Abstract
Most T cell progenitors develop into the αβ T cell lineage with an exception of a small fraction contributing to the γδ lineage throughout postnatal life. T cell progenitors usually commit to the αβ lineage upon the expression of a fully rearranged and functional TCRβ gene, and most cells that fail to produce a functional TCRβ chain will die instead of adopting the alternative γδ T cell fate. What prevents these cells from continuing TCRγ rearrangement and adopting the γδ T cell fate is not known. Here, we show that functional loss of Id3 results in a significant increase of γδ T cell production from progenitor cells undergoing TCRβ rearrangement. The enhanced γδ T cell development correlated with increased TCRγ gene rearrangement involving primarily Vγ1.1 in Id3 deficient mice. We further show that Id3 deficiency promotes γδ T cell production in dependent of TCRβ chain expression. Our data indicates that Id3 suppresses Vγ1.1 rearrangement and γδ lineage potential among T cell progenitors which have completed TCRβ gene rearrangement without producing a functional TCRβ protein.
Keywords: Thymus, VDJ recombination, E2A, TCR, DN3
Introduction
γδ and αβ T cells represent two functionally distinct groups of lymphocytes due to the expression of T cell receptors composed of either γδ or αβ hetero-dimers, respectively. T cell development begins in the fetal thymus with a wave of γδ T cells appearing first followed by the production of both γδ and αβ T cells. Throughout postnatal life, a majority of T cell progenitors adopt the αβ T cell fate whereas only a small proportion becomes γδ T cells. The molecular mechanism which limits the production of γδ T cells in postnatal life is not clear.
T cell development in the thymus occurs through consecutive developmental stages of CD4 and CD8 double negative (DN), double positive (DP), and single positive (SP). DN cells can be further separated into DN1-4 stages based on their developmental progression (1). DN1 cells contain the lymphoid progenitors which are also capable of giving rise to non-T lineage cells such as NK cells and dendritic cells. DN2 cells are mostly committed to the T cell lineage and the majority of DN2 cells retain developmental potential for either the αβ or γδ T cell lineages. DN3 cells are mostly en route to become αβ T cells, although a small fraction of DN3 cells are still capable of adopting the γδ T cell fate (2). Although the exact molecular mechanism influencing the αβ and γδ lineage fate at the DN3 stage is not clear, the expression of a TCRβ chain in the pre-TCR complex or the expression of a γδ TCR is a well documented critical checkpoint for the subsequent differentiation into the αβ or γδ lineage, respectively (3–5).
TCRβ gene rearrangement and TCRβ expression are subject to the rule of allelic exclusion, which ensures monoallelic expression of a functional TCRβ chain in each T cell. TCRβ locus rearrangement begins with D-J recombination in a bi-allelic manner and proceeds to V-DJ recombination on one allele at a time. If the first V-DJ rearrangement leads to an in-frame joining and expression of a functional TCRβ chain, the expressed TCRβ product blocks further rearrangement on the second allele. The second allele starts rearrangement only if the first V-DJ rearrangement results in an out-of-frame joining. Thus, a progenitor T cell only has an approximately 56% chance (1/3 + 2/3×1/3) to produce a functional TCRβ chain (6). The remaining cells that fail to produce a functional TCRβ chain are eliminated from the thymus by apoptosis, although the regulatory mechanism triggering apoptosis of these cells is currently unknown.
γδ T cells develop primarily from DN thymocytes and share common T cell progenitors with the αβ lineage. Both TCRδ and TCRγ genes undergo rearrangement in the DN2 and DN3 stages when the TCRβ gene is also actively rearranging. The δ gene has a single VDJC cluster that rearranges extensively at the DN2 stage (7, 8). The γ locus, which contains multiple VJC clusters, starts V-J rearrangement at the DN2 stage and continues to rearrange after entering the DN3 stage. Although the β locus begins D-J rearrangement at the DN2 stage, V-DJβ rearrangement does not occur until the DN3 stage. This delay in V-DJβ rearrangement suggests that most functional γδTCRs could be produced prior to the completion of TCRβ rearrangement. Therefore, γδ T cells may develop from the DN2 stage in the absence of competition with a functional TCRβ (7, 8). This model is further supported by a recent progenitor assay using the OP9-Delta1 culture system (2), which showed that DN2 to DN3 stage development is accompanied by a gradual loss of γδ progenitors and a gain of αβ progenitors.
Experimental evidence has shown that a significant fraction of γδ T cells develop at the DN2 stage in the absence of a functional TCRβ chain, though small fraction of DN3 cells are still capable of giving rise to γδ lineage T cells, indicating developmental potential at various developmental checkpoints. In fact, more than 10% of mature γδ T cells were found to express intracellular TCRβ, indicating that expression of a functional TCRβ chain does not preclude γδ lineage development. The mechanism of γδ T cell differentiation has long been debated, primarily between the selective versus instructive hypotheses. The selective hypothesis contends that T cell progenitor fate is determined prior to TCR rearrangement, while the instructive hypothesis states that lineage fate is determined via TCR signaling. More recent studies have indicated that the quantitative rather than qualitative difference between preTCR and γδ TCR signaling separate the two lineages (9, 10). A stronger signal is required for γδ T cell development whereas a weaker signal is sufficient to promote αβ lineage development. This signal strength model effectively explains the regulatory events post TCR expression. However, additional constrains must exist to limit the production of γδ T cells at the DN3 stage since γδ lineage development does not expand even in the absence of a functional TCRβ gene (11).
