiate a catalytic cycle, suggesting sumoylation may be most prevalent at sites of enzyme activity on DNA. In a separate HeLa cell study [121], PIASy was shown to be necessary for topo II chromosome dynamics. Following PIASy knockdown, topo II failed to accumulate at centromeres or axial cores and instead exhibited a more diffuse chromosomal staining pattern (Figure 2B). Thus, reminiscent of the situation in Xenopus, sumoylation appears to be required for topo II accumulation at centromeres in human cells.
The requirements for topo II SUMO modification have also been examined in mice, yielding some surprises [11]. RanBP2/Nup358 is a large protein that displays SUMO E3 ligase activity in vitro, localizes to the nuclear pore during interphase and functions at kinetochores in mitosis [172–174]. In a recent study [11], the RanBP2 E3 domain was found to stimulate topo II SUMO modification in vitro, and MEFs that were engineered to have reduced RanBP2 expression proved defective for topo II sumoylation in vivo. In contrast, PIASy knockout cells (PIASy is not essential in mice [175,176]) were proficient for topo II SUMO conjugation. Although MEFs appear to use a different E3 to target topo II, the role of topo II sumoylation appeared to be conserved, with down-regulation of RanBP2 leading to a failure of topo II to accumulate at mitotic centromeres. Overproduction of a topo II-SUMO N-terminal fusion protein restored topo II localization, suggesting the requirement for RanBP2 was directly mediated through topo II SUMO conjugation.
Strikingly, PIASy depletion in Xenopus extracts and human cells and RanBP2 deficiency in mice produces a uniform cellular phenotype: sister chromatids fail to disjoin and segregate properly. Time lapse imaging of this defect in HeLa cells showed chromosomes congress normally to the metaphase plate following PIASy knockdown, followed by extensive metaphase delay [10]. Similar to ICRF-193 treatment, this delay could be abolished by inhibition of Aurora B, resulting in extensive chromosome bridging and the appearance of lagging chromosomes. Co-depletion of both PIASy and the cohesin guardian Sgo1 showed that the absence of cohesin at centromeres did not rescue the chromatid non-disjunction phenotype, suggesting the residual linkage between the sisters was likely to be unresolved catenates. In RanBP2 deficient MEFs, chromatin bridging also appears to be a prominent defect, accompanied in this case by extensive chromosome mis-segregation [11]. Importantly, RanBP2 hypomorphic mice displayed an elevated incidence of both spontaneous and chemically-induced tumors, many of which presented with severe aneuploidy. There is as yet no direct evidence that these cancers are attributable to mis-regulation of topo II. Overall, however, the combined results from these studies strongly indicate that in higher eukaryotes there is likely to be a causal relationship between topo II sumoylation, topo II accumulation at centromeres, and the ability of sister chromatids to disjoin and segregate accurately.
Perhaps because chromosomes are less extensively linked by catenation in budding yeast, analysis of Top2 sumoylation in this organism has yielded a rather different picture. As noted in section 3., one issue concerns whether Top2 actually accumulates at yeast CENs in mitosis. A conserved role for sumoylation in directing Top2 to CENs is possible, as a Top2 fusion protein with one copy of Smt3 inserted into the CTD was found to cross-link robustly to CEN sequences [33]. Insertion of additional Smt3 copies resulted in a preferential concentration of Top2 within the nucleolus, suggesting different Top2 SUMO forms might be targeted to different chromosome compartments [177]. Despite this, the functional role of Top2 SUMO modification has been difficult to ascertain. top2ΔCTD, top2–3xKR and top2-SNM cells do not display overt growth defects and have at best relatively mild chromosome segregation defects [17,33]. Sister chromatids disjoin normally in top2-SNM strains, although there is a transient accumulation of bridged chromosomes [126]. There may therefore be a subtle perturbation to decatenation in top2-SNM mutants. If so, this defect does not obviously compromise the accuracy of chromosome segregation.
Overall, our characterization of top2-SNM mutants has suggested possible roles for Top2 sumoylation in three aspects of chromosome dynamics. First, top2-SNM strains are similar to top2–4 mutants in that the Top2 lesion can block chromatid separation in cohesin mutants [17]. As described in section 3.2., we think this may reflect stabilization of a catenate-independent linkage between sisters. Second, top2-SNM cells display extensive stretching of CEN chromatin during biorientation ([126]; Figure 3B). Since this is also observed in top2–4 mutants, it is possible that sumoylation promotes an aspect of Top2 function necessary to prevent CEN expansion. Finally, top2-SNM and top2–4 strains are similar in that the ability of Ipl1 to detach weakened kinetochores from the spindle is compromised in mtw1top2-SNM cells [126]. Using GFP-tagged circular minichromosomes we found that increased stretching and stabilization of minichromatid attachment could be observed in mtw1top2-SNM cells even in the absence of catenation. Thus, the relaxed requirement for Top2 sumoylation in decatenating yeast chromosomes may help reveal other roles for SUMO in Top2 function.
5. CONCLUDING REMARKS
As a final consideration, while it is becoming increasingly apparent that sumoylation acts as a determinant of topo II function, it is important to stress that we still have little understanding of the underlying mechanisms. What exactly is SUMO doing for topo II? Based on current evidence, however, there appear to be two non-exclusive ways in which topo II sumoylation is being conceptualized within the literature. In closing we compare and contrast these views.
