Summary
We have investigated the function of a cell envelope stress inducible gene, yvrI, which encodes a 22.5 kDa protein that includes a predicted σ70 region 4 domain, but lacks an apparent region 2 domain. YvrI interacts with RNA polymerase and overexpression of YvrI results in induction of OxdC, an oxalate decarboxylase maximally expressed under low pH conditions. We have used microarray-based analyses to define the YvrI regulon. YvrI is required for the transcription of three operons (oxdC-yvrL, yvrJ, and yvrI-yvrHa) each of which is preceded by a highly similar promoter sequence. Activation of these promoters requires both YvrI and the product of the second gene in the yvrI-yvrHa operon, YvrHa. YvrI and YvrHa together allow recognition of the oxdC promoter, stimulate DNA melting, and activate transcription by core RNA polymerase. Together, these results suggest that YvrI is a previously unrecognized σ factor in B. subtilis and that the 9.5 kDa YvrHa protein acts as a required co-activator of transcription. A yvrL deletion results in the upregulation of YvrI activity suggesting that YvrL is a negative regulator of YvrI-dependent transcription, possibly functioning as an anti-σ factor.
Keywords: acid stress, promoter, transcription, RNA polymerase, cell wall
Introduction
Bacillus subtilis encodes 17 known σ factors: 16 members of the σ70-family and one σ54-like regulator (Helmann, 2002). These include the vegetative σ (σA), eight group III factors (σB, σD, σE, σF, σG, σH, σI, and σK) and seven group IV (extracytoplasmic function; ECF) σ factors (σM, σV, σW, σX, σY, σZ, and σYlaC) and the σ54 family member σL. All σ factors function by associating reversibly with RNA polymerase (RNAP) core enzyme to form a holoenzyme competent for promoter recognition and transcription initiation (Paget and Helmann, 2003).
σ70 family members contain as many as four conserved regions (designated 1, 2, 3, and 4), with most identified members having at least regions 2 and 4 (Lonetto et al., 1992). Primary σ factors (group I) contain all four regions and are essential whereas group II factors are structurally similar, but non-essential. The alternative σ factors of group III contain only regions 2, 3 and 4, and the group IV (ECF) proteins contain only regions 2 and 4 (Paget and Helmann, 2003). Region 2 contains key determinants for core-binding, −10 recognition, and promoter melting and region 4 contains the helix-turn-helix motif that interacts with the −35 element and also provides a contact surface for many transcription activators (Gruber and Gross, 2003). During the automated annotation of newly sequenced bacterial genomes putative σ factors are typically defined as those predicted proteins containing, minimally, conserved regions 2 and 4.
In previous studies, we have characterized the effects of cell envelope-acting antibiotics on gene expression in B. subtilis (Butcher and Helmann, 2006; Cao et al., 2002b; Mascher et al., 2003). For example, exposure to the peptidoglycan synthesis inhibitor vancomycin induces the synthesis of genes controlled by several ECF σ factors (especially σW and σM), the two-component regulatory system LiaRS, as well as the yvrI and yvrL genes (Cao et al., 2002b). YvrI is an unknown function protein containing a predicted σ70-type region 4 and therefore possibly involved in regulating gene expression. As shown here, YvrI functions as an additional σ factor in B. subtilis.
Upstream of and divergently transcribed from yvrI is the gene oxdC which encodes an oxalate decarboxylase that has been extensively studied because of its unusual structure and reaction mechanism (Anand et al., 2002; Just et al., 2007; Svedruzic et al., 2007). Oxalate decarboxylases convert oxalate to formate and CO2 with the consumption of one proton. B. subtilis OxdC was the first oxalate decarboxylase discovered in a prokaryote (Tanner and Bornemann, 2000), the first member of the cupin-fold superfamily of enzymes shown to carry two metal (manganese) binding sites, and the first described hexameric bicupin (Anand et al., 2002). Oxalate decarboxylases are most commonly associated with fungi where their expression is induced by both low pH and the presence of oxalate salts (Gadd, 1999). In fungi, these enzymes are believed to play a role in degradative processes in soil and plant environments. Since OxdC expression in B. subtilis is induced under acidic conditions, but not by oxalate salts, it has been suggested that OxdC may play a role in pH homeostasis in these cells (Tanner and Bornemann, 2000). Recently, it was found that OxdC accumulates as one of the most abundant proteins in the cell wall when cells are grown in acidified media (Antelmann et al., 2007).
Here we show that oxdC is positively regulated by the YvrI σ factor and the YvrHa coactivator. Together, the YvrI-YvrHa regulatory proteins control the expression of at least five genes including their own operon, the oxdC-yvrL operon, and a newly annotated gene yvrJ. YvrL functions as a negative regulator of YvrI-dependent activation, possibly functioning as an anti-σ.
Results
Overexpression of YvrI induces OxdC
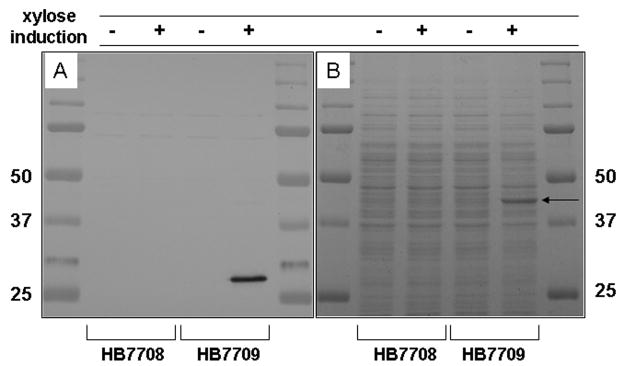
Since YvrI encodes a predicted DNA-binding protein with similarity to σ factors, we sought to monitor the effects of inducing YvrI on cell physiology. We fused an eight amino acid FLAG tag to the C-terminus of YvrI and expressed the YvrI-FLAG fusion protein using the PxylA-based expression vector pSWEET (Bhavsar et al., 2001). After growth in the presence of 2% xylose, the YvrI-FLAG protein could clearly be visualized by immunoblotting (Fig. 1A). Note that the 23.5 kDa YvrI-FLAG protein (confirmed using ESI-MS) migrates anomalously during SDS-PAGE, as also noted for many σ factors. Unexpectedly, when these same samples were stained with coomassie blue, a ~43 kDa protein was overproduced in those cells expressing YvrI-FLAG (Fig. 1B). This protein was identified as OxdC by MALDI-TOF MS analysis of tryptic peptides (data not shown). OxdC is encoded by a gene upstream of, and divergently oriented from, the yvrI-yvrHa operon (Fig. 2A).
Fig. 1. Conditional expression of YvrI induces OxdC.
A) Immunoblot showing xylose-dependent expression of YvrI-FLAG from integrated expression vector (strain HB7709) but not from strain carrying expression vector lacking yvrI-FLAG fusion (HB7708). B) Coomassie stained SDS-PAGE of samples in (A) showing YvrI-FLAG-dependent overexpression of OxdC (arrow).
Fig. 2. Identification of YvrI-activated promoters.
A) Genetic region surrounding the yvrI locus. Promoters identified in this study are indicated with angled arrows. Primer extension analysis was used to identify the (B) oxdC and (C) yvrJ transcriptional start sites from total RNA extracted from strain HB7709 cells grown without (lane 1) and with (lane 2) xylose induction of YvrI overexpression. YvrI-dependent transcripts are identified by arrows. D) sequence alignment of oxdC and yvrJ transcriptional start sites (angled arrow) and promoter regions showing all conserved nucleotides (shaded). The identification of the yvrI sequence was based on similarity to the experimentally determined promoter sequences. Number in parentheses refer to the distance (in nucleotides) between the known or predicted transcriptional start site and the start codon.
As shown herein, YvrI activates three clustered promoters in B. subtilis (Fig. 2A). The intergenic region between yvrI and oxdC is 428 bp in length and includes a predicted 153 bp open reading frame which we designate as yvrJ since this designation is not currently in use (although see (Wipat et al., 1998) and since the gene falls between yvrI and yvrK (oxdC) (Fig. 2A). Note that this ORF was annotated in B. licheniformis (BL00820) and more recently sequenced genomes, but not in B. subtilis. The annotation shown here, including the separation of yvrHa and yvrHb into two separate genes, differs from the original annotation (e.g. Subtilist Version 3.1, http://genolist.pasteur.fr/SubtiList/) and reflects the correction of several sequencing errors as described earlier (Kobayashi et al., 2001) and in this work (see details in Experimental Procedures).
