Summary
Brown fat is specialized in energy expenditure, a process that is principally controlled by the transcriptional co-activator PGC-1α. Here we describe a molecular network important for PGC-1α function and brown fat metabolism. We find that twist-1 is selectively expressed in adipose tissue, interacts with PGC-1α, and is recruited to the promoters of PGC-1α’s target genes to suppress mitochondrial metabolism and uncoupling. In vivo, transgenic mice expressing twist-1 in the adipose tissue are prone to high-fat diet induced obesity, whereas twist-1 heterozygous knockout mice are obesity-resistant. These phenotypes are attributed to their altered mitochondrial metabolism in the brown fat. Interestingly, the nuclear receptor PPARδ not only mediates the actions of PGC-1α, but also regulates twist-1 expression, suggesting a negative feedback regulatory mechanism. These findings reveal an unexpected physiological role for twist-1 in the maintenance of energy homeostasis and have important implications for understanding metabolic control and metabolic diseases.
Introduction
Obesity and its associated metabolic diseases are caused by a long-term imbalance between energy intake and energy expenditure. Adipose tissues serve as major sites to control energy balance. They are present in two functionally distinct types, white fat and brown fat. White fat stores excess energy in the form of triglycerides and releases them in times of energy need. By contrast, brown fat is specialized for energy expenditure by dissipating energy as heat, a process termed as adaptive thermogenesis (Cannon and Nedergaard, 2004; Lowell and Spiegelman, 2000). The unique metabolic property of brown fat is due to its high mitochondrial density and fuel oxidation capacity, and exclusive expression of uncoupling protein-1 (UCP1) in the inner mitochondrial membrane, which uncouples mitochondrial proton gradient from ATP production. Given the fundamental importance of adipose tissues in the maintenance of systematic energy homeostasis, their functions must be tightly regulated.
As a heat-generating organ, brown fat plays a key part in the regulation of energy balance and obesity, as evidenced in rodent studies. For instance, either ablation of brown fat through expression of a toxic transgene or knockout of UCP1 leads to high susceptibility to diet-induced obesity (Kontani et al., 2005; Lowell et al., 1993), whereas increase of UCP1 expression protects animals against diet-induced obesity (Kopecky et al., 1995). However, human adults, unlike rodents and human neonates, do not possess discrete brown fat depots, and brown fat cells are dispersed within white fat, casting doubt whether human brown fat cells are of physiological and/or pharmacological significance. On the other hand, it has long been observed that brown fat cells in humans have a remarkable capacity for recruitment and expansion in the presence of high sympathetic input or subject to prolonged cold exposure (Garruti and Ricquier, 1992; Huttunen et al., 1981; Lean et al., 1986). Moreover, recent tracer studies coupled with imaging technology demonstrated a much wider anatomic distribution of brown fat than was previously thought in several regions of the human body in normal individuals (reviewed in Nedergaard et al., 2007). Both cell culture and animal model studies also suggest that white fat cells show plasticity and can be induced to acquire brown fat features (Seale et al., 2007; Tiraby and Langin, 2003). Finally, only a small increase of brown fat activity appears to be sufficient to counteract obesity, as has been seen in mice that contain ectopic deposition of brown adipocytes in the skeletal muscle (Almind et al., 2007) or in mice that express UCP1 in the white fat at a very low level (Kopecky et al., 1995). These observations revive the idea that brown fat remains as an attractive therapeutic target tissue for obesity and associated diseases. Clearly, there is a strong need to understand the molecular basis underlying brown fat metabolism.
A central regulator in brown fat thermogenesis is the transcriptional co-activator PGC-1α (reviewed in Lin et al., 2005). PGC-1α is predominantly expressed in the brown fat and its expression is highly influenced by nutritional and environmental cues. Both overexpression and loss-of-function studies demonstrate that PGC-1α regulates the entire program of thermogenesis (Lin et al., 2005; Uldry et al., 2006). As might be expected, regulatory mechanisms must be in place to fine-tune PGC-1α function. Studies have suggested that PGC-1α activity is positively regulated by several transcriptional regulators; these include Src-1, Sirt1, LRP130 and PRDM16 (Cooper et al., 2006; Lagouge et al., 2006; Picard et al., 2002; Puigserver et al., 1999; Rodgers et al., 2005; Seale et al., 2007). Conversely, PGC-1α mRNA expression in the brown fat is inhibited by Rb protein and the orphan nuclear receptor SHP (Scime et al., 2005; Wang et al., 2005). Two factors, GCN5 and p160MBP, have been shown to suppress PGC-1α activity (Fan et al., 2004; Lerin et al., 2006); but their functional relevance in brown fat metabolism is unknown. Interestingly, among all the PGC-1α regulators identified so far, only PRDM16 displays a brown fat-selective expression pattern, with others being ubiquitously expressed in many tissues. Furthermore, the in vivo requirements for most of these factors in brown fat metabolism have not been addressed. Hence, despite these progresses, physiological regulators for PGC-1α in the brown fat largely remain to be identified. In particular, it is completely unclear how PGC-1α function is negatively modulated post-transcriptionally in the brown fat.
As a transcriptional co-activator, PGC-1α must dock to transcriptional factors to exert its biological function. In vitro, PGC-1α is found to interact with many transcriptional factors (Lin et al., 2005). Among them are nuclear receptor PPARs and ERRs, which themselves have been implicated in oxidative metabolism. However, the relative importance of these factors in brown fat metabolism is elusive. In the absence of PPARγ or ERRα, UCP1 can still be induced (Seale et al., 2007; Villena et al., 2007), indicating that neither PPARγ nor ERRα is essential for UCP1 expression. Thus, it remains to be determined what are the downstream major transcriptional factors that mediate PGC-1α function in the brown fat.
In this manuscript, we identified a transcriptional network present in the brown fat that regulates and coordinates with PGC-1α. Twist-1, a helix-loop-helix-containing transcriptional regulator, is expressed in embryonic stages and is important for early development (Chen and Behringer, 1995), anti-apoptosis (Sosic et al., 2003), osteoblast differentiation (Bialek et al., 2004). Recent work also shows that twist-1 is overexpressed in many human cancers and cancer cell lines, thereby driving tumor invasion and metastasis (Ansieau et al., 2008; Ma et al., 2007; Yang et al., 2004). Surprisingly, we find that twist-1 is mainly present in the adipose tissue and acts as a negative feedback regulator of PGC-1α/PPARδ-mediated brown fat metabolism. Our studies provide novel molecular insights into brown fat metabolism and have important implications in obesity and its associated metabolic diseases.