5.1. SUMO promotes topo II catalytic turnover
As has been proposed for thymine-DNA glycosylase [178], much of the data on topo II sumoylation could be explained by this moiety facilitating dissociation of the enzyme from DNA. Perhaps in some chromatin environments topo II occasionally encounters difficulty in completing its catalytic cycle, potentially stabilizing a link between DNA strands. A speculative idea is that topo II is specifically targeted for sumoylation under these conditions, with SUMO attachment to the CTD (assuming this is conserved) helping topo II to adopt an open conformation that releases the enzyme. It has also been suggested that SUMO modification of such a population could constitute a signal to topo II-responsive checkpoints [171]. There are at least two difficulties however. First, this model does not readily explain why sumoylation is required for topo II to accumulate at centromeres; it is necessary to postulate that the majority of the enzyme rapidly becomes stuck at non-centromeric sites without SUMO intervention. Second, if the role of SUMO is just to promote enzyme turnover there must be a distinct mechanism for centromere targeting. In theory, however, topo II might simply swarm its substrate, with the population exhibiting a collective tendency to concentrate at sites of catenation or torsional strain.
5.2. SUMO as a topo II localization tag
In this view, SUMO moieties directly mediate topo II recruitment to centomeres (and possibly to sites within axial cores), presumably through promoting protein-protein interactions with other chromatin components. This model should stimulate a search for centromeric proteins with SUMO binding motifs [179] that could act as topo II receptors. To explain how a modification that only affects a small fraction of topo II at steady state could direct such a large-scale re-distribution of the enzyme, it has been proposed that topo II remains attached to centromeres following SUMO removal [11]. Alternatively, since the majority of topo II actually appears to be exchangeable, rapid cycles of SUMO conjugation and removal throughout the population could presumably promote centromere enrichment. One weakness of this model, however, is that it does not readily explain the stimulation of sumoylation that occurs following topo II inhibition.
To summarize, an overview of the literature on topo II sumoylation reveals that this modification is responsive to enzyme activity and is closely coupled to topo II chromosome dynamics. In particular, in many cells SUMO modification appears to be important for allowing topo II to accumulate at mitotic centromeres. This almost certainly promotes a final step in decatenation so chromatids can disjoin efficiently. Intriguingly, recent evidence also suggests that topo II activity may be linked to centromere organization and mechano-chemistry, thereby influencing Ipl1/Aurora B error correction mechanisms. While the effects of specifically disrupting topo II sumoylation have yet to be examined in metazoans, topo II appears likely to be an important SUMO target in controlling genome stability. The field also appears poised to make significant inroads into understanding how SUMO is controlling topo II. It will be especially interesting to see if determining the molecular basis of this regulation provides useful insights into how sumoylation is modulating other chromosomal proteins.
Acknowledgments
The authors apologize to colleagues in the field for the many interesting articles that were not discussed or acknowledged due to space limitations. We thank Dr. D. Clarke for providing the micrographs in Figure 2, and we thank members of the Bachant laboratory for useful discussions. This work was supported by NIH grant GM66190 to J.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dasso M. Emerging roles of the SUMO pathway in mitosis. Cell Div. 2008;3:5. doi: 10.1186/1747-1028-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–51. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 3.Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell. 2008;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 6.Biggins S, Bhalla N, Chang A, Smith DL, Murray AW. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics. 2001;159:453–470. doi: 10.1093/genetics/159.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 2005;24:2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dieckhoff P, Bolte M, Sancak Y, Braus GH, Irniger S. Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Mol Microbiol. 2004;51:1375–1387. doi: 10.1046/j.1365-2958.2003.03910.x. [DOI] [PubMed] [Google Scholar]
- 10.Diaz-Martinez LA, Gimenez-Abian JF, Azuma Y, Guacci V, Gimenez-Martin G, Lanier LM, Clarke DJ. PIASgamma is required for faithful chromosome segregation in human cells. PLoS ONE. 2006;1:e53. doi: 10.1371/journal.pone.0000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R, van Deursen JM. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics. 2005;4:246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 15.Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem. 2004;279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- 16.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., 3rd Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 17.Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 18.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 19.Gadelle D, Filee J, Buhler C, Forterre P. Phylogenomics of type II DNA topoisomerases. Bioessays. 2003;25:232–242. doi: 10.1002/bies.10245. [DOI] [PubMed] [Google Scholar]