Identification of three YvrI-dependent operons
We used primer extension analysis to identify the start site for the YvrI-activated oxdC promoter (PoxdC) in cells induced for YvrI-FLAG expression from an ectopically integrated copy inserted in the amyE locus (Fig. 2B). Transcription initiates from an A residue 71 nt upstream of the oxdC start codon (Fig. 2D). To monitor the genetic requirements for activation of PoxdC, the oxdC-yvrI intergenic region was cloned upstream of lacZ and integrated ectopically. Induction of YvrI-FLAG led to activation of the PoxdC-lacZ fusion (Fig. 3A, first panel) in cells containing either an intact copy of the chromosomal yvrI-yvrHa operon (HB7717) or an in-frame yvrI deletion (HB7732). Induction was not observed in strains lacking yvrHa, suggesting that this gene encodes a required co-activator for oxdC expression (Fig. 3A). The yvrI and yvrHa genes (Fig. 2A) are believed to constitute a single transcriptional unit (Serizawa et al, 2005), an organization that we confirmed using northern hybridization (not shown). To confirm that both YvrI and YvrHa are required for transcriptional activation, we monitored oxdC transcription in a strain carrying a chromosomal yvrI-yvrHa deletion and independently integrated xylose-inducible expression plasmids encoding epitope-tagged YvrI or YvrHa variants (YvrI-FLAG or YvrHa-HA) (Fig. 3B). Co-expression of both proteins was required for PoxdC activation (strain HB7759). Based on immunoblot analysis (Fig. 3B, lower panel) we conclude that neither the size nor the accumulation of YvrI-FLAG was affected by coexpression with YvrHa.
Fig. 3. Promoter activation by YvrI and YvrHa.
A) activity of oxdC- and yvrI-lacZ fusions in the absence or presence of YvrI overexpression from an ectopically integrated copy (using a pSWEET-based plasmid integrated at amyE; see Supplementary Table S2). Induction was obtained by addition of 2% xylose for 60 min. Cells either carry the wildtype complement of yvrI and yvrHa genes (HB7717 and HB7716), a yvrI deletion (HB7732 and HB7728), a yvrHa deletion (HB7733 and HB7729) or a yvrI yvrHa deletion (HB7734 and HB7730). Note that measurements of the strong oxdC and weak yvrI promoter activities are reported using different scales. (B) oxdC promoter activation in a host strain carrying a yvrI-yvrHa deletion in its genomic locus requires co-expression from ectopically integrated yvrI and yvrHa genes (strain HB7759). Immunoblot detection demonstrates that neither the accumulation nor mobility of YvrI-FLAG is affected by co-expression with YvrHa.
A previous study demonstrated a 9-fold increase in oxalate decarboxylase activity in cells grown under constant acidic (pH 5) conditions (Tanner and Bornemann, 2000). When we measured oxdC expression at the transcriptional level using a lacZ reporter fusion under similar conditions we detected a reproducible, ~2–3-fold induction of oxdC-lacZ expression at pH 5.2 versus pH 7.5. The deletion of either yvrI or yvrHa abolished expression (data not shown). The basal activity of the oxdC promoter is weak and the yvrI promoter is even weaker, but both the induction and the negative effects of yvrI and yvrHa deletions are reproducible in liquid and plate-based assays (data not shown). Thus, acid induction of yvrI-yvrHa may contribute to the acid-inducible accumulation of oxalate decarboxylase activity (Tanner and Bornemann, 2000) and of OxdC itself as monitored in proteome studies (Antelmann et al., 2007).
YvrI also activates an autoregulatory promoter (PyvrI) upstream of the yvrI-yvrHa operon (Fig. 3A, right panel). As noted for the oxdC promoter, activation of the yvrI-yvrHa promoter was only observed in strains containing an intact copy of the yvrHa gene. We were unable to conclusively identify a start site for the weak PyvrI promoter using primer extension reactions or 5′RACE, probably because PyvrI is only weakly activated by YvrI-FLAG in liquid culture (although induction is readily apparent on a solid medium). Searching the oxdC-yvrI intergenic region for sequences similar to PoxdC revealed two highly similar sequences. The one predicted to initiate transcription ~11 nt upstream of yvrI presumably functions as the autoregulatory site as detected in reporter fusion studies. Indeed, a similar candidate promoter is present 14 bp upstream of the B. licheniformis yvrI gene.
During these studies we also noted a second, divergent promoter found more than 300 nt upstream of oxdC (Fig. 2D). This site (PyvrJ) precedes an open reading frame for which we propose the designation yvrJ. Primer extension analysis confirmed the presence of a YvrI-dependent transcript emanating from PyvrJ (Fig. 2C). A transcriptional fusion to lacZ revealed that, like PoxdC and PyvrI, activation of PyvrJ required both YvrI and YvrHa expression (not shown). As noted above, yvrJ was not previously annotated in the B. subtilis genome (Kunst et al., 1997) but yvrJ is preceded by a candidate ribosome-binding sequence and homologs of yvrJ are annotated and positionally conserved in B. licheniformis and B. thuringiensis. The function of YvrJ is not known.
Together, these results suggest that YvrI and YvrHa activate the expression of three operons all preceded by similar candidate promoter elements (Fig. 2D). These promoter sequences are highly conserved around regions −10 and −35, show similarity to σA-dependent promoters in their −35 elements, and have exceptionally T-rich linker regions. Nucleotide regions upstream of −40 and downstream of −8 indicate little conservation.
Biochemical characterization of YvrI as a σ factor
Sequence alignments between YvrI and known σ70-family proteins from B. subtilis (Fig. S1a) reveal that, like ECF σ factors, YvrI includes a predicted σ70 region 4 (Pfam family PF04545) but lacks most of region 3. Thus, from a primary sequence perspective, YvrI is most similar to the ECF σ factors. However, domain prediction programs fail to identify a σ70 region 2, usually the most strongly conserved region in σ factors (Lonetto et al., 1992). It is likely for this reason that YvrI has not been included among B. subtilis putative or known σ factors (Helmann, 2002).
Inspection of the aligned amino acid sequences hints that YvrI may retain at least some functions associated with σ region 2 (Fig. S1a). Residue D50 in YvrI is conserved amongst YvrI homologs (Fig. S1b) and is coincident with an acidic residue in all the B. subtilis σ factors (Fig. S1a). An acidic residue at this same position has been implicated in core binding by other σ factors (Sharp et al., 1999; Wilson and Lamont, 2006; Wong et al., 2003). Other points of conservation in region 2.2 are also evident since six of seven ECF σ factors carry aromatic residues coincident with F65 in YvrI and all of the ECF σ factors carry basic residues coincident with YvrI K58 (Fig S1a). In region 2.3, YvrI residue F77 is conserved amongst YvrI homologs and positionally coincides with aromatic amino acids that are conserved in almost all sigma factors. These same residues in E. coli σ70 (Y430) and B. subtilis σA (Y189) have been implicated in DNA melting (deHaseth and Helmann, 1995; Juang and Helmann, 1994). YvrI therefore displays some features reminiscent of σ70 region 2.