Id3 is one of few regulatory molecules known to be involved in both αβ and γδ T cell development (10, 12). Id3 encodes a small Helix-Loop-Helix (HLH) protein, which primarily functions as an inhibitor of E-proteins such as E2A (13). In the αβ T cell lineage Id3 is activated by the pre-TCR and TCR signaling pathways (14, 15). Expression of Id3 leads to inhibition of E2A function and consequently promotes cell survival and proliferation. A similar function for Id3 has been indicated downstream of γδ TCR signals during γδ T cell development in fetal thymus (10). It has been shown that Id3 is expressed and upregulated at the DN3 stage even before pre-TCR selection and γδ TCR selection (16). However, it is not known whether Id3 also plays a role before γδ TCR or β selection. Here, we show that loss of Id3 leads to a significant increase in numbers of γδ T cells. The enhanced γδ T cell development occurs primarily at the DN3 stage among progenitor T cells which have undergone extensive TCRβ gene V-DJ recombination. Our finding suggests that Id3 plays a critical role in limiting DN3 cells from progressing to the γδ lineage after non-productive TCRβ rearrangement.
MATERIALS AND METHODS
Mice
The Id3 knockout stain (17) has been backcrossed to C57Bl/6 11 generations before used in the present studies. This strain is now deposited in Jackson Labs (Bar Harbor). TCRβ knockout mice (11) were purchased from Jackson Labs. LAT−/− mice (18) and RAG1GFP mice (19) were obtained from Drs. Weiguo Zhang and Motonari Kondo, respectively. All animal procedures were approved by the Duke University Animal Use and Care Committee.
Flow cytometry analysis
Single-cell suspensions of lymphocytes from the thymus, spleen, bone marrow, and peripheral blood were prepared in ice-cold phosphate-buffered saline supplemented with 5% bovine calf serum. Erythrocytes were depleted by ammonium chloride lysis before use. Fetal thymus was harvested on day 17.5 or 18.5 of gestation. All suspensions were counted with a hemocytometer or coulter counter (Beckman coulter), and cells were stained immediately with a combination of a fluorescein isothiocyanate (FITC)-conjugated antibody, a phycoerythrin (PE)-conjugated antibody, an allophycocyanin (APC)-conjugated antibody, a TRI-COLOR® (TC)-conjugated antibody, and 7-amino-actinomycin D (7AAD) (Molecular Probes, Eugene, OR). Antibodies used in this study included FITC-conjugated anti-TCRγδ (GL3; Caltag Burlingame, Calif.), FITC-conjugated anti-B220 (RA3-6B2; Caltag), PE-conjugated anti-TCRβ (BD PharMingen), APC-conjugated anti-TCRβ (H57-597; eBioscience), APC-conjugated anti-B220 (RA3-6B2; Caltag), Biotin-conjugated anti-TCRγδ (GL3; Caltag), TC-conjugated anti-CD4 (CT-CD4; Caltag), anti-CD8a (CT-CD8a; Caltag), anti-B220 (RA3-6B2; Caltag), anti-CD11c (Caltag), anti-Ly6C/G (Gr-1) (Caltag). Dead and damaged cells were labeled with 7AAD, and were eliminated from the analysis. FITC-conjugated anti-Vγ1.1 TCR antibody, biotin- and FITC-conjugated anti-Vγ2 TCR antibody and Vγ3 TCR antibody were provided by Dr. David H. Raulet (University of California, Berkeley, CA). Cells were washed once with phosphate-buffered saline containing bovine calf serum and analyzed on a FACScalibur flow cytometer (Becton Dickinson, San Jose, Calif.). Data from 2 × 105 cells were collected and analyzed using the FlowJo program (TreeStar, Inc.).
In Vitro Stimulation
5 × 106 splenocytes were plated into 6 well plates with RPMI (GIBCO) supplemented with 5% FBS, P/S, L-glutamine, sodium pyruvate, and 2-ME. All wells were supplemented with 2ng/ml IL-2 and stimulated wells were supplemented with 1ug/ml Ionomycin and 10ng/ml PMA. Cells were incubated at 37°C for 2 hours. Cells were then supplemented with Monensin (Invitrogen) to a concentration of 3uM, and were incubated at 37°C for an additional 4 hours. Cells were then stained with CD8α, 7AAD, TCR-γδ-Biotin and B220 PECy5 for 20 minutes on ice. A secondary staining of SA-APC for 20 minutes on ice was then completed. Cells were then fixed in 2% paraformaldehyde in 1XPBS for 30 minutes on ice. Cells were then permeabilized in 0.5% saponin in FACS buffer for 20 minutes on ice. Cells were then resuspended in 0.5% saponin/FACS buffer for FACS analysis.
Intracellular TCRβ staining
5 × 107 splenocytes were stained with biotin-anti-TCR γδ (SA-APC added in the second step) and FITC- conjugated anti-B220 and TCRβ antibodies. B cells and αβ T cells were depleted with anti-FITC magnetic beads (Milteyi Biotech). The remaining cell population was then fixed with 2% paraformaldehyde, permeabilized with 0.5% saponin, and stained with PE-conjugated TCRβ.