Results
Twist-1 is selectively expressed in the adipose tissue but does not regulate adipogenesis
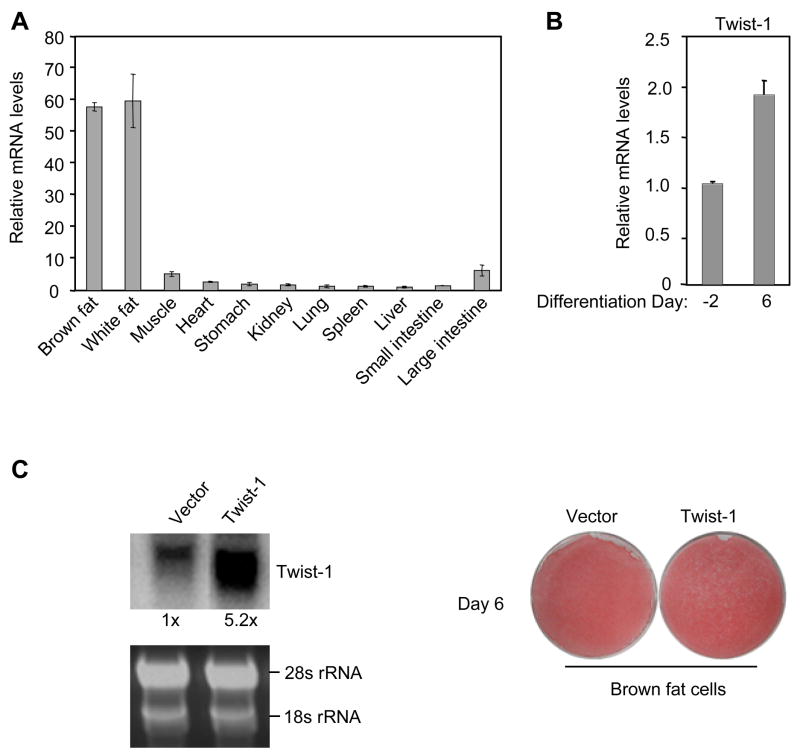
To understand tissue-specific molecular components and pathways that control energy metabolism, we queried the human and mouse gene expression databases (Su et al., 2002) to search for transcriptional regulators that are preferentially expressed in the adipose tissue. One factor identified from these databases is twist-1. As shown in Figure 1A, quantitative PCR analyses confirmed that twist-1 was most abundantly expressed in brown fat and white fat of adult mice, typically at a level more than10-fold higher than in other tissues. A similar expression profile was obtained with Northern blot analyses (data not shown). Moreover, twist-1 level was increased in mature adipocytes compared to preadipocytes in culture (Figure 1B). These data together led us to test whether twist-1 is involved in adipogenesis of brown fat or white fat. To this end, we infected a brown fat preadipocyte cell line we generated (see below) with twist-1 expression retrovirus. Cells with stable viral integration were pooled and then induced to adipogenic differentiation. Retroviral twist-1 expressed at a level of 5-fold relative to its endogenous level in mature adipocytes, which corresponded to a 10-fold difference in preadipocytes (Figure 1C). Oil-red O staining revealed no obvious difference in lipid accumulation between cells overexpressing twist-1 and cells with vector alone (Figure 1C). Similarly, retroviral overexpression of twist-1 in the white fat preadipocyte 3T3-L1 cells had no significant effect on adipogenic differentiation (Supplemental Figure S1). Taken together, we found that the transcriptional regulator twist-1 is selectively expressed in the adipose tissue and, interestingly, it does not participate in adipogenesis.
Figure 1.
Twist-1 is selectively expressed in the adipose tissue but does not regulate adipogenic differentiation. (A) Twist-1 expression in different tissues of adult C57BL6 mice (n=4). In this and all other figures, gene expression data were normalized with U36B4 levels. (B) Twist-1 expression before and after brown fat adipogenic differentiation in culture. (C) Oil red O staining of differentiated brown fat adipocytes overexpressing twist-1.
Twist-1 inhibits the transcriptional activity of PGC-1 α through their direct interaction
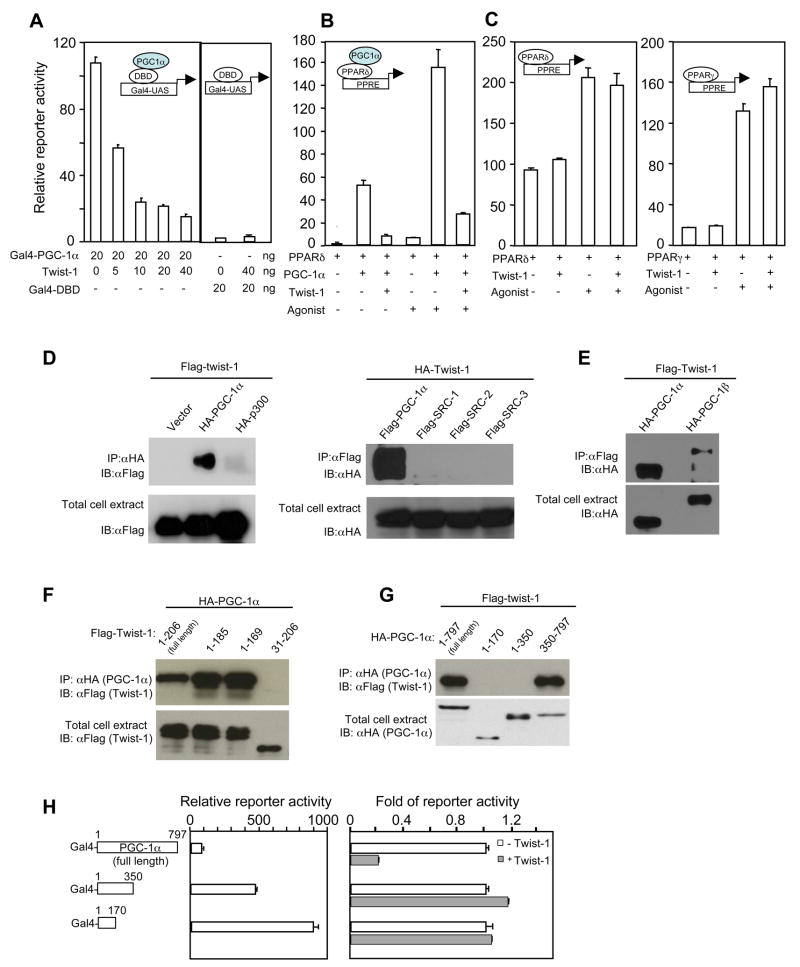
Given the lack of effect of twist-1 on adipogenesis, we considered whether twist-1 regulates energy dissipation, a predominant metabolic pathway in the brown fat controlled by PGC-1α. To begin testing this hypothesis, we examined whether twist-1 modulates the transcriptional activity of PGC-1α in cos-7 cells. Full-length PGC-1α fused with the DNA-binding domain (DBD) of yeast Gal4 had a robust transcription activity when assayed in a luciferase reporter driven by the Gal-UAS sequence (Puigserver et al., 1999). In the presence of twist-1, the activity of Gal4-PGC-1α was strongly inhibited (Figure 2A), while the activity of Gal4 DBD alone was unaffected. Twist-1 also inhibited PGC-1α-mediated activation of peroxisome proliferator-activated receptors (PPARs) (Figure 2B and data not shown). On the other hand, twist-1 had no effect on the basal or agonist-stimulated activity of PPARs (Figure 2C); this was due to the fact that, in cos-7 cells, there is little endogenous PGC-1α and the agonists-mediated activation of PPARs is presumably through recruitment of a different set of co-activators rather than PGC-1α. Thus, twist-1 is a specific inhibitor of PGC-1α.
Figure 2.
Twist-1 interacts with PGC-1α and suppresses its transcriptional activity. (A) Does-dependent inhibition of transcriptional activity of Gal4-PGC-1α by twist-1. (B and C) Twist-1 inhibits PGC-1α-mediated, but not agonist-mediated, activation of PPARs. PPARδ or PPARγ, 20 ng; PGC-1α, 20 ng; twist-1, 20 ng. (D and E) Interactions of twist-1 with co-activators. (F) PGC-1α interacts with the N-terminus of twist-1. (G) Twist-1 interacts with the C-terminal half of PGC-1α. (H) Suppression of PGC-1α activity by twist-1 requires their interaction. Left panel, Transcriptional activity of Gal4-PGC-1α fusion constructs was measured in the absence of twist-1. Right panel, the fold of transcriptional activity in the presence of twist-1 relative to the activity in the absence of twist-1 for Gal4-PGC-1α fusion constructs was measured.