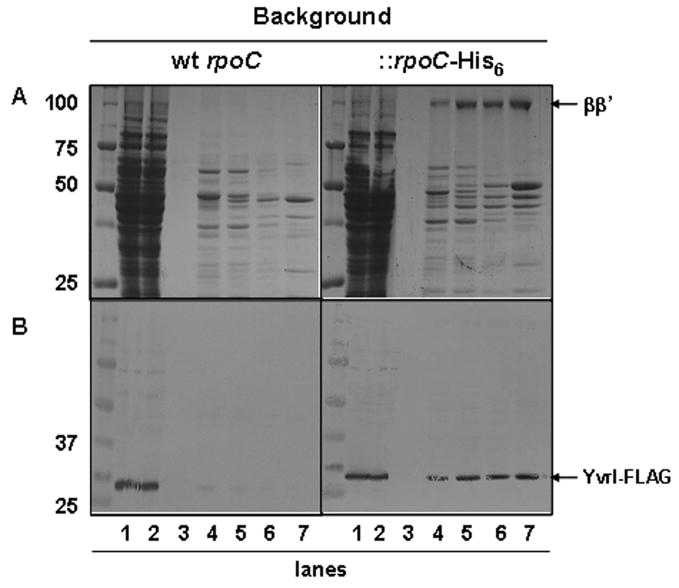
Bacterial σ factors associate reversibly with RNAP core enzyme to generate holoenzyme and direct promoter recognition and melting. To test whether YvrI displays σ factor activity we investigated (i) YvrI association with RNAP in vivo, (ii) activation of PoxdC transcription by YvrI in vitro, and (iii) the ability of YvrI to mediate open-complex formation at PoxdC. To monitor YvrI association with RNAP in vivo we expressed YvrI-FLAG in wildtype B. subtilis CU1065 cells and in a derivative of B. subtilis JH642 in which the β′ subunit was translationally fused to a C-terminal His tag (strain 1A774) to facilitate the one-step purification of holoenzyme from cells (Anthony et al., 2000). As expected, RNAP (detected by the presence of the high molecular mass β and β′ proteins; Fig. 4A) was only bound to the Ni-NTA column in extracts from the strain containing the rpoC-His6 gene. Immunoblot analysis (Fig. 4B) indicated that YvrI-FLAG co-eluted with RNAP in this strain. The YvrI-FLAG protein did not bind Ni-NTA in the extracts from cells from the strain lacking the His-tagged β′ consistent with the hypothesis that retention of YvrI is due to an interaction with RNAP. Thus, YvrI interacts with RNAP in vivo, as would be expected for a σ factor.
Fig. 4. Interaction of YvrI-FLAG with RNA polymerase.
A) SDS-PAGE separation of metal affinity purified proteins from strain HB7709 carrying wt rpoC and strain HB7711 carrying rpoC fused with a hexahistidine tag. YvrI-FLAG was overexpressed in each strain. Lanes 1, whole cell lysate; 2, flow-through; 3, 5 mM imidazole wash; 4, 10 mM imidazole elution; 5, 20 mM imidazole elution; 6, 30 mM imidazole elution, 7, 250 mM imidazole elution. Equivalent amounts of protein (5 μl for lanes 1 and 2 and 30 μl for lanes 3–7) were loaded onto each gel. B) Immunoblot analysis from gels identical to those shown in (A) using anti-FLAG polyclonal antibodies.
To determine whether or not YvrI could mediate promoter recognition, we reconstituted YvrI-dependent transcription in vitro. Purified B. subtilis RNAP holoenzyme (EσA) (Fig. 5A) was assayed using run-off transcription in the presence of either additional σA (σA-FLAG), YvrI-FLAG, YvrHa-HA, or both YvrI-FLAG and YvrHa-HA. Transcription from the PoxdC promoter to yield an ~82 nt run-off transcript was strongly stimulated by the presence of both YvrI and YvrHa, and a very weak band was also detected in reactions containing only YvrI (Fig. 5B). No recognition of this promoter site was observed in the σA-supplemented reaction. These results are consistent with the hypothesis that YvrI functions as a σ factor for promoter recognition and that this activity is strongly dependent on YvrHa as a co-activator. However, these results do not unequivocably establish that YvrI and YvrHa function directly with core enzyme since the RNAP used in these studies contains σA and is likely contaminated with trace amounts of other σ factors.
Fig. 5. Biochemical characterization of YvrI as a σ factor.
A) Purified RNAP preparations used in assays either with σA (EσA) or depleted for σA (E). B) YvrI and YvrHa-dependent multi-round transcription from PoxdC in vitro. Template DNA (10 nM) encoding PoxdC was included with 100 nM purified B. subtilis RNA polymerase (EσA) and 2 μM each of purified σA, YvrI, and YvrHa as indicated. Specific transcription from PoxdC is predicted to yield an 82 nt run-off transcript. The asterisk indicates the weak signal noted in reactions supplemented with YvrI alone. C) KMnO4 footprinting at the oxdC promoter (template strand) using 37 nM B. subtilis RNAP (E) shown in panel A or 100 nM E. coli core polymerase (Epicentre Biotechnologies) (E (Eco)). Reactions contained RNAP alone (lane 1), with YvrI (lane 2), both YvrI and YvrHa (lane 3), or YvrI, YvrHa and σA (lane 4) Nucleotide assignments are based upon an A/G ladder generated from the same strand (not shown).
To unambiguously establish that YvrI together with the YvrHa coactivator can provide σ factor function independent of any other B. subtilis protein we have reconstituted holoenzyme using commercially prepared E. coli core RNA polymerase (Epicentre Biotechnologies). The reconstituted holoenzyme weakly activated the PoxdC promoter as judged using in vitro transcription assays (data not shown), so we have instead monitored holoenzyme function using potassium permanganate footprinting to probe for open-complex formation in the DNA. As a control, YvrI was added to B. subtilis RNAP depleted for σA (E) (Fig. 5A) in the presence or absence of YvrHa and σA (Fig. 5C). Two thymine residues at position −4 and −5 of the template strand are particularly susceptible to permanganate modification. Modification (strand opening) only occurs in the presence of B. subtilis RNAP (E) supplemented with both YvrI and YvrHa (lane 3) and supplementation with additional amounts of σA does not affect the reactivity (lane 4). Significantly, the holoenzyme reconstituted with E. coli core RNAP together with YvrI and YvrHa yields a reactivity pattern identical to that using B. subtilis RNAP (right panel, lane 3). There is also weak reactivity detectable upon supplementation with only YvrI (lane 2), consistent with the weak transcription activity noted in Fig. 5B. Interestingly, the inclusion of σA (lane 4) abolished reactivity when using E. coli core. We speculate that this reflects a reduced affinity of YvrI and/or YvrHa for E. coli core and therefore more effective sequestration of available core by σA.
Defining the YvrI regulon
We have sought to define the complete set of genes that are activated by YvrI (the YvrI regulon). To this end, we examined (i) the transcriptional response to YvrI overexpression in vivo using DNA microarrays, (ii) monitored the effects of adding YvrI and YvrHa on promoter recognition in vitro using ROMA (run-off transcription microarray analysis) (Cao et al., 2002a), and (iii) used search string and position-weight matrix approaches to identify candidate YvrI-dependent promoter sequences.
In vivo microarray analyses of cells harvested either 10 or 20 min after induction of YvrI revealed a strong and reproducible induction (>500-fold) of oxdC, confirming that oxdC is a major target for YvrI-dependent transcription, and yvrL. There was also a strong signal for yvrI due to the induction of this gene by xylose in these strains (see Fig S2 for a scatterplot from a representative experiment). Neither yvrHa nor yvrJ are represented on these microarray slides (since neither gene was annotated as part of the original B. subtilis genome project). Interestingly, a number of other loci were also moderately, but reproducibly, upregulated after YvrI induction. These include the yxeLMNOPQ operon and the yngE, sacB, and glgD genes. These genes were not identified in the ROMA-based analysis (see below) nor are they preceded by obvious YvrI-like promoters. Although induction of these genes likely reflects secondary effects of YvrI induction, these effects are reproducible and may merit further attention.
For the ROMA analysis, purified B. subtilis RNAP was used to transcribe total genomic DNA both in the absence and presence of a 20-fold molar excess of YvrI and YvrHa. Transcribed RNA was analyzed using microarray slides and a depiction of the results from a representative experiment is shown in Fig. S3. In the absence of added YvrI and YvrHa, a number of genes displayed weak signals as expected based on the presence of σ factor in the purified RNAP preparation. Upon addition of YvrI and YvrHa these signals decrease in intensity, consistent with the known competitive binding of σ factors to RNAP, and several new signals appeared. The strongest signals (Fig. S3) corresponded to the oxdC, yvrL and yvrI genes corroborating the results from the in vivo-based microarray analyses and from our previous genetic experiments. All remaining signals that were significantly above background were mapped to this same region of the genome (Fig. S3) and correspond to genes co-directional with, and therefore likely resulting from read-through transcription from, the PoxdC and the PyvrI promoters. Since termination is relatively inefficient in vitro, read-through transcripts can create artifactual signals in ROMA experiments using high molecular weight genomic DNA as template, as also seen in recent experiments to define the σM regulon (Eiamphungporn and Helmann, 2008). None of these downstream genes were detected as induced in microarray studies using RNA prepared from cells overexpressing YvrI-FLAG.