PCR analysis of TCRβ gene D to J rearrangements
Total thymocytes and sorted TCRγδ or TCRαβ positive cells from spleen were lysed in a solution containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA (pH 8.0), 0.2 mg/ml proteinase K and 0.2% Triton X-100 for 30 min at 55°C and 10 min at 94°C. Touchdown PCR was performed with the following steps: 1) 5 min at 95°C; 2) 5 cycles of 30 s at 95°C, 30 s at 65°C and 2 min at 72°C; 3) 5 cycles of 30 s at 95°C, 30 s at 62°C, and 2 min at 72°C; 4) 5 cycles of 30 s at 95°C, 30 s at 60°C, and 2 min at 72°C; 5) 29 cycles of 30 s at 95°C, 30 s at 56°C, and 2 min at 72°C; and 6) 5 min at 72°C. The PCR primers are Dβ2 5’ (GGGACTGGGGGGGC) and Jβ2.7 3’(TGAGAGCTGTCTCCTACTATCGATT).
Quantitative PCR analysis of Dβ-Jβ, Vβ-DJβ, Jγ1, and Jγ4 germline DNA content
Quantitative PCR was performed on a Roche LightCycler using the FastStart DNA master SYBR green I kit (Roche) per manufacturer’s instructions. Each sample was first normalized by quantitative PCR amplification of the CD14 genomic locus. Thymocytes DNA from RAG2−/− mice was used to produce standard curve for all primer set. The primers for quantitative PCR are as follows. CD14 gene: CD14 for (GCTCAAACTTTCAGAATCTACCGAC) and CD14 rev (AGTCAGTTCGTGGAGGCCGGAAATC), DJβ2 germline: Dβ2GL for (CCTAGCAAGTTTCCCACGAG) and Dβ2GL rev (CTTCCTGTTGGTGTCCAGGT), V-DJβ germline: 2.4 UDBs (CTGCTTTGCCCAGTTCAGAG) and 2.4 UDBas (ACGAAACAGTGTCACCCTCC), Jγ1 germline: Jγ1GL for (ACCCAACTCCTCCGAACTTT) and Jγ1GL rev (CCACGGCTAGAGGAGACTTG), Jγ4 germline: for (TGGGTTCTATTTCCCCAGTG) and Jγ4GL rev (CTGTGGGCTTCCTGTCTCTC).
BrdU labeling assay
Id3−/− and WT control mice were injected with 1 mg BrdU via i.p. two times with a 2-hr interval. The thymus and spleen were taken at 6 hrs and 1 day after the first injection. BrdU incorporation was detected with a BrdU Flow kit (BD PharMingen) and analyzed on a FACScalibur flow cytometer (Becton Dickinson, San Jose, Calif.)
OP9-Dl1 culture
OP9-Dl1 stromal cells were cultured as described (16, 22). Monolayers of OP9-Dl1 cells were cultured in OP9 medium (MEM Alpha Medium supplemented with 20% FBS (HyClone), P/S, L-glutamine, and sodium pyruvate). Cocultures with OP9-Dl1 monolayer were maintained in IMDM supplemented with 5% FBS, P/S, L-glutamine, sodium pyruvate, and 2-ME with 1 × 102 sorted DN2 and DN3 thymocytes in the presence of 5 ng/ml Flt3 ligand (Peprotech) and 5 ng/ml IL-7 (BD Pharmingen). Cocultures were harvested by forceful pipetting at the indicated time points. Cell sorting was performed on a FACSVantage SE with a DiVa option (FACS facility of Duke Comprehensive Cancer Center). Cells used in bulk or single cell culture were collected after double sorting.
Results
Id3 knockout leads to a significant increase in numbers of γδ T cells
Rivera and colleagues have shown that αβ T cell development was partially blocked by disruption of the Id3 gene (12). Similar defect was observed in the study of a separate Id3 knockout allele (17). Further analysis showed that Id3 deficient (Id3−/−) mice exhibited a varying degree of elevated numbers of γδ T cells. After backcrossing the Id3 deficient allele to C57Bl/6 background for eleven generations, we found that the partial block of αβ T cell development remains unchanged. However, these backcrossed Id3−/− mice display a dramatic increase in numbers of γδ T cells in the thymus and spleen (Fig.1A &B). This γδ T cell population requires the T cell adaptor protein LAT (18), indicative that these γδ T cells are dependent on normal T cell signaling components (Fig. 1C). A functional assay using PMA and Ionomycin was performed to assess the responsiveness of these γδ T cells to stimulation, and the results show that not only are these cells responsive to stimulation as assessed by intracellular IFNγ production, but are hyper-responsive to stimulation (Fig. 1D). The physiological implications associated with this enhanced IFNγ production are yet unknown. The results of these experiments demonstrate that these γδ T cells likely represent a functional γδ T cell population, which signal through normal pathways.
Figure 1. An increase in numbers of γδ T cells in lymphoid tissues of Id3−/− mice.
(A) A representative flow cytometry analysis of γδ and αβ T cells in thymocytes and splenocytes from Id3−/− or wild type mice. Percentages of cells in the indicated quadrants are shown. (B) Absolute number of αβ and γδ T cells detected in thymus and spleen in control and Id3−/− mice are shown the bar graph with SEM (n=6 and 13, respectively). Id3−/− mice display on average an 8.4 and 2.0 fold increase of γδ T cells in thymus and spleen as compared to the wild type controls, respectively. (C) Representative flow cytometry analysis of γδ and αβ T cells in thymocytes from wild type, Id3−/−, Id3/LAT double deficient, and LAT−/− mice. Percentages of cells in the indicated quadrants are shown. (D) Histogram analysis of intracellular IFNγ from wild type and Id3−/− derived γδ T cells before and after stimulation with PMA/Ionomycin. Percentages of IFNγ positive cells are given in the brackets.