- 20.Caron PR, Watt P, Wang JC. The C-terminal domain of Saccharomyces cerevisiae DNA topoisomerase II. Mol Cell Biol. 1994;14:3197–3207. doi: 10.1128/mcb.14.5.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiozaki K, Yanagida M. Functional dissection of the phosphorylated termini of fission yeast DNA topoisomerase II. J Cell Biol. 1992;119:1023–1036. doi: 10.1083/jcb.119.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crenshaw DG, Hsieh T. Function of the hydrophilic carboxyl terminus of type II DNA topoisomerase from Drosophila melanogaster. II. In vivo studies. J Biol Chem. 1993;268:21335–21343. [PubMed] [Google Scholar]
- 23.Crenshaw DG, Hsieh T. Function of the hydrophilic carboxyl terminus of type II DNA topoisomerase from Drosophila melanogaster. I. In vitro studies. J Biol Chem. 1993;268:21328–21334. [PubMed] [Google Scholar]
- 24.McClendon AK, Dickey JS, Osheroff N. Ability of viral topoisomerase II to discern the handedness of supercoiled DNA: bimodal recognition of DNA geometry by type II enzymes. Biochemistry. 2006;45:11674–11680. doi: 10.1021/bi0520838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickey JS, Osheroff N. Impact of the C-terminal domain of topoisomerase IIalpha on the DNA cleavage activity of the human enzyme. Biochemistry. 2005;44:11546–11554. doi: 10.1021/bi050811l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardenas ME, Dang Q, Glover CV, Gasser SM. Casein kinase II phosphorylates the eukaryote-specific C-terminal domain of topoisomerase II in vivo. EMBO J. 1992;11:1785–1796. doi: 10.1002/j.1460-2075.1992.tb05230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells NJ, Addison CM, Fry AM, Ganapathi R, Hickson ID. Serine 1524 is a major site of phosphorylation on human topoisomerase II alpha protein in vivo and is a substrate for casein kinase II in vitro. J Biol Chem. 1994;269:29746–29751. [PubMed] [Google Scholar]
- 28.Daum JR, Gorbsky GJ. Casein kinase II catalyzes a mitotic phosphorylation on threonine 1342 of human DNA topoisomerase IIalpha, which is recognized by the 3F3/2 phosphoepitope antibody. J Biol Chem. 1998;273:30622–30629. doi: 10.1074/jbc.273.46.30622. [DOI] [PubMed] [Google Scholar]
- 29.Escargueil AE, Plisov SY, Filhol O, Cochet C, Larsen AK. Mitotic phosphorylation of DNA topoisomerase II alpha by protein kinase CK2 creates the MPM-2 phosphoepitope on Ser-1469. J Biol Chem. 2000;275:34710–34718. doi: 10.1074/jbc.M005179200. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Wang Y, Liu X. Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIalpha in cell cycle progression. J Biol Chem. 2008;283:6209–6221. doi: 10.1074/jbc.M709007200. [DOI] [PubMed] [Google Scholar]
- 31.Iida M, Matsuda M, Komatani H. Plk3 phosphorylates topoisomerase IIalpha at Thr(1342), a site that is not recognized by Plk1. Biochem J. 2008;411:27–32. doi: 10.1042/BJ20071394. [DOI] [PubMed] [Google Scholar]
- 32.Morrison C, Henzing AJ, Jensen ON, Osheroff N, Dodson H, Kandels-Lewis SE, Adams RR, Earnshaw WC. Proteomic analysis of human metaphase chromosomes reveals topoisomerase II alpha as an Aurora B substrate. Nucleic Acids Res. 2002;30:5318–5327. doi: 10.1093/nar/gkf665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi Y, Yong-Gonzalez V, Kikuchi Y, Strunnikov A. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics. 2006;172:783–794. doi: 10.1534/genetics.105.047167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung TD, Drake FH, Tan KB, Per SR, Crooke ST, Mirabelli CK. Characterization and immunological identification of cDNA clones encoding two human DNA topoisomerase II isozymes. Proc Natl Acad Sci U S A. 1989;86:9431–9435. doi: 10.1073/pnas.86.23.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakaguchi A, Kikuchi A. Functional compatibility between isoform alpha and beta of type II DNA topoisomerase. J Cell Sci. 2004;117:1047–1054. doi: 10.1242/jcs.00977. [DOI] [PubMed] [Google Scholar]
- 36.Akimitsu N, Adachi N, Hirai H, Hossain MS, Hamamoto H, Kobayashi M, Aratani Y, Koyama H, Sekimizu K. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIalpha. Genes Cells. 2003;8:393–402. doi: 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- 37.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 38.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 39.Lyu YL, Lin CP, Azarova AM, Cai L, Wang JC, Liu LF. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol Cell Biol. 2006;26:7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berger JM. Type II DNA topoisomerases. Curr Opin Struct Biol. 1998;8:26–32. doi: 10.1016/s0959-440x(98)80006-7. [DOI] [PubMed] [Google Scholar]
- 41.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q Rev Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 42.Salceda J, Fernandez X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu LF. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 44.Froelich-Ammon SJ, Osheroff N. Topoisomerase poisons: harnessing the dark side of enzyme mechanism. J Biol Chem. 1995;270:21429–21432. doi: 10.1074/jbc.270.37.21429. [DOI] [PubMed] [Google Scholar]
- 45.McClendon AK, Osheroff N. DNA topoisomerase II, genotoxicity, and cancer. Mutat Res. 2007;623:83–97. doi: 10.1016/j.mrfmmm.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zucchi R, Danesi R. Cardiac toxicity of antineoplastic anthracyclines. Curr Med Chem Anticancer Agents. 2003;3:151–171. doi: 10.2174/1568011033353434. [DOI] [PubMed] [Google Scholar]