Next, we searched the B. subtilis genome to identify candidate YvrI-dependent promoters based on a postion-weight matrix constructed from the known promoter sequences (Fig. 2D). The highest scoring hits belonged to the previously identified oxdC, yvrJ, and yvrI promoters. All other hits scored much lower than the oxdC, yvrJ, and yvrI promoters, and none contained a T-rich linker sequence similar to that seen in known YvrI-dependent promoters (Fig. 2D). Moreover, there was no correspondence between these additional candidate promoters and genes upregulated in either microarray-based assays. Thus, on the basis of two conceptionally different microarray-based analyses and sequence-based studies, we conclude that oxdC-yvrL, yvrJ, and yvrI-yvrHa constitute the entire YvrI regulon.
yvrL is in an operon with oxdC
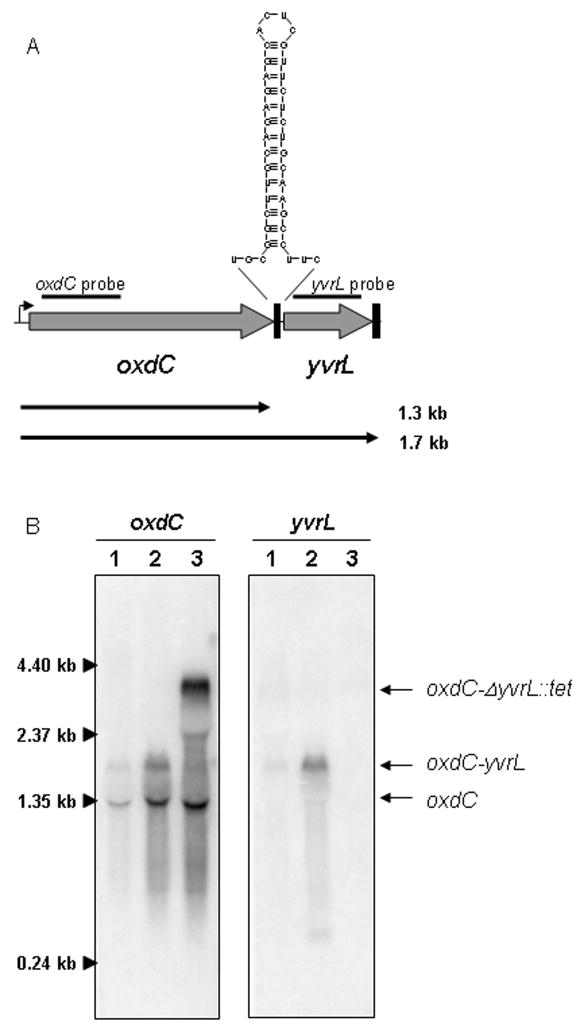
The yvrL gene was identified as a YvrI regulon member based on both in vivo- and in vitro-based microarray analyses. The oxdC-yvrL intergenic region contains a sequence encoding an inverted repeat that might function as a transcriptional terminator or attenuator, but there is no apparent YvrI-dependent promoter in this region (Fig. 6A). This suggests that yvrL may be co-transcribed with the upstream oxdC gene. Indeed, northern hybridization demonstrates that oxdC is transcribed primarily as a monocistronic mRNA, with a significant amount of readthrough transcript that also includes the yvrL gene (Fig. 6B). The oxdC probe hybridized to a ~1.3 kb band and a ~1.7 kb band while the yvrL probe hybridized only to the ~1.7 kb band demonstrating that it must be an operonic transcript. When the yvrL gene is entirely replaced with a tet cassette (lane 3), the 1.7 kb band increased in size (as detected by the oxdC probe) but was no longer detected by the yvrL probe, as expected. It was not determined from this analysis whether the smaller oxdC transcript results from transcription termination or increased stability of the 5′-end of a longer mRNA due, perhaps, to the presence of the noted stem-loop structure. Therefore, we fused DNA carrying PoxdC but ending either just before (HB7858) or just after (HB7857) the oxdC-yvrL intergenic region to a promoterless lacZ gene. Inclusion of the intergenic region resulted in an approximately 2-fold drop in β-galactosidase activity (not shown), consistent with the hypothesis that the stem-loop structure functions as an attenuator sequence. Regardless of mechanism, these results indicate that yvrL is expressed as part of a longer oxdC-yvrL transcript in cells in which YvrI is induced.
Fig. 6. Northern hybridization reveals operonic structure of oxdC-yvrL.
A) Diagram of oxdC-yvrl gene region showing predicted size of oxdC and oxdC-yvrL transcripts (arrows) and location of probes used for hybridization (Fig. 6B) below. Predicted structure of a stem-loop that may act as a transcriptional attenuator in the oxdC-yvrL intergenic region is shown. B) Northern hybridization using oxdC probe (panel 1) and yvrL probe (panel 2) against total RNA (5 μg) extracted from: lane 1, HB7709 cells (ectopic YvrI not induced); lane 2, HB7709 cells (ectopic YvrI induced); and lane 3, HB7813 cells (ectopic YvrI not induced). In HB7813, the yvrL gene has been replaced by a 2 kb tetracycline resistance cassette. Identification of bands indicated at right (arrows).
YvrL negatively regulates YvrI activity
In the course of the northern hybridization analysis, we observed that deletion of yvrL resulted in strongly increased transcription from PoxdC. Notably, in cells lacking yvrL the expression of oxdC was high even when YvrI was not induced (Fig. 6B; lane 3) whereas in the corresponding strain containing yvrL, expression was largely dependent on induction of YvrI (Fig. 6B; lane 1 vs. 2). Further evidence for a negative regulatory role of YvrL is apparent from studies using reporter fusion for all three YvrI-responsive promoters (PoxdC, PyvrI, PyvrJ) (Fig. S4). In these strains, YvrL represses the low basal promoter activity and reduces activity even under conditions where YvrI is induced as determined using β-galactosidase assays with an oxdC-lacZ fusion (Fig. S5). These results are consistent with the hypothesis that YvrL, a predicted membrane protein, negatively regulates YvrI-dependent transcription perhaps by acting as an anti-σ factor. Interestingly, overexpression of YvrI in a yvrL deletion strain results in cell lysis on plates (data not shown), a phenotype qualitatively similar to the effects of depleting cells of YhdL and YhdK, two negative regulators of the B. subtilis ECF sigma factor σM (Horsburgh and Moir, 1999). This phenotype is only observed in cells containing both YvrI and YvrHa, and is independent of OxdC overexpression, suggesting that it results from the unregulated activity of this alternative σ factor and perhaps competition for core RNAP.
Discussion
B. subtilis responds to cell envelope stresses by inducing the expression of a variety of protective enzymes, antimicrobial resistance proteins, and enzymes involved in cell wall and membrane functions (Jordan et al., 2008). Regulons induced by cell envelope stress include those controlled by the σB-dependent general stress response (Hecker et al., 2007), the σM (Eiamphungporn and Helmann, 2008; Jervis et al., 2007) and σW (Butcher and Helmann, 2006; Cao et al., 2002a) ECF σ factors, and the LiaRS TCS (Jordan et al., 2007; Mascher et al., 2004). Several of the cell envelope stress stimulons also include genes herein assigned to the YvrI/YvrHa regulon including both yvrI itself and yvrL. We have demonstrated that YvrI (in the presence of YvrHa) functions as a novel σ factor for RNAP to activate expression of three promoter sites controlling the expression of the yvrI-yvrHa, oxdC-yvrL, and yvrJ genes (Fig. 2A).
YvrI is about the size (22.5 kDa) of many ECF σ factors and carries a predicted region 4 in its C-terminus. There is no obvious region 2 in YvrI which is otherwise strongly conserved in other σ factors (Lonetto et al., 1992). However, a close inspection of alignments between YvrI homologs (Fig. S1b) and other σ factors (Fig. S1a) hints at what might be a vestigial region 2: a hypothesis currently being tested by site-directed mutagenesis. Alternatively, region 2 functions may be provided instead by the co-activator YvrHa. The size of YvrI and the fact that its primary regulatory target is an extracytoplasmic, stress-inducible protein (OxdC) indicates that this regulator may, in a physiological sense, most closely resemble ECF σ factors. Consistent with its role as a σ subunit for RNAP, YvrI co-purifies with RNAP (Fig. 4) and can, together with YvrHa, enable initiation at the oxdC promoter both in vitro and in vivo (Figs. 3 and 5B). We find no evidence of YvrI specifically binding to promoter DNA in the absence of RNAP (not shown). Holoenzyme reconstitution studies using E. coli core RNAP demonstrate that YvrI together with YvrHa are sufficient for site-specific strand opening at the oxdC promoter indicating that no other B. subtilis proteins are required for promoter recognition and melting (Fig. 5C). The three promoters activated by YvrI and YvrHa have similar sequences in the –35 and –10 elements and have a T-rich spacer element (Fig. 2D). Genome comparisons suggest that a related regulatory system is present in other Bacilli including B. licheniformis, B. thuringiensis, B. cereus, B. clausii, and Oceanobacillus iheyensis.