Increased population of Vγ1.1 cells and Vγ1.1Vγ3 double positive cells
This increase in numbers of γδ T cells seems to be restricted to postnatal life since numbers of γδ T cells in fetal thymus are similar between Id3−/− and wild type controls (Fig.2A). This observation suggests that different mechanisms may be involved in regulating fetal vs. postnatal development of the γδ cell lineage. Here, we will focus on further evaluation of the role of Id3 in postnatal γδ T cell development. Fetal and postnatal γδ T cell development are known to involve distinct sets of Vγ genes. For example, Vγ3 is almost exclusively used in the fetal stage to produce Vγ3 cells which migrate to and reside in the skin throughout postnatal life (23). In contrast, Vγ1.1 and Vγ2 are primarily used in postnatal life to produce γδ T cells which circulate to lymphoid tissues. Most γδ T cells present in Id3−/− mice were found to express Vγ1.1 (Fig.2B). The absolute number of Vγ2 expressing cells was comparable to that in wild type controls. A small fraction of γδ T cells in Id3−/− mice also expresses TCRVγ3. Double staining with Vγ1.1 and Vγ3 antibody demonstrated that essentially all Vγ3 positive cells in Id3−/− mice are also positive for Vγ1.1 (Fig.2C). This observation suggested that, in addition to increasing cell numbers, Id3 deletion may also disrupt the temporal control of Vγ gene usage.
Figure 2. Increased population of Vγ1.1 cells and Vγ1.1Vγ3 double positive cells.
(A) Percentages and absolute numbers of γδ T cells detected in fetal and neonatal thymus and neonatal spleen from wild type control and Id3−/− mice. (B) Flow cytometry analysis of thymocytes and splenocytes for expression of Vγ1.1 (top panel), Vγ2 (middle), and Vγ3 (bottom) among TCRγδ T cells. Non-lymphocytes and B220 positive B cells are eliminated by gating in the two-D plots. (C) Double staining shows that most Vγ3 positive cells from Id3−/−thymus and spleen are also positive for Vγ1.1.
Increased γδ T cells in Id3−/− mice is not due to proliferative expansion of mature γδ T cells
The increase in cell numbers may be caused by an expansion of existing γδ T cells or for other reasons such as enhanced generation of γδ T cells. To assess the proliferation rate of γδ T cells, we pulse labeled mice with BrdU and evaluated BrdU incorporation at 6 hours and 24 hours post BrdU injection (Fig.3). The percentage of BrdU incorporation among a given cell population at the 6 hour time point is used as an indicator for the proliferation status of the cell population. The results of the 24 hour chase may also be influenced by the influx of newly generated cells from precursors. Analysis of γδ T cells in thymus and spleen showed a lower percentage of BrdU positive cells in Id3−/− mice in comparison with wild type controls after a 6 hour pulse labeling (Fig.3A). This difference between the Id3−/− samples and wild type controls persisted for 24 hours after the initial labeling (Fig.3B). This result argues against the idea of an overall proliferative expansion of the existing γδ T cell pool. The absolute number of BrdU positive γδ T cells is higher in Id3−/− thymus and spleen than in wild type controls (Fig.3B), however this is most likely because the total amount of γδ T cell population is significantly larger in Id3−/− mice than in wild type controls. The dramatic difference in the population size of γδ T cells also makes it difficult to evaluate how much of BrdU labeling observed at the 24 hour time point is due to influx of newly generated γδ T cells. In the same analysis, the percentage of BrdU incorporation among αβ T cells is not significantly different between Id3−/−and wild type mice.
Figure 3. Increased incorporation of BrdU among γδ T cells in Id3−/− mice.
(A) Flow cytometry analysis of BrdU incorporation among gated γδ T cells from thymocytes and splenocytes 6 hrs after the first BrdU injection. Mice received two doses of BrdU injection with a 2 hr interval. (B) Percentage and numbers of BrdU positive γδ and αβ T cells in thymus and γδ T cells in spleen of Id3−/− or control mice at 6 hours or 1 day after the first BrdU injection. Each data point is the mean ± SEM of three independent experiments.
Id3−/− DN3 cells contain increased numbers of γδ T cell progenitors
As Id3 has been shown to be upregulated at the DN3 stage, we further evaluated γδ T cell development in OP9-Dl1 culture system, which is capable of supporting both αβ and γδ T cell development (2, 16). We tested DN2 and DN3 cells in culture and found a significant increase of γδ T cell development from Id3−/− mice as compared to wild type DN cells (Fig.4A). On average, Id3−/− DN2&3 cells gave rise to 3.1 times and 5.2 times more γδ T cells than wild type samples on day 5 and day 7, respectively. The ratio of γδ vs. αβ T cells was also 2.2 and 2.8 fold higher in Id3−/− samples than in wild type controls at day 5 and day 7, respectively. γδ T cell development from both wild type and Id3−/− DN2&3 progenitors was completely dependent on IL-7 (Fig.4B). This result argues against the possibility that Id3 knockout allows γδ T cell development to bypass a need for IL7 signaling, which is considered a limiting factor in γδ lineage development (24). All γδ T cells derived in this culture assay were negative for CD4 or CD8 markers (data not shown), indicating a developmental pathway independent of DN to DP transition. To further assess lineage specific progenitor activity at the DN3 stage, we performed a single cell assay (Fig.4C&D). The overall cloning efficiencies for TCR positive clones were 12.5% (48 out of 384 wells) and 6.6% (22 out of 334 wells) for wild type and Id3−/− DN3 cells, respectively. Among the TCR positive clones from the wild type mice, 67% were committed to the αβ cell lineage and only 13% committed to the γδ cell lineage, a result consistent with a recent report (2). In contrast, the frequency of αβ committed progenitors dropped to 45% while the frequency of γδ T cell progenitors rose to 32% in Id3−/− DN3 cells. The frequencies of αβ and γδ bi-potential progenitors were similar between the wild type (21%) and Id3−/− (23%) DN3 cells. Taken together, these data suggest that Id3 removal inhibits αβ and promotes γδ lineage development.