- 47.Lyu YL, Kerrigan JE, Lin CP, Azarova AM, Tsai YC, Ban Y, Liu LF. Topoisomerase IIbeta mediated DNA double-strand breaks: implications in doxorubicin cardiotoxicity and prevention by dexrazoxane. Cancer Res. 2007;67:8839–8846. doi: 10.1158/0008-5472.CAN-07-1649. [DOI] [PubMed] [Google Scholar]
- 48.Felix CA. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim Biophys Acta. 1998;1400:233–255. doi: 10.1016/s0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 49.Azarova AM, Lyu YL, Lin CP, Tsai YC, Lau JY, Wang JC, Liu LF. Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc Natl Acad Sci U S A. 2007;104:11014–11019. doi: 10.1073/pnas.0704002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larsen AK, Escargueil AE, Skladanowski A. Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol Ther. 2003;99:167–181. doi: 10.1016/s0163-7258(03)00058-5. [DOI] [PubMed] [Google Scholar]
- 51.Andoh T, Ishida R. Catalytic inhibitors of DNA topoisomerase II. Biochim Biophys Acta. 1998;1400:155–171. doi: 10.1016/s0167-4781(98)00133-x. [DOI] [PubMed] [Google Scholar]
- 52.Roca J, Ishida R, Berger JM, Andoh T, Wang JC. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc Natl Acad Sci U S A. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Classen S, Olland S, Berger JM. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc Natl Acad Sci U S A. 2003;100:10629–10634. doi: 10.1073/pnas.1832879100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Germe T, Hyrien O. Topoisomerase II-DNA complexes trapped by ICRF-193 perturb chromatin structure. EMBO Rep. 2005;6:729–735. doi: 10.1038/sj.embor.7400465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carpenter AJ, Porter AC. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol Biol Cell. 2004;15:5700–5711. doi: 10.1091/mbc.E04-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 57.Sundin O, Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981;25:659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 58.Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 60.Buchenau P, Saumweber H, Arndt-Jovin DJ. Consequences of topoisomerase II inhibition in early embryogenesis of Drosophila revealed by in vivo confocal laser scanning microscopy. J Cell Sci. 1993;104(Pt 4):1175–1185. doi: 10.1242/jcs.104.4.1175. [DOI] [PubMed] [Google Scholar]
- 61.DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci U S A. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uemura T, Tanagida M. Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: uncoordinated mitosis. EMBO J. 1986;5:1003–1010. doi: 10.1002/j.1460-2075.1986.tb04315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 64.Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 65.Downes CS, Mullinger AM, Johnson RT. Inhibitors of DNA topoisomerase II prevent chromatid separation in mammalian cells but do not prevent exit from mitosis. Proc Natl Acad Sci U S A. 1991;88:8895–8899. doi: 10.1073/pnas.88.20.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gorbsky GJ. Cell cycle progression and chromosome segregation in mammalian cells cultured in the presence of the topoisomerase II inhibitors ICRF-187 [(+)-1,2-bis(3,5-dioxopiperazinyl-1-yl)propane; ADR-529] and ICRF-159 (Razoxane) Cancer Res. 1994;54:1042–1048. [PubMed] [Google Scholar]
- 67.Shamu CE, Murray AW. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clarke DJ, Johnson RT, Downes CS. Topoisomerase II inhibition prevents anaphase chromatid segregation in mammalian cells independently of the generation of DNA strand breaks. J Cell Sci. 1993;105:563–569. doi: 10.1242/jcs.105.2.563. [DOI] [PubMed] [Google Scholar]
- 69.Ishida R, Sato M, Narita T, Utsumi KR, Nishimoto T, Morita T, Nagata H, Andoh T. Inhibition of DNA topoisomerase II by ICRF-193 induces polyploidization by uncoupling chromosome dynamics from other cell cycle events. J Cell Biol. 1994;126:1341–1351. doi: 10.1083/jcb.126.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hardy CD, Crisona NJ, Stone MD, Cozzarelli NR. Disentangling DNA during replication: a tale of two strands. Philos Trans R Soc Lond B Biol Sci. 2004;359:39–47. doi: 10.1098/rstb.2003.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holm C. Coming undone: how to untangle a chromosome. Cell. 1994;77:955–957. doi: 10.1016/0092-8674(94)90433-2. [DOI] [PubMed] [Google Scholar]
- 72.Warburton PE, Earnshaw WC. Untangling the role of DNA topoisomerase II in mitotic chromosome structure and function. Bioessays. 1997;19:97–99. doi: 10.1002/bies.950190203. [DOI] [PubMed] [Google Scholar]
- 73.Earnshaw WC, Heck MM. Localization of topoisomerase II in mitotic chromosomes. J Cell Biol. 1985;100:1716–1725. doi: 10.1083/jcb.100.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gasser SM, Laroche T, Falquet J, Boy de la Tour E, Laemmli UK. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986;188:613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- 75.Taagepera S, Rao PN, Drake FH, Gorbsky GJ. DNA topoisomerase II alpha is the major chromosome protein recognized by the mitotic phosphoprotein antibody MPM-2. Proc Natl Acad Sci U S A. 1993;90:8407–8411. doi: 10.1073/pnas.90.18.8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saitoh Y, Laemmli UK. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell. 1994;76:609–622. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 77.Sumner AT. The distribution of topoisomerase II on mammalian chromosomes. Chromosome Res. 1996;4:5–14. doi: 10.1007/BF02254938. [DOI] [PubMed] [Google Scholar]