In addition to its own operon (yvrI-yvrHa), YvrI activates the oxdC-yvrL and yvrJ genes. The function of yvrJ is presently unknown, and yvrL appears to function, at least in part, as a membrane-bound negative feedback regulator. The overall physiological role of the YvrI/YvrHa regulatory system appears to be focused on induction of OxdC, an abundant manganese-dependent oxalate decarboxylase that accumulates in the cell wall in cells grown in acidic medium (Antelmann et al., 2007; Tanner and Bornemann, 2000; Tanner et al., 2001). It is postulated that OxdC, in the presence of oxalate, may help buffer the medium by proton consumption. However, we have been unable to demonstrate an acid-sensitive growth phenotype for oxdC or yvrI mutant strains.
The pathways leading to activation of the YvrI-YvrHa regulatory system, in response to cell envelope stress and low pH, are not yet understood. A previous analysis (Serizawa et al., 2005) demonstrated that a yvrI-yvrHa transcript appeared late in exponential growth and appeared to depend upon the adjacent two-component regulatory system (yvrHb-yvrG, Fig. 2A). However, we did not observe any effect of deletion of yvrHb-yvrG on activation of PoxdC or PyvrI whether in response to overexpression of YvrI or growth under low pH conditions (data not shown).
An intriguing aspect of YvrI function concerns the role of the YvrHa coactivator. Although the functional role of YvrHa is unclear, it is reminiscent of other σ factor systems where a co-activator is implicated (Browning and Busby, 2004). YvrHa might function as a conventional DNA-binding activator. However, YvrHa lacks obvious DNA-binding domains and we have been unable to detect an interaction with promoter DNA. In E. coli, the activation of several σS-specific promoters is partially dependent upon Crl (Bougdour et al., 2004; Pratt and Silhavy, 1998). Crl binds to σS (Bougdour et al., 2004) and plays a role in recruiting this σ factor to form holoenzyme. In vitro the protein appears to have a stimulatory effect on other σ factors as well but in vivo seems to bias holoenzyme formation towards RpoS (Gaal et al., 2006; Typas et al., 2007). Transcription of bacteriophage T4 late genes is also dependent upon two proteins: gp55, a σ factor distantly related to the σ70 family, and gp33 (Gribskov and Burgess, 1986; Kassavetis and Geiduschek, 1984; Williams et al., 1989; Winkelman et al., 1994). gp33 interacts with the β flap of RNAP (Nechaev et al., 2004), an interaction site also contacted by region 4 in σ70-family regulators. gp55 and gp33 are therefore thought to constitute a two-subunit phage σ factor (Nechaev et al., 2004). In several cases, σ factors are synthesized as inactive pro-proteins that are activated by cleavage of an inhibitory amino-terminal domain (Cutting et al., 1990; LaBell et al., 1987; Lu et al., 1990). A processing role for YvrHa seems unlikely since YvrI mobility is unchanged when expressed in cells containing or lacking YvrHa. Therefore, we currently favor a model in which YvrHa interacts directly with either YvrI or RNAP core enzyme (or both) to enable YvrI function.
The presence of OxdC as one of the most highly abundat cell wall proteins in acid-grown cells suggests that this protein plays an important, although still ill-defined, role. While OxdC has been thoroughly studied in terms of reaction mechanism and structure (Anand et al., 2002; Just et al., 2004), the regulation of OxdC expression had not previously been explored. Our results suggest that expression of this protein relies on a previously undescribed and unusual σ factor (YvrI), together with a coactivator protein (YvrHa). The regulatory processes that contribute to the high level accumulation of OxdC are unclear as is its mechanism of translocation to the cell wall. The pathways, and ultimately the significance, of the observed induction of yvrI, yvrL, and oxdC by cell envelope stress also remain to be elucidated.
Experimental Procedures
Bacterial strains and growth conditions
All strains (Table S1) were grown at 37°C in LB broth or this medium solidified with 1.5% agar. Supplements as required were added to the following concentrations: ampicillin (100 μg ml−1), erythromycin (1 μg ml−1), lincomycin (25 μg ml−1), spectinomycin (100 μg ml−1), tetracycline (10 μg ml−1), chloramphenicol (10 μg ml−1), kanamycin (10 μg ml−1), 5-bromo-4-chloro-3-indoyl-β-D-galactopyranoside (80 μg ml−1), isopropyl-β-D-thiogalactopyranoside (1 mM), and D-xylose (2%). Antibiotics in broth were used at half the concentration above.
Genetic methods
Unmarked in-frame gene deletions were generated using the conditionally replicating plasmid pMAD as described (Arnaud et al., 2004) (Table S2). Briefly, 700–800 bp regions upstream and downstream of each targeted gene were amplified by PCR and fused using respective 3′ and 5′ SalI sites. The distal primers were used to amplify the fusion product prior to cloning into pMAD. Primers (Table S3) used to amplify the upstream and downstream fragments (respectively) were: ΔyvrI (2849/2850 and 2851/2852); ΔyvrHa (2853/2854 and 2855-2856) and ΔyvrI-yvrHa (2849/2850 and 2855/2856). Gene replacement with an antibiotic resistance gene was conducted using LFH-PCR (long-flanking-homology-PCR) as described (Butcher and Helmann, 2006; Mascher et al., 2003). Site-directed mutagenesis was conducted using a protocol based upon the Stratagene Quik-Change Mutagenesis kit with the oligomers listed in Table S3. All other genetic manipulations were conducted using standard methodologies.
Primer extension
Primer extension analysis was conducted on 40 μg total bacterial RNA isolated using an RNeasy kit (Qiagen) from strain HB7709 or this strain induced with 2% xylose. RNA and 32P-end-labeled primers (~ 1 ×105 cpm) were mixed, heated to 80°C and allowed to cool slowly to room temperature. cDNA was reverse transcribed using Multiscribe reverse transcriptase (Applied Biosystems) as previously described (MacLellan et al., 2005). Primers used for oxdC, yvrI and yvrJ reactions were #2836, 2835, and 2962, respectively (Table S3). A ladder was generated using Sequenase version 2 (USB) sequencing reactions on plasmid pSM003. Reaction products were separated on a 6% denaturing polyacrylamide sequencing gel and visualized using a phosphoimaging cassette.
Northern hybridization
Internal fragments of the oxdC and yvrL genes were amplified, purified and approximately 50 ng of each product was labeled with [α-32P]dATP using the DECAprime II Random Priming DNA Labeling Kit (Ambion). Unincorporated [α-32P]dATP was removed by NucAway spin columns (Ambion). For the hybridization, 5 or 10 μg of total RNA from uninduced HB7709 or HB7813 cells or HB7709 cells induced with 2% xylose for 15 minutes was loaded on a formaldehyde Agarose gel. After electrophoresis, the RNA was transferred to a Zeta-Probe Blotting Membrane in a downward transfer using 10x SSC (1.5 M NaCl, 0.15 M sodium citrate) as transfer buffer and the RNA was crosslinked to membrane with UV light. The blot was prehybridized at 42°C for at least 30 min with Ultrahyb buffer (Ambion) and the labeled probe was added after denaturing at 95°C for 10 min. The hybridization was performed overnight at 42°C. On the next day, the membrane was washed twice with low-stringency buffer (2x SSPE (Ambion) plus 0.1% SDS) at room temperature for 5 min, followed by two high-stringency washes (0.1% SSPE (Ambion) plus 0.1% SDS) at 42°C for 15 min. The membrane was wrapped in plastic wrap, exposed to a phosphor screen, and analyzed using a PhosphorImager. oxdC and yvrL probes were amplified with primers 3422-3423 and 3424-3425, respectively.