Figure 4. Id3−/− DN3 cells show increased progenitor activity in OP9-Dl1 culture assay.
(A) Analysis of 100 sorted DN2&3 thymocytes from wild type and Id3−/− mice after 5 or 7 days of culture on OP9-Dl1 stromal layer in the presence of Flt3 and IL7. Total thymocytes recovered, γδ T cells, and the ratio of γδ vs. αβ are shown. Results are the mean ± SEM of six independent wells for each genotype at each time point. (B) Representative FACS plots of double staining of CD4 and CD8a (left panel) or TCRγδ and TCRβ (right panel) at day 7 of OP9-Dl1 culture with or without IL7. Percentage of cells in each quadrant is given in the plot. (C) Representative FACS plots of three classes of clonal expansion from single DN3 cells cultured in OP9-Dl1 for 7 days. (D) Bar graph summary of relative percentage of each clone type among the TCR positive clones from wild type and Id3−/− DN3 cells. The total clones scored for wild type and Id3−/− samples are 48 and 22 from 384 and 334 individually seeded DN3 cells, respectively.
Most Id3−/− γδ T cells develop before the preTCR checkpoint
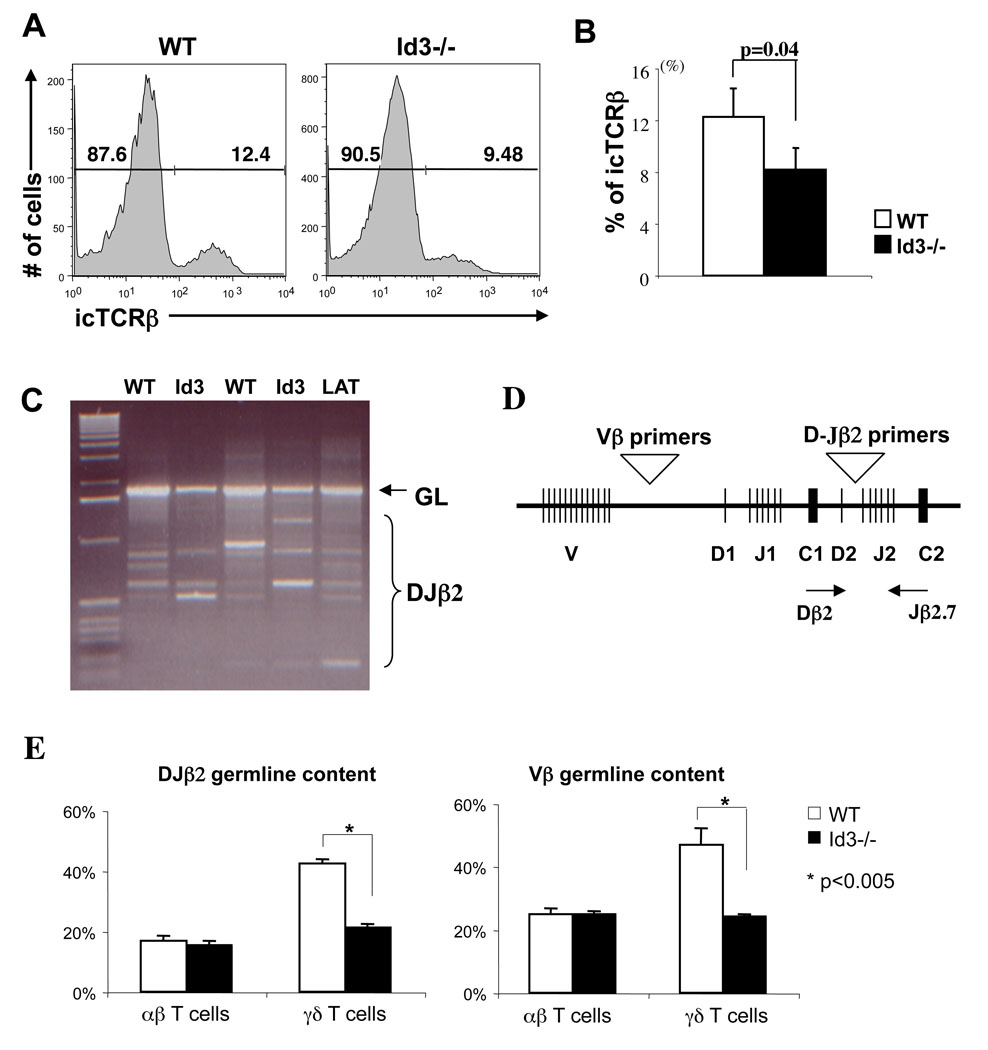
Id3 has been implicated to play an important role downstream of the pre-TCR signal leading to αβ lineage commitment and expansion (15). It is possible that a block in αβ lineage development at this stage may divert cells into γδ lineage. This has been shown to be the case in pre-Tα knockout mice, in which T cell development is diverted to the γδ lineage after successful TCRβ rearrangement (25). This conclusion is supported by the fact that γδ T cells that develop in pre-Tα knockout mice show an increased frequency of intracellular TCRβ expression. To determine if a similar mechanism is responsible for enhanced γδ T cell development in Id3−/− mice, we evaluated intracellular TCRβ expression among Id3−/− γδ T cells. In contrast to pre-Tα knockout mice, we found that Id3−/− γδ T cells displayed a slightly reduced frequency of intracellular TCRβ expression as compared to wild type controls (Fig.5A&B). This result indicates that majority of γδ T cells in Id3−/− mice are derived before the pre-TCR checkpoint.