- 78.Meyer KN, Kjeldsen E, Straub T, Knudsen BR, Hickson ID, Kikuchi A, Kreipe H, Boege F. Cell cycle-coupled relocation of types I and II topoisomerases and modulation of catalytic enzyme activities. J Cell Biol. 1997;136:775–788. doi: 10.1083/jcb.136.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tavormina PA, Come MG, Hudson JR, Mo YY, Beck WT, Gorbsky GJ. Rapid exchange of mammalian topoisomerase II alpha at kinetochores and chromosome arms in mitosis. J Cell Biol. 2002;158:23–29. doi: 10.1083/jcb.200202053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Null AP, Hudson J, Gorbsky GJ. Both alpha and beta isoforms of mammalian DNA topoisomerase II associate with chromosomes in mitosis. Cell Growth Differ. 2002;13:325–333. [PubMed] [Google Scholar]
- 81.Boy de la Tour E, Laemmli UK. The metaphase scaffold is helically folded: sister chromatids have predominantly opposite helical handedness. Cell. 1988;55:937–944. doi: 10.1016/0092-8674(88)90239-5. [DOI] [PubMed] [Google Scholar]
- 82.Swedlow JR, Sedat JW, Agard DA. Multiple chromosomal populations of topoisomerase II detected in vivo by time-lapse, three-dimensional wide-field microscopy. Cell. 1993;73:97–108. doi: 10.1016/0092-8674(93)90163-k. [DOI] [PubMed] [Google Scholar]
- 83.Christensen MO, Larsen MK, Barthelmes HU, Hock R, Andersen CL, Kjeldsen E, Knudsen BR, Westergaard O, Boege F, Mielke C. Dynamics of human DNA topoisomerases IIalpha and IIbeta in living cells. J Cell Biol. 2002;157:31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gimenez-Abian JF, Clarke DJ, Mullinger AM, Downes CS, Johnson RT. A postprophase topoisomerase II-dependent chromatid core separation step in the formation of metaphase chromosomes. J Cell Biol. 1995;131:7–17. doi: 10.1083/jcb.131.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moens PB, Earnshaw WC. Anti-topoisomerase II recognizes meiotic chromosome cores. Chromosoma. 1989;98:317–322. doi: 10.1007/BF00292383. [DOI] [PubMed] [Google Scholar]
- 86.Cobb J, Miyaike M, Kikuchi A, Handel MA. Meiotic events at the centromeric heterochromatin: histone H3 phosphorylation, topoisomerase II alpha localization and chromosome condensation. Chromosoma. 1999;108:412–425. doi: 10.1007/s004120050393. [DOI] [PubMed] [Google Scholar]
- 87.Rattner JB, Hendzel MJ, Furbee CS, Muller MT, Bazett-Jones DP. Topoisomerase II alpha is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J Cell Biol. 1996;134:1097–1107. doi: 10.1083/jcb.134.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ishida R, Takashima R, Koujin T, Shibata M, Nozaki N, Seto M, Mori H, Haraguchi T, Hiraoka Y. Mitotic specific phosphorylation of serine-1212 in human DNA topoisomerase IIalpha. Cell Struct Funct. 2001;26:215–226. doi: 10.1247/csf.26.215. [DOI] [PubMed] [Google Scholar]
- 89.Swedlow JR, Hirano T. The making of the mitotic chromosome: modern insights into classical questions. Mol Cell. 2003;11:557–569. doi: 10.1016/s1097-2765(03)00103-5. [DOI] [PubMed] [Google Scholar]
- 90.Belmont AS. Mitotic chromosome scaffold structure: new approaches to an old controversy. Proc Natl Acad Sci U S A. 2002;99:15855–15857. doi: 10.1073/pnas.262672799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Belmont AS. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 92.Andersen CL, Wandall A, Kjeldsen E, Mielke C, Koch J. Active, but not inactive, human centromeres display topoisomerase II activity in vivo. Chromosome Res. 2002;10:305–312. doi: 10.1023/a:1016571825025. [DOI] [PubMed] [Google Scholar]
- 93.Agostinho M, Rino J, Braga J, Ferreira F, Steffensen S, Ferreira J. Human topoisomerase IIalpha: targeting to subchromosomal sites of activity during interphase and mitosis. Mol Biol Cell. 2004;15:2388–2400. doi: 10.1091/mbc.E03-08-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Floridia G, Zatterale A, Zuffardi O, Tyler-Smith C. Mapping of a human centromere onto the DNA by topoisomerase II cleavage. EMBO Rep. 2000;1:489–493. doi: 10.1093/embo-reports/kvd110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spence JM, Critcher R, Ebersole TA, Valdivia MM, Earnshaw WC, Fukagawa T, Farr CJ. Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array. EMBO J. 2002;21:5269–5280. doi: 10.1093/emboj/cdf511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spence JM, Fournier RE, Oshimura M, Regnier V, Farr CJ. Topoisomerase II cleavage activity within the human D11Z1 and DXZ1 alpha-satellite arrays. Chromosome Res. 2005;13:637–648. doi: 10.1007/s10577-005-1003-8. [DOI] [PubMed] [Google Scholar]
- 97.Kallio M, Lahdetie J. Fragmentation of centromeric DNA and prevention of homologous chromosome separation in male mouse meiosis in vivo by the topoisomerase II inhibitor etoposide. Mutagenesis. 1996;11:435–443. doi: 10.1093/mutage/11.5.435. [DOI] [PubMed] [Google Scholar]
- 98.Kas E, Laemmli UK. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992;11:705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 100.Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 102.Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, Foiani M. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21:1921–1936. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Porter AC, Farr CJ. Topoisomerase II: untangling its contribution at the centromere. Chromosome Res. 2004;12:569–583. doi: 10.1023/B:CHRO.0000036608.91085.d1. [DOI] [PubMed] [Google Scholar]