Protein Purification
RNAP holoenzyme was purified essentially as described (Helmann, 2003) with some modifications. Briefly, holoenzyme was purified from 200 g B. subtilis strain MH5636 cells. Cells were lysed by four passes through a French pressure cell and nucleic acids were precipitated from the clarified cell lysate using 0.5% polyethyleneimidine (PEI) (pH 7.9). Bound proteins were eluted from the PEI pellet using 1.0 M NH4Cl, precipitated with ammonium sulphate, dialyzed, and applied to a 5 ml heparin sepharose column. Protein was eluted using a 0–0.8 M NaCl gradient and then precipitated using four volumes of 3.9 M NH4SO4 (pH 7.9). The precipitate was resuspended in a minimal volume of TGED buffer (10 mM Tris-HCl (pH 8.0) containing 0.1 mM EDTA, 0.1 mM DTT, 10% glycerol) with 500 mM NaCl, dialyzed overnight, and applied to a superdex 200 chromotography column equilibriated in the same buffer. Eluant fractions containing RNAP were pooled and dialyzed into TGED buffer modified to contain 50% glycerol and 50 mM NaCl and stored at −20°C. The amount of σA co-purifying with RNAP varied dramatically between preparations and between fractions and are designated as EσA and E for those fractions containing near stoichiometric amounts of σA and depleted for σA, respectively (see Figure 5A).
YvrI-FLAG was purified from E. coli pLysS cells after induction with 1 mM IPTG. Cells were resuspended in TGED and lysed by freeze-thaw and passage through a French pressure cell. Inclusion bodies and cellular debris were collected by centrifugation and washed three times in TGED containing 0.5% Triton-X100 (TGEDX) and one wash with TGED containing 2% sodium deoxycholate. Inclusion bodies were resuspended in 5 ml TGEDX and solubilized by gradually adding 1.5% sarkosyl. After centrifugation, the supernatant containing solubilized protein was slowly diluted to 50 ml with TGEDX and dialyzed overnight in 4 l TGEDX. Re-natured YvrI-FLAG was captured from solution by passing the dialysate through a 5 ml S sepharose column. Protein was eluted with 0.1 to 0.8 M NaCl gradient. Fractions containing YvrI-FLAG were pooled, dialyzed into TGED containing 50% glycerol, and stored at −20°C.
YvrHa-HA was purified from E. coli BL21 pLysS cells after induction with 1 mM IPTG for 2.5 h. The cell pellet was subjected to freeze-thaw, resuspended in 25 ml TGED containing 200 mM NaCl, and passed through a French pressure cell four times. Clarified lysate was loaded onto a 5 ml DEAE-sepharose column and proteins were eluted with a 60 ml 0.2–0.7 M NaCl gradient plus an additional 30 ml 0.7 M NaCl in TGED. YvrHa-HA eluted late in the gradient and fractions containing the protein were pooled and subjected to precipitation with 2 volumes of 3.9 M NH4SO4 (pH 7.9). The re-dissolved pellet was dialyzed, bound to a 10 ml monoQ column and eluted with and 0 to 1.0 M NaCl. Proteins were further separated upon a Superdex 75 column. The fractions containing YvrHa were pooled and dialyzed into TGED containing 50% glycerol.
Induction of FLAG-tagged SigA in E. coli BL21 with 1 mM IPTG resulted in overexpressed protein that was approximately 50% soluble. After cell lysis, soluble proteins were precipitated with solid ammonium sulphate (to 80% saturation). The protein precipitate was resuspended in TGED, dialyzed and applied to a 10 ml DEAE-sepharose column. Bound proteins were eluted with a NaCl step gradient. Relevant fractions were desalted by dialysis and applied to a 10 ml monoQ column. Protein was eluted with a 0 to 1.0 M NaCl gradient. Relevant fractions were pooled and dialyzed into TGED containing 50% glycerol before storage at −20°C.
RNAP pull-down assays
YvrI-FLAG under the control of the xylA promoter was integrated into strain CU1065 (to form strain HB7709) or strain JH642 PolHis (to form strain HB7711). Overnight cultures of both strains were used to inoculate fresh 100 ml volumes of LB broth. Cultures were grown to an O.D. of 0.4, induced with 2% xylose and grown for a further 1 h. Cells were harvested and subjected to lysis with a French pressure cell. Clarified lysate was supplemented with 5 U DNase I and incubated for 30 min at room temperature. Each lysate was further supplemented with 0.5 ml of Ni-NTA metal affinity beads and incubated with slow rotation for 1 h at 4°C. Beads were recovered and washed with 10 column volumes of 50 mM phosphate buffer (pH 8.0) containing 100 mM NaCl. To both columns were applied successive 0.5 ml washes using the buffer above but with increasing concentrations (5, 10, 20, 30, and 250 mM) of imidazole. Each wash eluant was collected as it has been our experience that tagged RNAP binds rather weakly to Ni-NTA beads and substantial elution of the protein occurs even at low imidazole concentrations. Volumeteric equivalents of the crude lysate, flow through, and each eluant for each of the two strains were run on duplicate 10% SDS-PAGE gel. For both strains, one gel was subjected to coomassie staining and the other was subjected to western immunoblotting using anti-FLAG antibodies in order to visualize YvrI-FLAG.
Potassium permanganate footprinting
Open complex formation at the oxdC promoter was detected using permanganate footprinting (Sasse-Dwight and Gralla, 1989) and plasmid pSM106 (Table S2). Purified RNAP was pre-mixed with 20-fold excess of σ factors or YvrHa as required in reconstitution buffer (10 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 10 mM KCl, 50 mM NaCl, 50% glycerol, 0.1 mM DTT) and incubated on ice for 30 minutes. Each 20 μl footprinting reaction contained 1 μg pSM106 DNA and RNAP (37 nM or 100 nM of either B. subtilis or E. coli core, respectively) in a buffer containing 10 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 10 mM KCl, 10 mM NaCl, 5% glycerol, 0.1 mM DTT, and 100 ug ml−1 BSA. After the protein mixture and DNA were added, the reactions were incubated at 37°C for 20 minutes to allow for holoenzyme:promoter binding. The reactions were started by the addition of 1 μl of 40 mM KMnO4. After incubation for 3 minutes at 37°C, reactions were stopped by the addition of 1 μl 14.3 M β-mercaptoethanol and placed on ice. Primer extension was used to visualize modified bases in the template strand DNA. Each 20 μl reaction contained 7.5 μl Thermo-start Master mix (Thermo Scientific), 1 μl end-labeled primer 3891 (~ 105 cpm), and 5 μl of the footprinting reaction mixture. Extension reactions were carried out in a thermocycler for 29 cycles. Each reaction was supplemented with an equal volume of formamide dye and separated upon a 6% polyacrylamide denaturing (8 M urea) sequencing gel. An A/G ladder was used to calibrate positions on the gel.
ROMA analysis
ROMA (run off in vitro transcription/microarray analysis) is a method that involves detection of in vitro transcription products after reacting holoenzyme with sheared genomic DNA (Cao et al., 2002a). A typical reaction (50 μl) contained 100 nM RNA polymerase, 2 μM each YvrI and YvrHa (20-fold molar excess relative to RNA polymerase), 1 μg chromosomal DNA of W168 (sheared by pipetting and vortexing), 40 nmol CTP, GTP, ATP and UTP, and 5 μl 10× transcription buffer (180 mM Tris, pH 8.0, 100 mM NaCl, 100 mM KCl, 100 mM MgCl2, 50% glycerol, 10 mM DTT and 100 μg ml−1 acetylated BSA). A control reaction contained RNA polymerase, but not YvrI and YvrHa. Reconstitution of holoenzyme occurred on ice for 15 min. After adding DNA and a further incubation for 10 minutes at 37°C, the reaction was started by the addition of NTPs and was allowed to proceed for 20 minutes at 37°C. The reaction was stopped by adding 200 μl stop solution (2.5 M NH4OAc, 10 mM EDTA and linear acrylamide (15 μg ml−1). For each treatment, three individual reactions were combined, extracted with phenol/chloroform and the RNA was precipitated overnight at −80°C. The pellet was dissolved in 20 μl DEPC-treated water and the template DNA was removed by a 30 min treatment with TURBO DNase (Ambion). The mRNA was used to generate fluorescence labeled cDNA using the SuperScript Indirect cDNA Labeling System (Invitrogen) following the provided instructions. The cDNA from the control reaction without σ factor was labeled with Alexa Fluor 555 and the cDNA from the reaction containing YvrI and YvrHa was labeled with Alexa Fluor 647. Both cDNA populations were hybridized overnight on an oligonucleotide-based microarray slide and scanned and analyzed using the software GenePix Pro 4.0 (Axon Instruments) as described below.