We examined the possibility whether co-expression of twist-1 causes any decrease of PGC-1α protein level and/or perturbation of its nuclear localization, thus could account for its inhibition of PGC-1α transcriptional activity. Flag-tagged twist-1 and HA-tagged PGC-1α were co-expressed in HEK293 cells. In contrary to what would be expected, PGC-1α protein was surprisingly stabilized in the presence of twist-1 (Figure S2A). Consistent with this observation, twist-1 blocked the ubiquitination of PGC-1α (data not shown). Immunofluorescence staining further demonstrates that expression of twist-1 stabilized PGC-1α and did not perturb its cellular localization pattern (Figure S2B).
We next determined whether twist-1 and PGC-1α physically associate. An interaction between these two proteins was readily detected (Figure 2D and E). There was also a weak association between twist-1 and PGC-1β (Figure 2E). On the other hand, interactions of twist-1 with other co-activators, such as p300 or src members, were almost undetectable (Figure 2D). We found that the N-terminal first 30 amino acid of twist-1, a small region that is conserved in the closely related member twist-2, was required for its interaction with PGC-1α, while the conserved C-terminus was dispensable (Figure 2F). Not surprisingly, twist-2 also interacted with PGC-1α and suppressed its transcriptional activity (Figure S3), but twist-2 expression in the brown fat and white fat is extremely low. The C-terminal half (350 aa -end) of PGC-1α mediated its interaction with twist-1 (Figure 2G). Importantly, deletion of this C-terminal half substantially increased its transcriptional activity compared to the full length (Figure 2H, left panel) and abolished twist-1’s inhibitory effects (Figure 2H, right panel). These data demonstrate that inhibition of PGC-1α activity by twist-1 depends on their interaction, despite that this interaction renders PGC-1α protein more stable.
Twist-1 specifically suppresses PGC-1 α-mediated mitochondrial oxidative metabolism and uncoupling
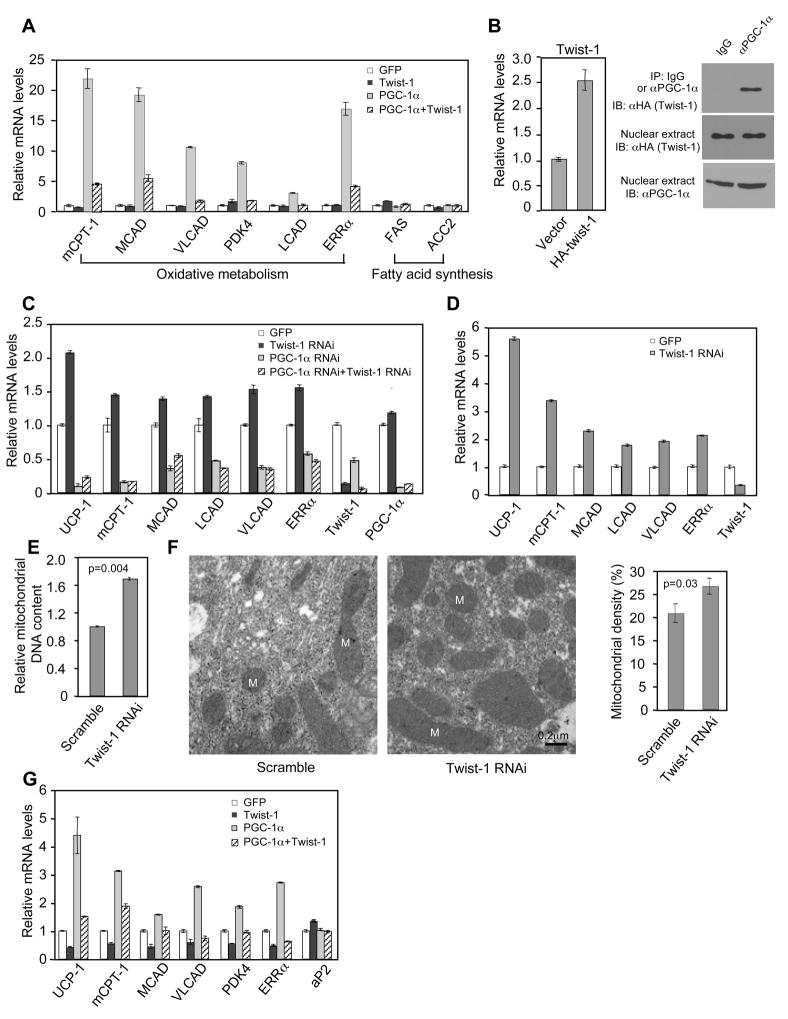
We asked whether the inhibition of PGC-1α transcriptional activity by twist-1 is of functional significance. We first addressed this question in differentiated C2C12 myotubules; these cells express little endogenous levels of PGC-1α and twist-1, therefore providing us a system to directly and unambiguously assess their functional interaction by ectopic expression. As shown by others and in Figure 3A, adenovirus expression of PGC-1α in the myotubules induced the expression of an array of genes involved in mitochondrial oxidative metabolism. Co-expression of twist-1 strongly negated the induction of these PGC-1α target genes (Figure 3A). Yet, twist-1 had little effect on the basal levels of these genes when PGC-1α was not present, nor had any effects on non-PGC-1α target genes, such as those involved in fatty acid synthesis. Moreover, the PGC-1α-stimulated oxygen consumption and fatty acid oxidation was reduced by co-expression of twist-1, while basal oxygen consumption and fatty acid oxidation was unaffected (Figure S4). Twist-1 also inhibited PGC-1β-induced gene expression (Figure S5), but was less robust, consistent with their weak physical association. Taken together, these results indicate that twist-1 specifically targets PGC-1 to modulate oxidative metabolism.
Figure 3.
Twist-1 regulates PGC-1α-mediated mitochondrial oxidative metabolism and uncoupling. (A) Differentiated C2C12 muscle cells infected with PGC-1α and twist-1 adenovirus. (B) Twist-1 and PGC-1α interact at physiological levels in brown fat adipocytes. (C) Brown fat preadipocytes expressing a lentivirus twist-1 RNAi construct or control construct were infected with PGC-1α RNAi or control adenovirus during differentiation. (D) Gene expression in differentiated brown fat adipocytes infected with twist-1 knockdown adenovirus. (E) Mitochondrial DNA content indicated by the level of mitochondrial genome-encoded CoxII gene. (F) Left, representative electron microscopy micrographs from differentiated brown fat adipocytes. Right, Quantification of mitochondrial density from thirteen micrographs. M, mitochondria. (G) Gene expression in differentiated brown fat adipocytes infected with adenovirus expressing twist-1, PGC-1α, or both.
Twist-1 belongs to a class of transcriptional regulators that contain a basic helix-loop-helix domain. The basic region recognizes a consensus DNA element called E-box. To rule out the possibility that the inhibitory effects of twist-1 requires its binding to potential E-boxes on the promoters of PGC-1α target genes, we generated a twist-1 point mutation in residue E121, a conserved residue located in the basic region important for DNA-binding (Atchley and Fitch, 1997). Twist-1 binds to the E-box of the promoter of the GLI-Krupped family member GLI1 and activates its expression (Villavicencio et al., 2002). The E121K mutant lost its ability to induce GLI1 expression (Figure S6A). However, this mutant was still capable of suppressing both the activity of PGC-1α in reporter assays and the expression of PGC-1α target genes (Figure S6B and C). These results demonstrate that suppression of PGC-1α target gene expression by twist-1 does not rely on its DNA binding activity. This is consistent with the data that twist-1 has a widespread inhibitory effect on every PGC-1α target gene we examined (Figure 3A and data not shown).