- 104.Baumann C, Korner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 105.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spence JM, Phua HH, Mills W, Carpenter AJ, Porter AC, Farr CJ. Depletion of topoisomerase IIalpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J Cell Sci. 2007;120:3952–3964. doi: 10.1242/jcs.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang LH, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008;117:123–135. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Toyoda Y, Yanagida M. Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol Biol Cell. 2006;17:2287–2302. doi: 10.1091/mbc.E05-11-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Skoufias DA, Lacroix FB, Andreassen PR, Wilson L, Margolis RL. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol Cell. 2004;15:977–990. doi: 10.1016/j.molcel.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 110.Mikhailov A, Cole RW, Rieder CL. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr Biol. 2002;12:1797–1806. doi: 10.1016/s0960-9822(02)01226-5. [DOI] [PubMed] [Google Scholar]
- 111.Iwai M, Hara A, Andoh T, Ishida R. ICRF-193, a catalytic inhibitor of DNA topoisomerase II, delays the cell cycle progression from metaphase, but not from anaphase to the G1 phase in mammalian cells. FEBS Lett. 1997;406:267–270. doi: 10.1016/s0014-5793(97)00282-2. [DOI] [PubMed] [Google Scholar]
- 112.Holm C, Stearns T, Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989;9:159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bhalla N, Biggins S, Murray AW. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol Biol Cell. 2002;13:632–645. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Murray AW, Szostak JW. Chromosome segregation in mitosis and meiosis. Annu Rev Cell Biol. 1985;1:289–315. doi: 10.1146/annurev.cb.01.110185.001445. [DOI] [PubMed] [Google Scholar]
- 115.Diaz-Martinez LA, Gimenez-Abian JF, Clarke DJ. Chromosome cohesion - rings, knots, orcs and fellowship. J Cell Sci. 2008;121:2107–2114. doi: 10.1242/jcs.029132. [DOI] [PubMed] [Google Scholar]
- 116.Dewar H, Tanaka K, Nasmyth K, Tanaka TU. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- 117.Vagnarelli P, Morrison C, Dodson H, Sonoda E, Takeda S, Earnshaw WC. Analysis of Scc1-deficient cells defines a key metaphase role of vertebrate cohesin in linking sister kinetochores. EMBO Rep. 2004;5:167–171. doi: 10.1038/sj.embor.7400077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 119.Pinsky BA, Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 120.Gimenez-Abian JF, Clarke DJ, Gimenez-Martin G, Weingartner M, Gimenez-Abian MI, Carballo JA, Diaz de la Espina SM, Bogre L, De la Torre C. DNA catenations that link sister chromatids until the onset of anaphase are maintained by a checkpoint mechanism. Eur J Cell Biol. 2002;81:9–16. doi: 10.1078/0171-9335-00226. [DOI] [PubMed] [Google Scholar]
- 121.Diaz-Martinez LA, Gimenez-Abian JF, Clarke DJ. Cohesin is dispensable for centromere cohesion in human cells. PLoS ONE. 2007;2:e318. doi: 10.1371/journal.pone.0000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stanvitch G, Moore LL. cin-4, a gene with homology to topoisomerase II, is required for centromere resolution by cohesin removal from sister kinetochores during mitosis. Genetics. 2008;178:83–97. doi: 10.1534/genetics.107.075275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Aguilar C, Davidson C, Dix M, Stead K, Zheng K, Hartman T, Guacci V. Topoisomerase II suppresses the temperature sensitivity of Saccharomyces cerevisiae pds5 mutants, but not the defect in sister chromatid cohesion. Cell Cycle. 2005;4:1294–1304. doi: 10.4161/cc.4.9.1997. [DOI] [PubMed] [Google Scholar]
- 124.Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS. Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J Cell Biol. 2004;166:775–785. doi: 10.1083/jcb.200406049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Warsi TH, Navarro MS, Bachant J. DNA topoisomerase II is a determinant of the tensile properties of yeast centromeric chromatin and the tension checkpoint. Mol Biol Cell. 2008;19:4421–4433. doi: 10.1091/mbc.E08-05-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- 128.Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Coelho PA, Queiroz-Machado J, Carmo AM, Moutinho-Pereira S, Maiato H, Sunkel CE. Dual role of topoisomerase II in centromere resolution and aurora B activity. PLoS Biol. 2008;6:e207. doi: 10.1371/journal.pbio.0060207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Loncarek J, Kisurina-Evgenieva O, Vinogradova T, Hergert P, La Terra S, Kapoor TM, Khodjakov A. The centromere geometry essential for keeping mitosis error free is controlled by spindle forces. Nature. 2007;450:745–749. doi: 10.1038/nature06344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chang CJ, Goulding S, Earnshaw WC, Carmena M. RNAi analysis reveals an unexpected role for topoisomerase II in chromosome arm congression to a metaphase plate. J Cell Sci. 2003;116:4715–4726. doi: 10.1242/jcs.00797. [DOI] [PubMed] [Google Scholar]
- 132.Haraguchi T, Kaneda T, Hiraoka Y. Dynamics of chromosomes and microtubules visualized by multiple-wavelength fluorescence imaging in living mammalian cells: effects of mitotic inhibitors on cell cycle progression. Genes Cells. 1997;2:369–380. doi: 10.1046/j.1365-2443.1997.1280326.x. [DOI] [PubMed] [Google Scholar]
- 133.Clarke DJ, Vas AC, Andrews CA, Diaz-Martinez LA, Gimenez-Abian JF. Topoisomerase II checkpoints: universal mechanisms that regulate mitosis. Cell Cycle. 2006;5:1925–1928. doi: 10.4161/cc.5.17.3200. [DOI] [PubMed] [Google Scholar]
- 134.Wang L, Eastmond DA. Catalytic inhibitors of topoisomerase II are DNA-damaging agents: induction of chromosomal damage by merbarone and ICRF-187. Environ Mol Mutagen. 2002;39:348–356. doi: 10.1002/em.10072. [DOI] [PubMed] [Google Scholar]