Figure 5. Expression and rearrangement status of the TCRβ locus in peripheral γδ T cells.
(A) Histograms of intra cellular TCRβ expression in splenic γδ T cells from Id3−/− and control mice. Percentages of icTCRβ negative and positive cells are indicated in the plot. (B) Bar graph shows the percentage of icTCRβ positive cells. Results are mean ± SEM of three independent experiments as shown in (A). P value is calculated using paired two tailed student t-test. (C) PCR analysis of Dβ2-Jβ2.7 rearrangement in splenic γδ T cells from two Id3−/− and two wild type mice. Thymocytes from LAT−/− mice were included as a positive control. Positions of the germline product and rearranged products are indicated on the side of the gel. (D) A diagram of the TCRβ locus highlighting the positions of primers used for the analysis of D-Jβ2 rearrangement and V-DJβ and D-Jβ2 germline retention. (E) Realtime PCR analysis of D-Jβ2 and V-DJβ germline retention among αβ and γδ T cells from wild type and Id3−/− mice. Y-axis indicates the percentage of germline DNA retained relative to RAG2−/− thymocyte DNA.
Increased TCRβ gene rearrangement associated with Id3−/− γδ T cells
The assay for intracellular TCRβ expression only detects functional TCRβ gene rearrangement. To further evaluate the overall rearrangement status of TCRβ locus among γδ T cells, we carried out a PCR assay to detect various Dβ2 to Jβ2 rearrangements (Fig.5C). Extensive D-Jβ2 rearrangement was detected in γδ T cells from both wild type and Id3−/− mice. The TCR DJβ2 germline band appeared to be underrepresented in Id3−/− cells as compared to wild type cells. To further quantify germline TCRβ DNA content among mature T cells, we carried out a realtime PCR assay for germline DNA configuration at two separate locations within the TCRβ locus (Fig.5D). DNA from RAG2 knockout mice was used as the baseline for 100% germline retention. We found that Vβ germline DNA content in αβ T cells is indistinguishable between wild type and Id3−/− mice (Fig.5E). In contrast, TCRβ germline DNA content in Id3−/− γδ T cells is significantly lower than that in wild type controls and is reduced to a level similar to that in αβ T cells. The reduction of germline DNA in Id3−/− γδ T cells was found for both D-Jβ and V-DJβ specific primers. This result suggests that a significant fraction of γδ T cells in Id3−/− mice is produced either during or after TCRβ gene rearrangement.