Both twist-1 and PGC-1α are mainly expressed in brown fat. To investigate the functional role of twist-1 in a more physiological setting, we generated an immortalized preadipocyte brown fat cell line (δf/f) from newborn mice in which the PPARδ alleles are floxed by the loxp sites. After differentiation, the δf/f cells exhibited the characteristics of brown fat, including robust induction of PGC-1α and uncoupling protein 1 (UCP1) gene expression (Figure S7A and B). We tested whether twist-1 and PGC-1α associate at endogenous levels in brown fat cells. Because of the poor quality of the commercially available twist-1 antibody, we stably expressed HA-tagged twist-1 in δf/f cells at a level that was 1.5-fold of its endogenous level. As shown in Figure 3B, PGC-1α antibody, but not control IgG, co-immunoprecipitated HA-twist-1 from nuclear extracts of differentiated brown fat cells, suggesting that an interaction between twist-1 and PGC-1α occurs at physiological levels in vivo.
To test the functional requirement of endogenous twist-1 in the brown fat cells, we used lentivirus to stably express shRNA against twist-1 in brown fat preadipocytes. After differentiation into mature adipocytes, we analyzed the expression of PGC-1α target genes. Stable knockdown of twist-1 led to a systematic increased expression of PGC-1α target genes including UCP1 (Figure 3C and Figure S8), while simultaneous knockdown of PGC-1α by RNAi adenovirus largely eliminated these effects (Figure 3C). Similar results were obtained when twist-1 was acutely knocked down with RNAi adenovirus in differentiated brown fat cells (Figure 3D), further suggesting that the observed effects of twist-1 is unrelated to adipogenesis. Moreover, depletion of twist-1 increased mitochondrial biogenesis, as judged by mitochondrial DNA content and electron microscopy (Figure 3E and F). Conversely, overexpression of twist-1 suppressed the expression of PGC-1α target genes (Figure 3G) and mitochondrial biogenesis (Figure S9). These results demonstrate an important role of endogenous twist-1 in the regulation of mitochondrial oxidative metabolism in brown fat cells, most likely through negative modulation of endogenous PGC-1α function.
Transgenic expression of twist-1 in the adipose tissue suppresses brown fat metabolism
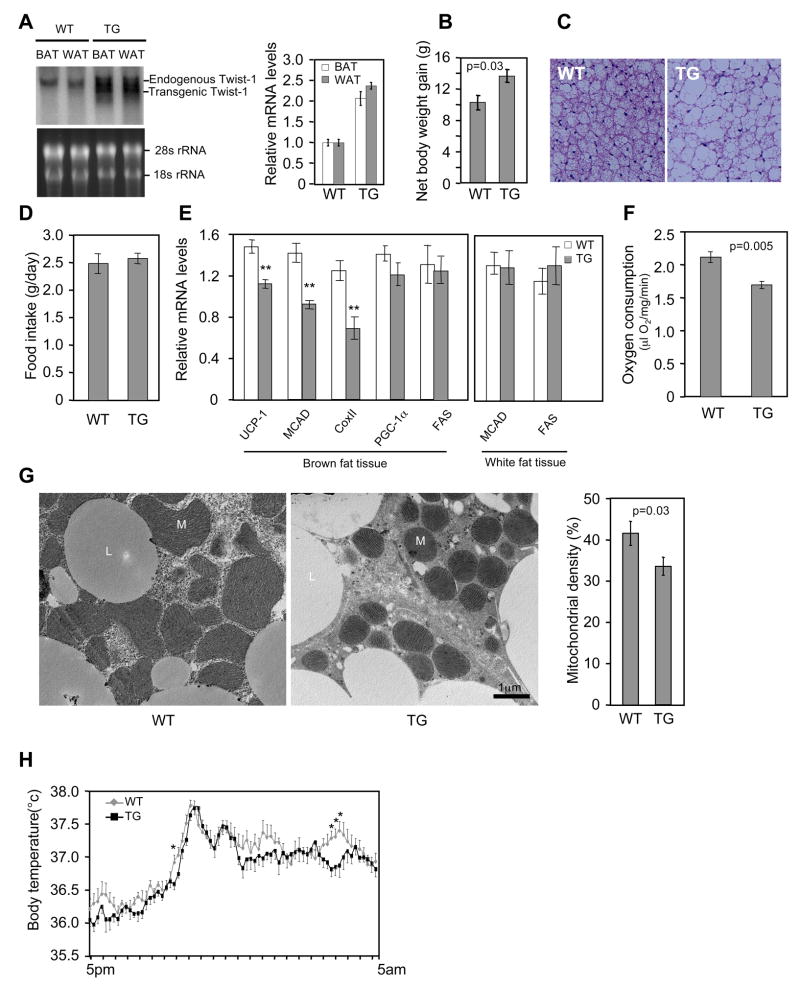
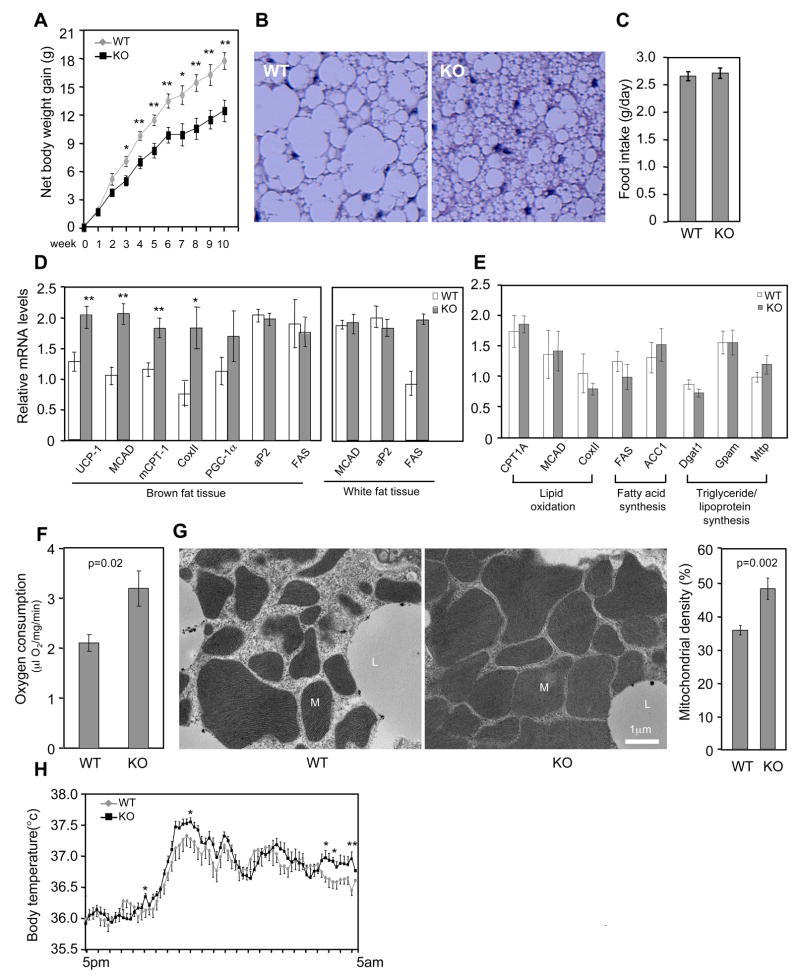
To investigate whether the in vitro identified regulatory role of twist-1 is relevant to in vivo fat metabolism, we generated twist-1 transgenic mice, driven by the aP2 promoter. The transgene was expressed approximately 2.5-fold of its endogenous level (Figure 4A). Under normal chow diet, there was no difference in body weight between the transgenic mice and control littermates. However, when placed on a high-fat diet, the transgenic mice gained more body weight (Figure 4B) and accumulated substantially more lipids in the brown fat (Figure 4C), although they consumed similar amount of food as controls (Figure 4D). Thus, a modest overexpression of twist-1 in the adipose tissue is sufficient to render the animals susceptible to high-fat diet induced obesity. Indeed, the transgene decreased the expression of UCP1 and fatty acid oxidation genes in the brown fat (Figure 4E). No gene expression difference was observed in the white fat (Figure 4E), consistent with the fact that there is little PGC-1α in this tissue. Next, we isolated brown fat tissue and measured oxygen consumption ex vivo. The rate of oxygen consumption was significantly lower in transgenic mice than in the control littermates (Figure 4F). Electron microscopy also revealed a clear reduction of mitochondria density in the brown fat of transgenic mice (Figure 4G). Finally, we monitored their body temperature when the mice were fed a high-fat diet. While no difference were observed at day time, the body temperature of the transgenic mice appeared to be lower at night time than their control littermates (Figure 4H), probably reflecting an insufficiency of fat burning and heat production in the brown fat. These results together pinpoint a compromised brown fat function in the twist-1 transgenic mice.