- 135.Hajji N, Pastor N, Mateos S, Dominguez I, Cortes F. DNA strand breaks induced by the anti-topoisomerase II bis-dioxopiperazine ICRF-193. Mutat Res. 2003;530:35–46. doi: 10.1016/s0027-5107(03)00135-0. [DOI] [PubMed] [Google Scholar]
- 136.Park I, Avraham HK. Cell cycle-dependent DNA damage signaling induced by ICRF-193 involves ATM, ATR, CHK2, and BRCA1. Exp Cell Res. 2006;312:1996–2008. doi: 10.1016/j.yexcr.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 137.Huang KC, Gao H, Yamasaki EF, Grabowski DR, Liu S, Shen LL, Chan KK, Ganapathi R, Snapka RM. Topoisomerase II poisoning by ICRF-193. J Biol Chem. 2001;276:44488–44494. doi: 10.1074/jbc.M104383200. [DOI] [PubMed] [Google Scholar]
- 138.Downes CS, Clarke DJ, Mullinger AM, Gimenez-Abian JF, Creighton AM, Johnson RT. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature. 1994;372:467–470. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- 139.Gimenez-Abian JF, Weingartner M, Binarova P, Clarke DJ, Anthony RG, Calderini O, Heberle-Bors E, Moreno Diaz de la Espina S, Bogre L, De la Torre C. A topoisomerase II-dependent checkpoint in G2-phase plant cells can be bypassed by ectopic expression of mitotic cyclin B2. Cell Cycle. 2002;1:187–192. [PubMed] [Google Scholar]
- 140.Deming PB, Cistulli CA, Zhao H, Graves PR, Piwnica-Worms H, Paules RS, Downes CS, Kaufmann WK. The human decatenation checkpoint. Proc Natl Acad Sci U S A. 2001;98:12044–12049. doi: 10.1073/pnas.221430898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Deming PB, Flores KG, Downes CS, Paules RS, Kaufmann WK. ATR enforces the topoisomerase II-dependent G2 checkpoint through inhibition of Plk1 kinase. J Biol Chem. 2002;277:36832–36838. doi: 10.1074/jbc.M206109200. [DOI] [PubMed] [Google Scholar]
- 142.Kaufmann WK, Campbell CB, Simpson DA, Deming PB, Filatov L, Galloway DA, Zhao XJ, Creighton AM, Downes CS. Degradation of ATM-independent decatenation checkpoint function in human cells is secondary to inactivation of p53 and correlated with chromosomal destabilization. Cell Cycle. 2002;1:210–219. [PubMed] [Google Scholar]
- 143.Mikhailov A, Shinohara M, Rieder CL. Topoisomerase II and histone deacetylase inhibitors delay the G2/M transition by triggering the p38 MAPK checkpoint pathway. J Cell Biol. 2004;166:517–526. doi: 10.1083/jcb.200405167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Skoufias DA, Andreassen PR, Lacroix FB, Wilson L, Margolis RL. Mammalian Mad2 and Bub1/BubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc Natl Acad Sci U S A. 2001;98:4492–4497. doi: 10.1073/pnas.081076898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shannon KB, Canman JC, Salmon ED. Mad2 and BubR1 function in a single checkpoint pathway that responds to a loss of tension. Mol Biol Cell. 2002;13:3706–3719. doi: 10.1091/mbc.E02-03-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Taylor SS, Hussein D, Wang Y, Elderkin S, Morrow CJ. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J Cell Sci. 2001;114:4385–4395. doi: 10.1242/jcs.114.24.4385. [DOI] [PubMed] [Google Scholar]
- 147.Andrews CA, Vas AC, Meier B, Gimenez-Abian JF, Diaz-Martinez LA, Green J, Erickson SL, Vanderwaal KE, Hsu WS, Clarke DJ. A mitotic topoisomerase II checkpoint in budding yeast is required for genome stability but acts independently of Pds1/securin. Genes Dev. 2006;20:1162–1174. doi: 10.1101/gad.1367206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Clarke DJ, Bachant J. Kinetochore structure and spindle assembly checkpoint signaling in the budding yeast, Saccharomyces cerevisiae. Front Biosci. 2008;13:6787–6819. doi: 10.2741/3189. [DOI] [PubMed] [Google Scholar]
- 149.Mao Y, Desai SD, Liu LF. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J Biol Chem. 2000;275:26066–26073. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- 150.Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. Proc Natl Acad Sci U S A. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Desai SD, Liu LF, Vazquez-Abad D, D’Arpa P. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J Biol Chem. 1997;272:24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- 152.Mo YY, Yu Y, Shen Z, Beck WT. Nucleolar delocalization of human topoisomerase I in response to topotecan correlates with sumoylation of the protein. J Biol Chem. 2002;277:2958–2964. doi: 10.1074/jbc.M108263200. [DOI] [PubMed] [Google Scholar]
- 153.Desai SD, Li TK, Rodriguez-Bauman A, Rubin EH, Liu LF. Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 2001;61:5926–5932. [PubMed] [Google Scholar]
- 154.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 155.Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem. 2001;276:40652–40658. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- 156.Rallabhandi P, Hashimoto K, Mo YY, Beck WT, Moitra PK, D’Arpa P. Sumoylation of topoisomerase I is involved in its partitioning between nucleoli and nucleoplasm and its clearing from nucleoli in response to camptothecin. J Biol Chem. 2002;277:40020–40026. doi: 10.1074/jbc.M200388200. [DOI] [PubMed] [Google Scholar]
- 157.Christensen MO, Krokowski RM, Barthelmes HU, Hock R, Boege F, Mielke C. Distinct effects of topoisomerase I and RNA polymerase I inhibitors suggest a dual mechanism of nucleolar/nucleoplasmic partitioning of topoisomerase I. J Biol Chem. 2004;279:21873–21882. doi: 10.1074/jbc.M400498200. [DOI] [PubMed] [Google Scholar]
- 158.Horie K, Tomida A, Sugimoto Y, Yasugi T, Yoshikawa H, Taketani Y, Tsuruo T. SUMO-1 conjugation to intact DNA topoisomerase I amplifies cleavable complex formation induced by camptothecin. Oncogene. 2002;21:7913–7922. doi: 10.1038/sj.onc.1205917. [DOI] [PubMed] [Google Scholar]