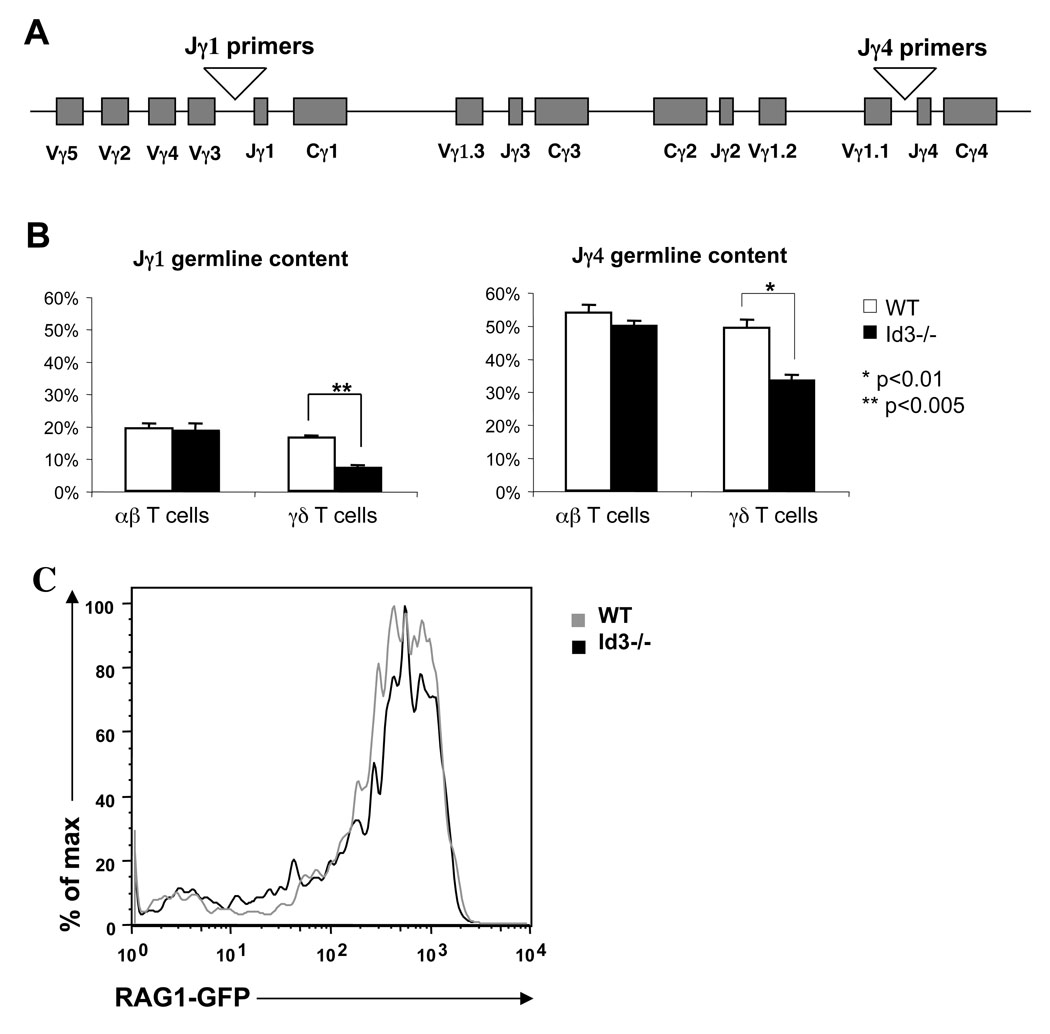
Increased TCRγ gene rearrangement in Id3−/− mice
The unique organization of TCRγ locus permits multiple trials for successful V-J recombination (Fig.6A). Thus, an increase in γδ T cell numbers in Id3 knockout mice could be due to increased opportunities for additional rounds of TCRγ rearrangement. We compared TCRγ locus germline DNA content among mature γδ T cells of wild type and Id3−/− mice (Fig.6B). Both Jγ1 and Jγ4 germline DNA were significantly underrepresented in γδ T cells isolated from Id3−/− mice as compared to wild type mice. In contrast, the germline TCRγ DNA content in αβ T cells remained the same in Id3−/− mice and wild type mice. To assess whether the increase in germline rearrangement was a result of enhanced RAG activity, Id3−/− mice were crossed with a RAG1GFP knockin mouse (19) to determine the level of RAG activity. Id3 knockout did not alter RAG1 expression among DN2/3 thymocytes, indicating a separate mechanism is likely responsible for the extensive germline rearrangement (Fig. 6C).
Figure 6. Germline DNA content at the Vγ locus.
(A) Diagram of the TCRγ gene locus and positions of primers used in the analysis of Jγ1 and Jγ4 germline DNA. (B) Quantitative PCR analysis of V-Jγ1 and V-Jγ4 germline retention. CD14 was used as loading control to normalize the amount of DNA in each sample. Percentage of germline DNA retention was calculated by comparing with total thymic DNA from Rag2−/− mice, which was set as 100%. (C) Histogram analysis of relative GFP expression of DN2 and DN3 populations from wild type and Id3−/− mice.
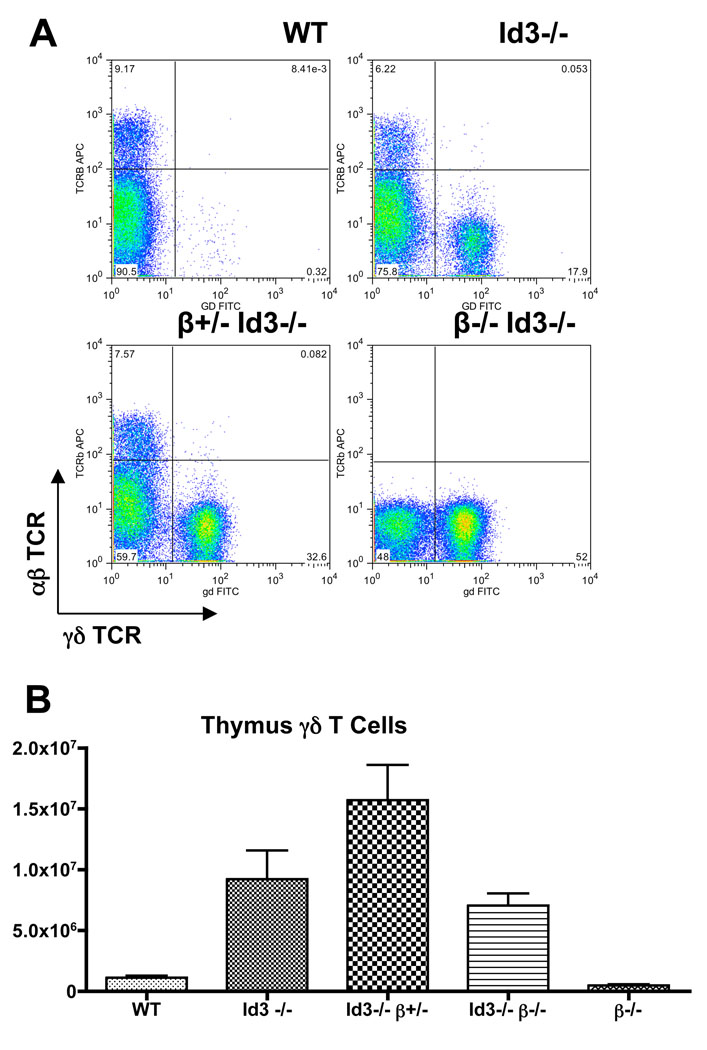
Id3 deficiency promotes γδ T cell development in the absence of any functional TCRβ expression