Figure 4.
Transgenic expressing twist-1 in adipose tissue inhibits brown fat metabolism. (A) Twist-1 transgene (TG) expression in the brown and white fat tissue. (B) Net body weight gain in wild-type control littermates (WT) and transgenic (TG) mice after a 2-month high fat diet (n=7–12 mice per group). Their initial body weights were similar, 16.65 ± 0.21 g (WT) vs. 16.80 ± 0.43 g (TG) (p=0.73). (C) Representatives of brown fat histology after a 7-week high fat diet. (D) Consumption of high fat diet (n=5 mice per group). (E) Gene expression in brown fat and white fat (n=6 mice per group). **p<0.01. (F) Ex vivo oxygen consumption of brown fat (n=4 mice per group). (G) Mitochondrial density. Left, representative electron microscopy micrographs. Right panel, quantification from twelve micrographs of four mice per group. M, mitochondria; L, lipid droplets. (H) Body temperature was recorded in a 10-min interval (n=8 mice per group). *p<0.05
Heterozygous twist-1 knockout mice have increased brown fat metabolism and are obesity resistant
We asked whether twist-1 is indispensable for the regulation of energy homeostasis in vivo. Twist-1 homozygous knockout is embryonically lethal. We thus compared the metabolic phenotype of twist-1 heterozygous mice and their body weight matched control littermates. When place on a high fat diet, the knockout animals showed a remarkable obesity resistant phenotype (Figure 5A), accompanied by substantial less lipid accumulation in the brown fat (Figure 5B) and yet similar food consumption (Figure 5C). At the molecular level, expression of UCP1 and oxidation genes in the brown fat, but not in the white fat, was elevated (Figure 5D). As in the transgenic mice, the level of PGC-1α in the brown fat of the knockout mice was not significantly changed, consistent with the idea that the effects of twist-1 is due to its direct modulation of PGC-1α activity, not PGC-1α expression. Expression of liver genes important for fatty acid oxidation or fatty acid and triglyceride synthesis was also not affected (Figure 5E). Moreover, opposite to what observed in the transgenic mice, the brown fat of heterozygous knockout mice displayed elevated oxygen consumption (Figure 5F) and mitochondrial biogenesis (Figure 5G), and their body temperature tended to be higher at night time (Figure 5H). These results suggest that the level of twist-1 in vivo is quantitatively important for the control of energy homeostasis, providing compelling in vivo evidence supporting our model that twist-1 is a critical negative regulator of PGC-1α-mediated mitochondrial metabolism and uncoupling in the brown fat.
Figure 5.
Increased brown fat metabolism in twist-1 heterozygous knockout mice. (A) Body weight gain in wild-type control littermates (WT) and heterozygous knockout (KO) mice during high fat diet (n=5–6 mice per group). *p<0.02. **p<0.01. Their initial body weights were similar, 20.68 ± 2.09 g (WT) vs. 19.60 ± 1.26 g (KO) (p=0.47). (B) Representative of brown fat histology after a 9-week high fat diet. Note, here more lipid accumulation in the control mice (C56BL6 background) is observed than in the control mice (mixed background) in Figure 4C, probably due to a longer period of high fat diet and/or genetic background. (C) Consumption of high fat diet (n=5–6 mice per group). (D) Gene expressions in brown fat and white fat (n=5–7 mice per group). *p<0.05. **p<0.01. (E) Gene expression in liver (n=5–6 mice per group). (F) Ex vivo oxygen consumption of brown fat (n=5 mice per group). (G) Mitochondrial density. Left, representative electron microscopy micrographs. Right panel, quantification from ten micrographs of four mice per group. M, mitochondria; L, lipid droplets. (H) Body temperature was recorded in a 10-min interval (n=7–10 per group). *p<0.05. **p<0.01.
Twist-1 inhibits Histone H3 acetylation on the promoters of PGC-1 α target genes
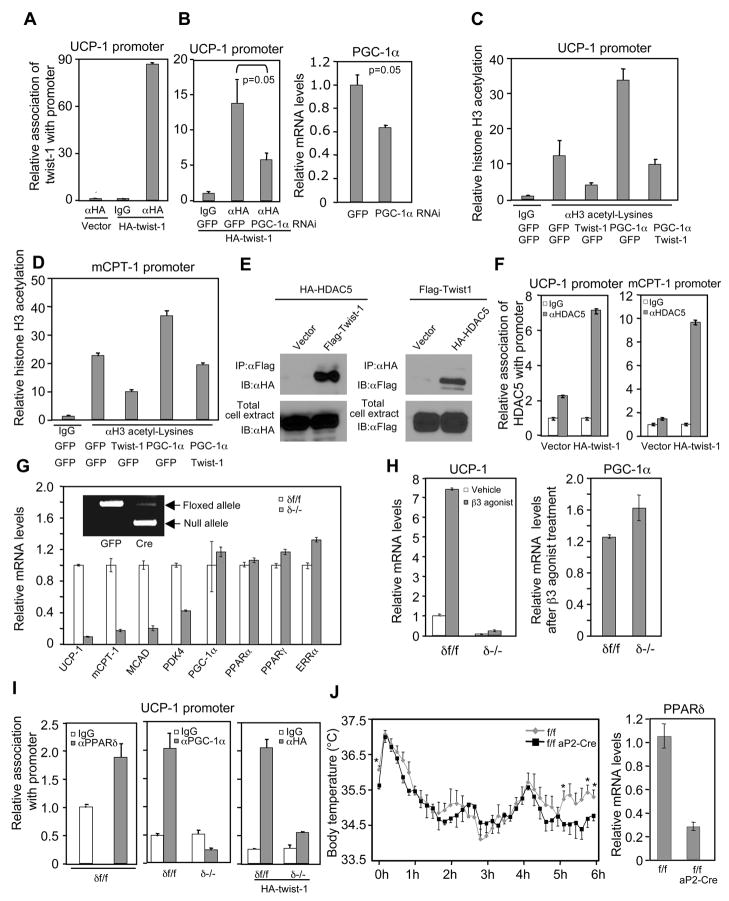
We performed chromatin immunoprecipitation assays in brown fat cells to determine the molecular basis by which twist-1 suppresses PGC-1α function. We focused on the UCP1 promoter, as this promoter contains a well-defined PPAR-binding site that may underlie the induction of UCP1 by PGC-1α (Collins et al., 2004). Both PGC-1α and twist-1 at their endogenous levels associated with the PPAR-binding site of the UCP1 promoter (Figure 6A, and middle panel of Figure 6I). Importantly, twist-1’s association was markedly decreased when PGC-1α was knocked down by 40% (Figure 6B), supporting the notion that twist-1 is recruited to the promoter by its interaction with PGC-1α. Consistent with the PGC-1α target gene expression data, expression of twist-1 suppressed PGC-1α-elicited Histone H3 acetylation (Figure 6C). Similar results were obtained with the carnitine palmitoyltransferase 1 (mCPT1) gene promoter (Figure S10 and Figure 6D). We found that twist-1 associated with the class II Histone deacetylase HDAC5 (Figure 6E) and recruits HDAC5 to the promoters of UCP1 and mCPT1 (Figure 6F), which could at least in part explain the loss of Histone H3 acetylation caused by twist-1.