Microarray analyses
The strains used for this analysis were HB7777 and HB7799. For RNA isolation, 5 ml LB inoculated with 50 μl of an overnight culture of these strains were grown at 37°C to an OD600 of 0.3. These pre-cultures were used to inoculate fresh 40 ml volumes of LB medium. Upon growth to OD600 of 0.4, each culture was split into 20 ml volumes and induced with 2% xylose for 10 or 20 min, respectively. After induction, the 20 ml culture were mixed with 4 ml stop solution (95:5 ethanol:phenol), and the cells were harvest by centrifugation at 5000 rpm for 5 min. The pellet was washed with 5 ml TE-buffer (3 mM Tris-HCl, pH 8.0, 1 mM EDTA), resuspended in 750 μl lysozyme-TE (30 mM Tris-HCl, 1 mM EDTA, pH 8.0, 15 mg ml−1 lysozyme), and incubated with shaking at 37°C for 25 min. After the first 15 minutes, 20 μl of Proteinase K (20 mg ml−1) was added and incubated for an additional 10 min. After lysis, 4 ml RLT buffer supplemented with β-mercaptoethanol was added, and RNA isolation was performed with the RNeasy Midi kit (Qiagen) according to the manufacturer’s protocol. The RNA was eluted with 150 μl of RNase-free water and contaminating genomic DNA was removed by a 30 min treatment with TURBO DNase (Ambion). After ethanol precipitation, 20 μg of total RNA was used for first-strand cDNA synthesis which was then labelled using the SuperScript Indirect cDNA Labeling System (Invitrogen). The cDNA derived from strain HB7777 was labeled with Alexa Fluor 647 and the cDNA from HB7799 with Alexa Fluor 555. Approximately 100 pmol of each dye were used to hybridize the labeled cDNA to a 65-mer oligonucleotide-based microarray representing each annotated B. subtilis W168 gene in duplicate. The hybridization was carried out overnight at 42°C. The slides were washed, scanned and the data was normalized using the software GenePix Pro 4.0 (Axon Instruments). The average of the median intensities of the spot duplicates was calculated after background subtraction and the data was filtered to remove (i) genes that were not significantly expressed under both conditions (average of sum of medians < 10) or (ii) genes where the deviation of the two spots was >30% of the average.
Enzyme assays
Multiple-round in vitro transcription reactions were conducted at 37°C for 20 minutes in 10 μl reaction volumes. DNA templates were prepared using PCR and the primers described in Table S3 and were used at a concentration of 10 nM in each reaction. Purified RNAP was used at a final concentration of 40 nM and purified YvrI, YvrHa, or SigA were added as required to a concentration of 200 nM. The reaction buffer contained: 10 mM Tris-HCl (pH 7.5), 0.15 M KCl, 0.01 M MgCl2, 1 mM DTT, acetylated BSA (100 ng ml−1), 1 mM of ATP and GTP, 0.5 mM CTP, 0.05 mM UTP, and 20–50 μCi 32P-[αUTP]. Radio-labeled Decade markers (Ambion Inc) were used as standards and all products were separated upon 6% denaturing (7 M Urea) polyacrylamide sequencing gels. Beta-galactosidase assays were conducted using standard procedures.
Mass spectrometry
OxdC was identified using trypsin digestion followed by matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) mass spectrometry. The relevant protein band was excised from a coomassie blue stained and destained SDS-PAGE gel and washed twice in 20 μl volumes of 50 mM ammonium bicarbonate containing 50% acetonitrile for 10 minutes at 37°C. After drying, gel slice was re-hydrated in a 40 mM ammonium bicarbonate solution containing 9% acetonitrile and 20 ng μl−1 trypsin and incubated at 37°C overnight. The sample was analyzed using an Applied Biosystems 4700 mass spectrometer. Confirmation of the expected size of YvrI-FLAG was conducted by dialyzing 5 μg purified protein into 0.1% formic acid using drop dialysis and subjecting the sample to ESI-MS analysis.
Bioinformatic and other computational methods
Sequence alignments were conducted using ClustalW. Protein domain predictions were carried out with CD-search (Marchler-Bauer and Bryant, 2004) on the conserved domain database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) or Pfam (http://pfam.sanger.ac.uk/) and predictionsof helix-turn helix motifs were by a program from Network Protein Sequence Analysis (http://npsa-pbil.ibcp.fr/NPSA/npsa_hth.html) using a previously published prediction method (Dodd and Egan, 1990). Search strings constructed from the YvrI-dependent promoter consensus was used to screen the B. subtilis genome on the Subtilist website. The program Virtual Footprint (http://www.prodoric.de/vfp/) was used to construct a position weight matrix based on B. subtilis and B. licheniformis known or predicted promoters and to screen the B. subtilis genome for similar sequences. For some figures, digital images were manipulated with Adobe photoshop to improve visual presentation without altering critical information content.
Genome sequence corrections and amendments
The B. subtilis 168 genome as originally annotated describes a gene order of yvrI, yvrH, and yvrG where yvrHG were predicted to encode a two-component regulatory system. During their analysis of YvrHG function, Kobayashi et al. (2001) discovered a sequencing error (single G insertion at nucleotide number 336) in the original sequence that generated a shift in the YvrH open reading frame (Kobayashi et al., 2001). Correction of this error resulted in the annotation of two separate ORFs that they re-named YvrHa and YvrHb. Using northern hybridization, Serizawa et al (2005) deduced that yvrI and yvrHa constitute an operon and that yvrHb and yvrG comprise a separate operon likely encoding a two-component regulatory system (Serizawa et al., 2005). This new assignment closely matches the ORF prediction in B. licheniformis. In the course of our analysis we detected another frame shifting single nucleotide insertion (C at nucleotide position 239) in yvrHa. The effect of this sequencing error was to increase the predicted length of YvrHa to 113 amino acids before a fortuitous stop codon was encountered. Correction of this error results in a predicted length of 79 amino acids for YvrHa, which renders the length of the protein consistent with homolog predictions in other bacilli. Finally, our sequencing also revealed that the third to last amino acid codon in the yvrI gene should be GGG instead of AGG resulting in an R to G amino acid change at this position.
Acknowledgments
We thank Anna-Barbara Hachmann for guidance with the DNA microarray and ROMA experiments and for critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health (GM-047446).