- 159.Haluska P, Jr, Saleem A, Rasheed Z, Ahmed F, Su EW, Liu LF, Rubin EH. Interaction between human topoisomerase I and a novel RING finger/arginine-serine protein. Nucleic Acids Res. 1999;27:2538–2544. doi: 10.1093/nar/27.12.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hammer E, Heilbronn R, Weger S. The E3 ligase Topors induces the accumulation of polysumoylated forms of DNA topoisomerase I in vitro and in vivo. FEBS Lett. 2007;581:5418–5424. doi: 10.1016/j.febslet.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 161.Chen XL, Silver HR, Xiong L, Belichenko I, Adegite C, Johnson ES. Topoisomerase I-dependent viability loss in saccharomyces cerevisiae mutants defective in both SUMO conjugation and DNA repair. Genetics. 2007;177:17–30. doi: 10.1534/genetics.107.074708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jacquiau HR, van Waardenburg RC, Reid RJ, Woo MH, Guo H, Johnson ES, Bjornsti MA. Defects in SUMO (small ubiquitin-related modifier) conjugation and deconjugation alter cell sensitivity to DNA topoisomerase I-induced DNA damage. J Biol Chem. 2005;280:23566–23575. doi: 10.1074/jbc.M500947200. [DOI] [PubMed] [Google Scholar]
- 163.Mo YY, Yu Y, Ee PL, Beck WT. Overexpression of a dominant-negative mutant Ubc9 is associated with increased sensitivity to anticancer drugs. Cancer Res. 2004;64:2793–2798. doi: 10.1158/0008-5472.can-03-2410. [DOI] [PubMed] [Google Scholar]
- 164.Isik S, Sano K, Tsutsui K, Seki M, Enomoto T, Saitoh H, Tsutsui K. The SUMO pathway is required for selective degradation of DNA topoisomerase IIbeta induced by a catalytic inhibitor ICRF-193(1) FEBS Lett. 2003;546:374–378. doi: 10.1016/s0014-5793(03)00637-9. [DOI] [PubMed] [Google Scholar]
- 165.Huang RY, Kowalski D, Minderman H, Gandhi N, Johnson ES. Small ubiquitin-related modifier pathway is a major determinant of doxorubicin cytotoxicity in Saccharomyces cerevisiae. Cancer Res. 2007;67:765–772. doi: 10.1158/0008-5472.CAN-06-2839. [DOI] [PubMed] [Google Scholar]
- 166.Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, Rubin EH, Liu LF. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol Cell Biol. 2003;23:2341–2350. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wu J, Liu LF. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 1997;25:4181–4186. doi: 10.1093/nar/25.21.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xiao H, Mao Y, Desai SD, Zhou N, Ting CY, Hwang J, Liu LF. The topoisomerase IIbeta circular clamp arrests transcription and signals a 26S proteasome pathway. Proc Natl Acad Sci U S A. 2003;100:3239–3244. doi: 10.1073/pnas.0736401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang A, Lyu YL, Lin CP, Zhou N, Azarova AM, Wood LM, Liu LF. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J Biol Chem. 2006;281:35997–36003. doi: 10.1074/jbc.M604149200. [DOI] [PubMed] [Google Scholar]
- 170.Lin CP, Ban Y, Lyu YL, Desai SD, Liu LF. A ubiquitin-proteasome pathway for the repair of topoisomerase I-DNA covalent complexes. J Biol Chem. 2008;283:21074–21083. doi: 10.1074/jbc.M803493200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Agostinho M, Santos V, Ferreira F, Costa R, Cardoso J, Pinheiro I, Rino J, Jaffray E, Hay RT, Ferreira J. Conjugation of human topoisomerase 2 alpha with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer Res. 2008;68:2409–2418. doi: 10.1158/0008-5472.CAN-07-2092. [DOI] [PubMed] [Google Scholar]
- 172.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 173.Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14:611–617. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 174.Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 175.Wong KA, Kim R, Christofk H, Gao J, Lawson G, Wu H. Protein inhibitor of activated STAT Y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life. Mol Cell Biol. 2004;24:5577–5586. doi: 10.1128/MCB.24.12.5577-5586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Roth W, Sustmann C, Kieslinger M, Gilmozzi A, Irmer D, Kremmer E, Turck C, Grosschedl R. PIASy-deficient mice display modest defects in IFN and Wnt signaling. J Immunol. 2004;173:6189–6199. doi: 10.4049/jimmunol.173.10.6189. [DOI] [PubMed] [Google Scholar]
- 177.Takahashi Y, Strunnikov A. In vivo modeling of polysumoylation uncovers targeting of Topoisomerase II to the nucleolus via optimal level of SUMO modification. Chromosoma. 2007 doi: 10.1007/s00412-007-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lindsley JE, Wang JC. Proteolysis patterns of epitopically labeled yeast DNA topoisomerase II suggest an allosteric transition in the enzyme induced by ATP binding. Proc Natl Acad Sci U S A. 1991;88:10485–10489. doi: 10.1073/pnas.88.23.10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shiozaki K, Yanagida M. A functional 125-kDa core polypeptide of fission yeast DNA topoisomerase II. Mol Cell Biol. 1991;11:6093–6102. doi: 10.1128/mcb.11.12.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Austin CA, Marsh KL, Wasserman RA, Willmore E, Sayer PJ, Wang JC, Fisher LM. Expression, domain structure, and enzymatic properties of an active recombinant human DNA topoisomerase II beta. J Biol Chem. 1995;270:15739–15746. doi: 10.1074/jbc.270.26.15739. [DOI] [PubMed] [Google Scholar]
- 183.Meczes EL, Gilroy KL, West KL, Austin CA. The impact of the human DNA topoisomerase II C-terminal domain on activity. PLoS ONE. 2008;3:e1754. doi: 10.1371/journal.pone.0001754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 186.Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]