Figure 6.
Molecular basis of twist-1 inhibition of PGC-1α function and PPARδ acts downstream of PGC-1α and twist-1. (A and B) Twist-1 association with the PPAR-binding site in the UCP1 promoter. Brown fat preadipocytes expressing a physiological level of HA-twist-1 or HA vector alone were differentiated. Cells were then untreated (A) or infected with indicated adeneovirus (B, left panel). The PGC-1α mRNA level in a parallel knockdown experiment was shown (B, Right panel). (C and D) Histone H3 acetylation on the promoters of PGC-1α target genes. (E) Twist-1 interacts with HDAC5. (F) Association of endogenous HDAC5 with promoters of UCP1 and mCPT1. (G) Gene expression in differentiated, PPARδ-deleted brown fat cells. (H) UCP1 expression in differentiated brown fat adipocytes treated with 10 μM β3-adrenergic receptor agonist CL316243 for 5 h. (I) PGC-1α and twist-1 association with the UCP1 promoter was abolished in differentiated, PPARδ-deleted brown fat adipocytes. (J) Left, body temperature during cold exposure was recorded in a 10-min interval (n=5 mice per group). *p<0.05. Right, PPARδ mRNA level.
PPAR δ mediates the actions of PGC-1 α and twist-1 in brown fat cells
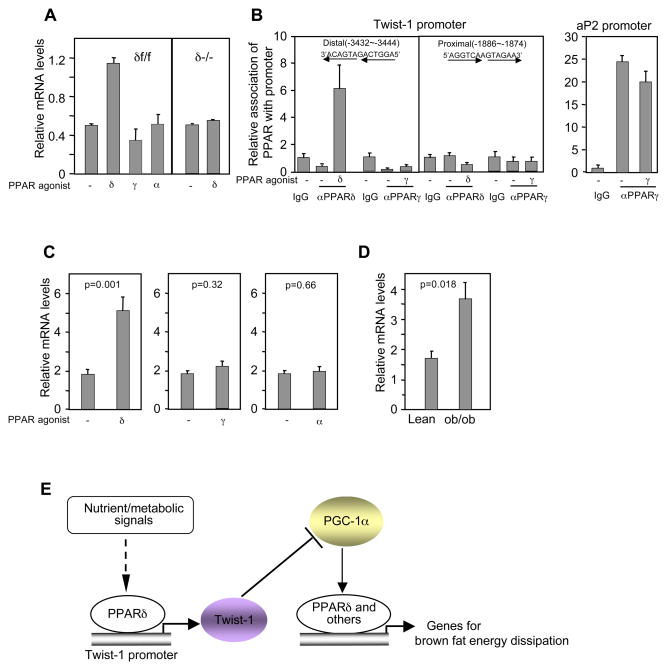
To further understand the molecular framework of this PGC-1α-twist-1 pathway, we sought to determine what transcription factor mediates the effects of PGC-1α in the brown fat and therefore, could be linked to twist-1’s suppressive function. As activation of PPARδ promotes mitochondrial metabolism and uncoupling (Wang et al., 2003), we considered PPARδ as a candidate. To genetically test this possibility, we first examined whether there is a phenotypic overlap among loss of PPARδ, PGC-1α acute knockdown and twist-1 overexpression. We knocked out the PPARδ gene (floxed alleles) in brown fat cells by cre recombinase adenovirus-mediated excision. Compared to the cells that were infected by GFP control virus, the PPARδ-deficient cells (δ−/−) displayed decreased expression of mitochondrial metabolic genes (Figure 6G) but normal adipogenesis (Figure S11), a phenotype remarkably similar to those of PGC-1α acute knockdown (Figure 3C and Figure S12) and twist-1 overexpression (Figure 3G). In particular, loss of PPARδ caused a dramatic reduction of UCP1gene expression by more than 10-fold. The levels of PGC-1α, along with those of PPARγ, PPARα and ERRα, which are also involved in mitochondrial metabolism, remained unchanged (Figure 6G). This indicates an essential, cell-autonomous requirement for PPARδ in the expression of some, but not all, of PGC-1α target genes. To further test whether PPARδ acts downstream of PGC-1α, we measured UCP1 induction by the β3-adrenergic receptor agonist in PPARδ-deficient brown fat cells. The β3-adrenergic receptor agonist stimulates UCP1 expression through increasing both the activity and expression level of PGC-1α (Collins et al., 2004). Although the PPARδ-deficient cells had similar levels of PGC-1α as in the wild type cells, they were almost completely unresponsive to the agonist in the induction of UCP1 (Figure 6H). Indeed, in the absence of PPARδ, PGC-1α was no longer able to associate with the UCP1 promoter, which consequently resulted in the loss of association of twist-1 with the promoter (Figure 6I). Finally, we placed floxed PPARδ mice carrying aP2-cre transgene at 4°C for 6 hours. These animals were compromised in maintaining their body temperature during the last hour of the cold exposure compared to their control littermates without aP2-cre (Figure 6J), further indicating a functional convergence of PPARδ and PGC-1α. There was about 30% of PPARδ expression remaining in the brown fat of the knockout mice (Figure 6J; and Barak et al., 2002); this might account for the normal cold response during early time points. Together, these lines of genetic and biochemical evidence suggest that PPARδ functions as a key mediator downstream of PGC-1α and twist-1.
Twist-1 is induced by PPAR δ activation in brown fat
We explored how twist-1 expression might be regulated. Treatment of differentiated brown fat cells with the PPARδ agonist, but not the agonist for PPARγ or PPARα, induced twist-1 expression (Figure 7A). This induction was PPARδ-dependent as the agonist had no effect when the PPARδ gene was deleted. Interestingly, similar levels of twist-1 were observed in PPARδ-deficient cells relative to wild-type cells (Figure 7A); this pattern differs from the target genes involved in metabolism described in Figure 6G, indicating that PPARδ is not required for the maintenance of basal twist-1 expression. There are two probable PPAR-binding sites in twist-1 promoter, located in the −3.43 kb and −1.88 kb region, respectively. In the basal conditions, neither PPARδ nor PPARγ was associated with them. Upon addition of their respective agonists, PPARδ, but not PPARγ, was recruited to the distal site (Figure 7B), although PPARγ evidently associated with the PPAR-binding site in its target gene aP2 promoter in the same experiments; these data are consistent with the agonist-dependent regulation of twist-1 by PPARδ. To determine whether activation of PPARδ induces twist-1 expression in animals, we analyzed tissue RNA samples from wild-type C57BL6 mice treated with vehicle or the PPARδ agonist. The level of twist-1 in brown fat tissue was induced 2.8-fold by the agonist (Figure 7C). However, no significant changes were observed in animals treated with agonists for PPARγ or PPARα (Figure 7C). Our data suggest that twist-1 serves as a negative feedback regulatory loop to tightly modulate the PGC-1α/PPARδ-controlled brown fat metabolism (Figure 7E). Intriguingly, twist-1 level was elevated in the brown fat of obese mice (Figure 7D). This, coupled with decreases of PGC-1α (Kakuma et al., 2000) and PPARδ (Figure S13) in these animals, may further aggravate the state of obesity.