References
- Anand R, Dorrestein PC, Kinsland C, Begley TP, Ealick SE. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 Å resolution. Biochemistry. 2002;41:7659–7669. doi: 10.1021/bi0200965. [DOI] [PubMed] [Google Scholar]
- Antelmann H, Towe S, Albrecht D, Hecker M. The phosphorus source phytate changes the composition of the cell wall proteome in Bacillus subtilis. J Proteome Res. 2007;6:897–903. doi: 10.1021/pr060440a. [DOI] [PubMed] [Google Scholar]
- Anthony LC, Artsimovitch I, Svetlov V, Landick R, Burgess RR. Rapid purification of His(6)-tagged Bacillus subtilis core RNA polymerase. Protein Expr Purif. 2000;19:350–354. doi: 10.1006/prep.2000.1272. [DOI] [PubMed] [Google Scholar]
- Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar AP, Zhao X, Brown ED. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl Environ Microbiol. 2001;67:403–410. doi: 10.1128/AEM.67.1.403-410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougdour A, Lelong C, Geiselmann J. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J Biol Chem. 2004;279:19540–19550. doi: 10.1074/jbc.M314145200. [DOI] [PubMed] [Google Scholar]
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- Butcher BG, Helmann JD. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol Microbiol. 2006;60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- Cao M, Kobel PA, Morshedi MM, Wu MF, Paddon C, Helmann JD. Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J Mol Biol. 2002a;316:443–457. doi: 10.1006/jmbi.2001.5372. [DOI] [PubMed] [Google Scholar]
- Cao M, Wang T, Ye R, Helmann JD. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol Microbiol. 2002b;45:1267–1276. doi: 10.1046/j.1365-2958.2002.03050.x. [DOI] [PubMed] [Google Scholar]
- Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- deHaseth PL, Helmann JD. Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA. Mol Microbiol. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- Dodd IB, Egan JB. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W, Helmann JD. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol Microbiol. 2008 doi: 10.1111/j.1365-2958.2007.06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T, Mandel MJ, Silhavy TJ, Gourse RL. Crl facilitates RNA polymerase holoenzyme formation. J Bacteriol. 2006;188:7966–7970. doi: 10.1128/JB.01266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd GM. Fungal production of citric and oxalic acid: importance in metal speciation, physiology and biogeochemical processes. Adv Microb Physiol. 1999;41:47–92. doi: 10.1016/s0065-2911(08)60165-4. [DOI] [PubMed] [Google Scholar]
- Gribskov M, Burgess RR. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986;14:6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Hecker M, Pane-Farre J, Volker U. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–236. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- Helmann JD. Purification of Bacillus subtilis RNA polymerase and associated factors. Methods Enzymol. 2003;370:10–24. doi: 10.1016/S0076-6879(03)70002-0. [DOI] [PubMed] [Google Scholar]
- Horsburgh MJ, Moir A. Sigma M, an ECF RNA polymerase sigma factor of Bacillus subtilis 168, is essential for growth and survival in high concentrations of salt. Mol Microbiol. 1999;32:41–50. doi: 10.1046/j.1365-2958.1999.01323.x. [DOI] [PubMed] [Google Scholar]
- Jervis AJ, Thackray PD, Houston CW, Horsburgh MJ, Moir A. SigM-responsive genes of Bacillus subtilis and their promoters. J Bacteriol. 2007;189:4534–4538. doi: 10.1128/JB.00130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S, Rietkotter E, Strauch MA, Kalamorz F, Butcher BG, Helmann JD, Mascher T. LiaRS-dependent gene expression is embedded in transition state regulation in Bacillus subtilis. Microbiology. 2007;153:2530–2540. doi: 10.1099/mic.0.2007/006817-0. [DOI] [PubMed] [Google Scholar]
- Jordan S, Hutchings MI, Mascher T. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol Rev. 2008;32:107–146. doi: 10.1111/j.1574-6976.2007.00091.x. [DOI] [PubMed] [Google Scholar]
- Juang YL, Helmann JD. A promoter melting region in the primary sigma factor of Bacillus subtilis. Identification of functionally important aromatic amino acids. J Mol Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- Just VJ, Stevenson CE, Bowater L, Tanner A, Lawson DM, Bornemann S. A closed conformation of Bacillus subtilis oxalate decarboxylase OxdC provides evidence for the true identity of the active site. J Biol Chem. 2004;279:19867–19874. doi: 10.1074/jbc.M313820200. [DOI] [PubMed] [Google Scholar]
- Just VJ, Burrell MR, Bowater L, McRobbie I, Stevenson CE, Lawson DM, Bornemann S. The identity of the active site of oxalate decarboxylase and the importance of the stability of active-site lid conformations. Biochem J. 2007;407:397–406. doi: 10.1042/BJ20070708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Geiduschek EP. Defining a bacteriophage T4 late promoter: bacteriophage T4 gene 55 protein suffices for directing late promoter recognition. Proc Natl Acad Sci U S A. 1984;81:5101–5105. doi: 10.1073/pnas.81.16.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ogura M, Yamaguchi H, Yoshida K, Ogasawara N, Tanaka T, Fujita Y. Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J Bacteriol. 2001;183:7365–7370. doi: 10.1128/JB.183.24.7365-7370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Codani JJ, Connerton IF, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- LaBell TL, Trempy JE, Haldenwang WG. Sporulation-specific sigma factor sigma 29 of Bacillus subtilis is synthesized from a precursor protein, P31. Proc Natl Acad Sci U S A. 1987;84:1784–1788. doi: 10.1073/pnas.84.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Halberg R, Kroos L. Processing of the mother-cell sigma factor, sigma K, may depend on events occurring in the forespore during Bacillus subtilis development. Proc Natl Acad Sci U S A. 1990;87:9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLellan SR, Smallbone LA, Sibley CD, Finan TM. The expression of a novel antisense gene mediates incompatibility within the large repABC family of alpha-proteobacterial plasmids. Mol Microbiol. 2005;55:611–623. doi: 10.1111/j.1365-2958.2004.04412.x. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol. 2003;50:1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- Mascher T, Zimmer SL, Smith TA, Helmann JD. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother. 2004;48:2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Kamali-Moghaddam M, Andre E, Leonetti JP, Geiduschek EP. The bacteriophage T4 late-transcription coactivator gp33 binds the flap domain of Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A. 2004;101:17365–17370. doi: 10.1073/pnas.0408028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LA, Silhavy TJ. Crl stimulates RpoS activity during stationary phase. Mol Microbiol. 1998;29:1225–1236. doi: 10.1046/j.1365-2958.1998.01007.x. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S, Gralla JD. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989;264:8074–8081. [PubMed] [Google Scholar]
- Serizawa M, Kodama K, Yamamoto H, Kobayashi K, Ogasawara N, Sekiguchi J. Functional analysis of the YvrGHb two-component system of Bacillus subtilis: identification of the regulated genes by DNA microarray and northern blot analyses. Biosci Biotechnol Biochem. 2005;69:2155–2169. doi: 10.1271/bbb.69.2155. [DOI] [PubMed] [Google Scholar]
- Sharp MM, Chan CL, Lu CZ, Marr MT, Nechaev S, Merritt EW, Severinov K, Roberts JW, Gross CA. The interface of sigma with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 1999;13:3015–3026. doi: 10.1101/gad.13.22.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedruzic D, Liu Y, Reinhardt LA, Wroclawska E, Cleland WW, Richards NG. Investigating the roles of putative active site residues in the oxalate decarboxylase from Bacillus subtilis. Arch Biochem Biophys. 2007;464:36–47. doi: 10.1016/j.abb.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner A, Bornemann S. Bacillus subtilis YvrK is an acid-induced oxalate decarboxylase. J Bacteriol. 2000;182:5271–5273. doi: 10.1128/jb.182.18.5271-5273.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner A, Bowater L, Fairhurst SA, Bornemann S. Oxalate decarboxylase requires manganese and dioxygen for activity. Overexpression and characterization of Bacillus subtilis YvrK and YoaN. J Biol Chem. 2001;276:43627–43634. doi: 10.1074/jbc.M107202200. [DOI] [PubMed] [Google Scholar]
- Typas A, Barembruch C, Possling A, Hengge R. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of sigmas activity and levels. Embo J. 2007;26:1569–1578. doi: 10.1038/sj.emboj.7601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KP, Muller R, Ruger W, Geiduschek EP. Overproduced bacteriophage T4 gene 33 protein binds RNA polymerase. J Bacteriol. 1989;171:3579–3582. doi: 10.1128/jb.171.6.3579-3582.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MJ, Lamont IL. Mutational analysis of an extracytoplasmic-function sigma factor to investigate its interactions with RNA polymerase and DNA. J Bacteriol. 2006;188:1935–1942. doi: 10.1128/JB.188.5.1935-1942.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman JW, Kassavetis GA, Geiduschek EP. Molecular genetic analysis of a prokaryotic transcriptional coactivator: functional domains of the bacteriophage T4 gene 33 protein. J Bacteriol. 1994;176:1164–1171. doi: 10.1128/jb.176.4.1164-1171.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipat A, Brignell SC, Guy BJ, Rose M, Emmerson PT, Harwood CR. The yvsA-yvqA (293 degrees–289 degrees) region of the Bacillus subtilis chromosome containing genes involved in metal ion uptake and a putative sigma factor. Microbiology. 1998;144(Pt 6):1593–1600. doi: 10.1099/00221287-144-6-1593. [DOI] [PubMed] [Google Scholar]
- Wong K, Kassavetis GA, Leonetti JP, Geiduschek EP. Mutational and functional analysis of a segment of the sigma family bacteriophage T4 late promoter recognition protein gp55. J Biol Chem. 2003;278:7073–7080. doi: 10.1074/jbc.M211447200. [DOI] [PubMed] [Google Scholar]