Figure 7.
Twist-1 is induced upon PPARδ activation in the brown fat. (A) Twist-1 expression in differentiated brown fat adipocytes treated with vehicle or PPAR agonists for 24 h. (B) PPARδ associates with the twist-1 promoter. (C) Twist-1 expression in the brown fat of C57BL6 mice treated with the PPAR agonists (n=5 mice per group). (D) Twist-1 expression in the brown fat of lean and obese mice (n=3 mice per group). (E) A coordinated transcriptional regulatory network in brown fat metabolism.
Discussion
It has now become increasingly appreciated that brown fat cells may play an important part in human energy homeostasis. A better understanding of the molecular basis underlying its unique metabolic property is obviously the first step in order to take full advantage of this cell type as a target for obesity and associated diseases. While previous work has demonstrated a central role for PGC-1α in governing brown fat metabolism, what are other factors involved, in particular, how these potential factors functionally and mechanistically interact and coordinate with PGC-1α, is poorly understood. Our results presented here reveal a transcriptional network that provides insights into this question. We demonstrate that twist-1 is physiologically critical in brown fat metabolism by directly antagonizing PGC-1α. Moreover, twist-1 constitutes part of a feedback regulatory loop, in which the nuclear receptor PPARδ not only acts downstream to mediate, at least in part, the function of PGC-1α, but also controls twist-1 expression. Interestingly, this brown fat-specific regulatory network appears to be exclusively involved in energy dissipation but not in fat cell formation.
Twist-1 is highly expressed in embryonic stage and is induced in cancer cells, consistent with its roles in early development and tumor metastasis, respectively. Unexpectedly, at postnatal stage, twist-1 is mainly expressed in the adipose tissue. Our studies performed both in vitro and in vivo elucidate that twist-1 specifically regulates PGC-1α-controlled oxidative metabolism and uncoupling in the brown fat. In particular, adipose twist-1 transgenic mice and heterozygous knockout display completely opposite body weight gain and body temperature profiles. These phenotypes can be largely explained by their altered PGC-1α target gene expression profiles, oxygen consumption, and mitochondrial biogenesis in the brown fat. Our results clearly establish twist-1 as a metabolic brake for energy expenditure in the brown fat and that its expression level is critically important for the maintenance of whole body energy homeostasis. At present, the function of twist-1 in the white fat is unclear. It is possible that twist-1 may regulate additional pathways related to adipocyte biology.
One question is how the interaction of twist-1 and PGC-1α leads to suppression of PGC-1α function. Twist-1 neither destabilizes PGC-1α nor perturbs its nuclear localization. We also found no evidence that the interaction displaces PGC-1α from its target gene promoters. Instead, twist-1 is recruited by PGC-1α to the PGC-1α target genes promoters and thus exerts its inhibitory function locally. This mode of action is clearly different from the one employed in the suppression of Runx2 during bone development; there, twist-1 blocks Runx2 association with the target gene promoter through its interaction with the DNA-binding domain of Runx2 (Bialek et al., 2004). Interestingly, the regions of twist-1 that interact with PGC-1α and Runx2 appear to be different, located in the N-terminus and C-terminus, respectively; this further indicates a versatile nature of twist-1 in gene regulation. The recruitment of twist-1 to PGC-1α target gene promoters causes a dramatic decrease of Histone H3 acetylation, likely due to a subsequent recruitment of Histone deacetylase HDAC5. These results together outline a plausible underlying mechanism by which twist-1 modulates brown fat metabolism. Other potential mechanisms are also possible. For example, twist-1 was shown to inhibit p300 acetyltransferase activity in vitro (Hamamori et al., 1999). However, we were unable to detect any association of p300 with the UCP1 promoter. Another acetyltransferase, SRC-1, has been shown to regulate UCP1 expression (Picard et al., 2002). Indeed, we find that SRC-1 is present on the UCP1 promoter, but this association is not affected by twist-1 (our unpublished data), consistent with the data that these two proteins do not interact.
Our studies further uncovered PPARδ as an integral component in coordinating the actions of PGC-1α and twist-1 in brown fat metabolism. The requirement for PPARδ on expression of PGC-1α target genes occurs in the context of normal expression of PPARγ, PPARα and ERRα. Previous work in other cell systems suggests that the transcription factor YY1 and ERRα act downstream of PGC-1α in mitochondrial oxidative metabolism (Cunningham et al., 2007; Mootha et al., 2004; Schreiber et al., 2004). Those studies and ours presented here indicate that how PGC-1α is mediated downstream is more complex, and is likely to be cellular context- and/or promoter-dependent. While by no means excluding the involvement of other transcription factors, our genetic analyses suggest that in brown fat cells PPARδ is an important mediator, in particular for UCP1 expression. This idea is strengthened by our observation that fat-specific PPARδ knockout mice are compromised in maintaining body temperature during cold exposure. Furthermore, we find that PPARδ, but not PPARα or PPARγ, binds to the twist-1 promoter, and directs twist-1 expression both in brown fat cell culture and in whole animals, suggesting a feedback regulatory mechanism. As PPARδ is a lipid sensor that is activated by fatty acid (Chawla et al., 2003), the transcriptional network described here would presumably allow brown fat to response to body’s nutrients status to ensure energy homeostasis.
In summary, our results provide important molecular insights into how the PGC-1α-controlled energy expenditure pathway is modulated and coordinated in the brown fat. Pharmacologically increasing PGC-1α activity and/or expression is considered a therapeutic strategy for metabolic diseases (Baur et al., 2006; Feige et al., 2008; Lagouge et al., 2006). Our data indicate that decreasing twist-1 level or disrupting its interaction with PGC-1α may offer an alternative strategy.
Experimental Procedures
Viruses
Adenoviruses for overexpression and knockdown were generated using the AdEasy-1 system (He et al., 1998). PGC-1α RNAi adenovirus construct was kindly provided by Dr. Marc Montminy (Koo et al., 2004). All adenoviruses were purified with cesium chloride ultracentrifugation and virus titers were determined in HEK293 cells by scoring GFP positive cells. Twist-1 RNAi lentivirus constructs (Yang et al., 2004) was obtained from Addgene.
Adenovirus infection was performed in differentiated cells unless otherwise indicated. GFP control adenovirus was added, if needed, to ensure a similar degree of infection in different samples. Assays were performed two days after infection.
Generation and differentiation of brown fat cells
Immortalized brown fat preadipocytes (δf/f) were generated as described (Tseng et al., 2004). We used mice containing floxed PPARδ alleles (Barak et al., 2002) that have been backcrossed with C57BL6 for 9 generations. Details were available in Supplemental Experimental Procedures.
Luciferase reporter assays, co-immunoprecipitation, and chromatin immunoprecipitation
These were described in Supplemental Experimental Procedures.
Animals
aP2-promoter directed twist-1 transgenic mouse line was generated in a SV129/C57BL6 mixed background and was backcrossed with C57BL6 for four generations. Heterozygous twist-1 knockout mice in a C57BL6 background were purchased from Jackson Laboratory and cohorts were generated by crossing with wild-type C57BL6. Conditional PPARδ floxed mice (Barak et al., 2002) and aP2-Cre transgenic mice (He et al., 2003) were described. Littermate controls were used in all the experiments. Normal diet containing 4% (w/w) fat was obtained from Harlan Teklad. A high fat diet containing 35% (w/w) fat content was obtained from Bioserv (Product# F3282). Food consumption was measured daily for individually caged mice for two weeks. The University of Massachusetts Medical School’s Institutional Animal Care and Use Committee approved all the animal studies.
Body temperature
Mice were implanted with paraffin-coated iButtons (DS1922T-F5, Embedded data systems) into their peritoneal cavities as described (Davidson et al., 2003). Mice were allowed to recover for 2 weeks. Body temperature was then recorded at a 10-min sampling interval. The data were retrieved with a data reader.
Oxygen consumption assays
Brown fat was isolated and chopped into small pieces. Equal amount (30 mg) was used in 5 ml DPBS supplemented with 25mM glucose, 1mM pyruvate, and 2% BSA. Respiration was measured with a Clark-type electrode (YSI model 5300).
Experiments with PPAR agonists
In brown fat adipocyte culture, differentiated cells were treated with vehicle, PPARδ agonist GW501516 (0.1 μM), or PPARγ agonist rosiglitazone (1 μM) for 24 hours. For in vivo experiments, total RNA samples from wild-type C57BL6 mice treated with GW501516 were generated previously (Wang et al., 2004). Wild-type C57BL6 mice were gavaged daily with rosiglitazone (30 mg/kg per day), PPARα agonist GW7647 (3 mg/kg per day), or vehicle for one week.
Electron microscopy
Details were available in Supplemental Experimental Procedures. We used ImageJ software to calculate mitochondrial area and cellular area in individual micrographs, and mitochondrial density was expressed as a percentage of cellular area. In data presented in Figure 4G and Figure 5G of animal experiments, area of lipid droplet was not included in the cellular area to avoid exaggerating the difference of mitochondrial density.
Statistical analysis
Student’s t test (two-tailed) was used for statistical analysis. p<0.05 was considered significant. Data are presented as mean ± SEM.
Supplementary Material
Acknowledgments
We thank Drs. Michael Green, Michael Czech and Silvia Corvera for insightful comments on this work; Drs. Ron Evans, Marc Montminy, Heonjoong Kang and Brian Lewis for reagents; Drs. Hong-Ping Guan, Yasuyuki Okawa and Claude Gazin for help on CHIP assays; Dr. Greg Hendricks and Tracy Levin for electron microscopy; Alison Burkart for help on oxygen consumption, Drs. Matthew Paul and Robert Dallmann for help on use of iButtons; Sue Wheeler, Heidi Chandler and Dr. Denice Godfrey for animal surgery. These studies were funded by grants from American Diabetes Association, Smith Family Foundation, and NIH/NIDDK (R01DK076118).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almind K, Manieri M, Sivitz WI, Cinti S, Kahn CR. Ectopic brown adipose tissue in muscle provides a mechanism for differences in risk of metabolic syndrome in mice. Proc Natl Acad Sci U S A. 2007;104:2366–2371. doi: 10.1073/pnas.0610416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci U S A. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Y, Liao D, He W, Ong ES, Nelson MC, Olefsky JM, Boland R, Evans RM. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer. Proc Natl Acad Sci U S A. 2002;99:303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Chawla A, Lee CH, Barak Y, He W, Rosenfeld J, Liao D, Han J, Kang H, Evans RM. PPARdelta is a very low-density lipoprotein sensor in macrophages. Proc Natl Acad Sci U S A. 2003;100:1268–1273. doi: 10.1073/pnas.0337331100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- Collins S, Cao W, Robidoux J. Learning new tricks from old dogs: beta-adrenergic receptors teach new lessons on firing up adipose tissue metabolism. Mol Endocrinol. 2004;18:2123–2131. doi: 10.1210/me.2004-0193. [DOI] [PubMed] [Google Scholar]
- Cooper MP, Qu L, Rohas LM, Lin J, Yang W, Erdjument-Bromage H, Tempst P, Spiegelman BM. Defects in energy homeostasis in Leigh syndrome French Canadian variant through PGC-1alpha/LRP130 complex. Genes Dev. 2006;20:2996–3009. doi: 10.1101/gad.1483906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Aujard F, London B, Menaker M, Block GD. Thermochron ibuttons: an inexpensive method for long-term recording of core body temperature in untethered animals. J Biol Rhythms. 2003;18:430–432. doi: 10.1177/0748730403256066. [DOI] [PubMed] [Google Scholar]
- Fan M, Rhee J, St-Pierre J, Handschin C, Puigserver P, Lin J, Jaeger S, Erdjument-Bromage H, Tempst P, Spiegelman BM. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 Activation Mimics Low Energy Levels and Protects against Diet-Induced Metabolic Disorders by Enhancing Fat Oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Garruti G, Ricquier D. Analysis of uncoupling protein and its mRNA in adipose tissue deposits of adult humans. Int J Obes Relat Metab Disord. 1992;16:383–390. [PubMed] [Google Scholar]
- Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu HY, Wang JY, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci U S A. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen P, Hirvonen J, Kinnula V. The occurrence of brown adipose tissue in outdoor workers. Eur J Appl Physiol Occup Physiol. 1981;46:339–345. doi: 10.1007/BF00422121. [DOI] [PubMed] [Google Scholar]
- Kakuma T, Wang ZW, Pan W, Unger RH, Zhou YT. Role of leptin in peroxisome proliferator-activated receptor gamma coactivator-1 expression. Endocrinology. 2000;141:4576–4582. doi: 10.1210/endo.141.12.7804. [DOI] [PubMed] [Google Scholar]
- Kontani Y, Wang Y, Kimura K, Inokuma KI, Saito M, Suzuki-Miura T, Wang Z, Sato Y, Mori N, Yamashita H. UCP1 deficiency increases susceptibility to diet-induced obesity with age. Aging Cell. 2005;4:147–155. doi: 10.1111/j.1474-9726.2005.00157.x. [DOI] [PubMed] [Google Scholar]
- Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R, Evans RM, Olefsky J, Montminy M. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat Med. 2004;10:530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- Kopecky J, Clarke G, Enerback S, Spiegelman B, Kozak LP. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J Clin Invest. 1995;96:2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lean ME, James WP, Jennings G, Trayhurn P. Brown adipose tissue in patients with phaeochromocytoma. Int J Obes. 1986;10:219–227. [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- Lowell BBVSS, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- Picard F, Gehin M, Annicotte J, Rocchi S, Champy MF, O’Malley BW, Chambon P, Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B, Spiegelman BM. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scime A, Grenier G, Huh MS, Gillespie MA, Bevilacqua L, Harper ME, Rudnicki MA. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab. 2005;2:283–295. doi: 10.1016/j.cmet.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosic D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiraby C, Langin D. Conversion from white to brown adipocytes: a strategy for the control of fat mass? Trends Endocrinol Metab. 2003;14:439–441. doi: 10.1016/j.tem.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Tseng YH, Kriauciunas KM, Kokkotou E, Kahn CR. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol Cell Biol. 2004;24:1918–1929. doi: 10.1128/MCB.24.5.1918-1929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Villavicencio EH, Yoon JW, Frank DJ, Fuchtbauer EM, Walterhouse DO, Iannaccone PM. Cooperative E-box regulation of human GLI1 by TWIST and USF. Genesis. 2002;32:247–258. doi: 10.1002/gene.10078. [DOI] [PubMed] [Google Scholar]
- Villena JA, Hock MB, Chang WY, Barcas JE, Giguere V, Kralli A. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc Natl Acad Sci U S A. 2007;104:1418–1423. doi: 10.1073/pnas.0607696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu J, Saha P, Huang J, Chan L, Spiegelman B, Moore DD. The orphan nuclear receptor SHP regulates PGC-1alpha expression and energy production in brown adipocytes. Cell Metab. 2005;2:227–238. doi: 10.1016/j.cmet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Wang YX, Lee CH, Tiep S, Yu RT, Ham J, Kang H, Evans RM. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell. 2003;113:159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.