It has been shown that deletion of the constant regions of the TCRβ gene blocks αβ lineage development without any significant enhancement of γδ T cell development (11). If Id3 knockout promotes γδ lineage development independent of any functional TCRβ gene product, we predict that removal of Id3 on the TCRβ deficient background should lead to a significant enhancement of γδ lineage development similarly to that observed in Id3 knockout on TCRβ sufficient background. Therefore, we examined γδ T cell development in Id3 and TCRβ double deficient mice. Indeed, we find that numbers of γδ T cells were increased dramatically, from an average of 0.5 × 106 γδ T cells in TCRβ−/− mice to 7 × 106 γδ T cells in Id3−/−TCRβ−/− mice (Fig.7). This result supports the notion that Id3 knockout promotes γδ T cell development from progenitors which are incapable of producing a functional TCRβ product. In the cross between the Id3 and TCRβ mutant alleles, we observed that Id3−/−TCRβ+/−mice rather than Id3−/−TCRβ−/− mice produced the highest number of γδ T cells among all the genotype groups analyzed. This outcome could be attributed to the lack of the αβ lineage cells on TCRβ−/− background, which are thought to promote γδ T cell development in trans (Silva-Santos, 2005 and see discussion).
Figure 7. Additional enhancement of γδ T cell development by preventing the expression of a functional TCRβ chain.
A. Representative FACS plots showing the relative ratio of αβ vs. γδ T cells in the thymus of C57Bl/6 wild type control (n=13), Id3−/− mice (n=12), Id3−/−TCRβ+/− mice (n=5), Id3−/−TCRβ−/− mice (n−4), and TCRβ−/− mice (n=3).
B. Summary of absolute numbers of γδ T cells from thymus of each relevant genotype.
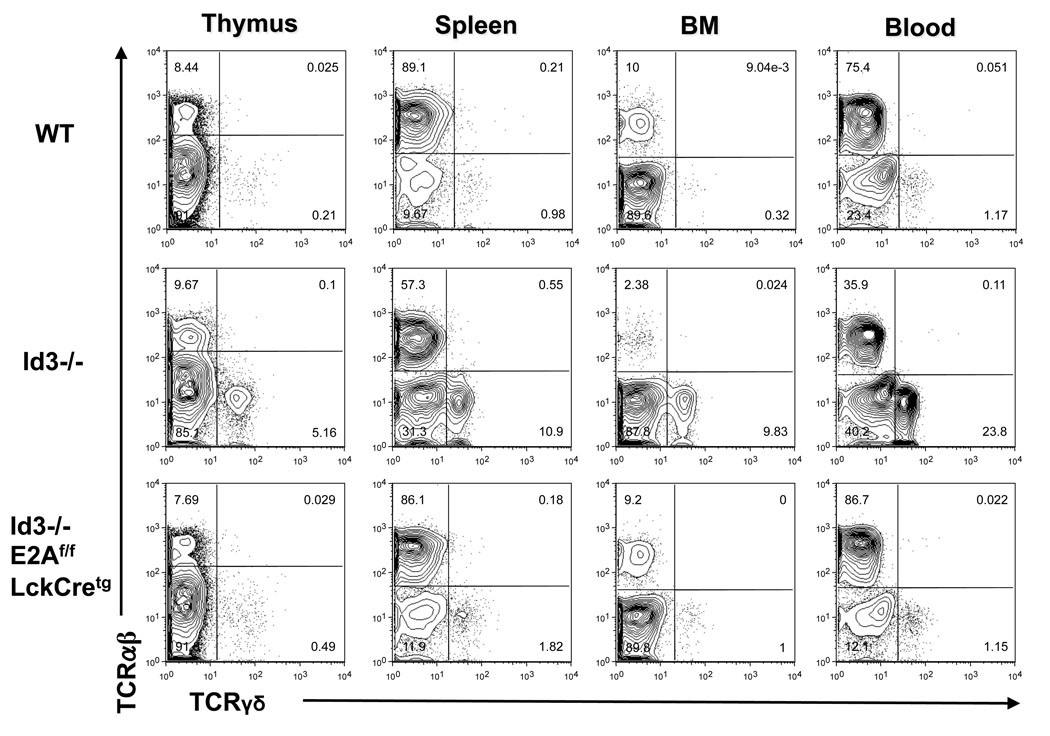
Enhanced γδ T cell development in Id3−/− mice is dependent on E2A
Id3 is known to inhibit E2A activity through competitive dimerization. E2A has been shown to regulate rearrangement of TCRβ (26)and TCRγ (27)genes. Therefore, it is conceivable that enhanced E2A activity may account for the phenotype described above. To test this idea and to further verify that the γδ phenotype is an intrinsic property of the developing T cells, we evaluated γδ T cell development after removing E2A exclusively in the T lineage on the Id3 knockout background. We have shown previously that conditional disruption of E2A in T cell lineage in mice carrying E2A floxed allele and a LckCre transgene does not have any significant effect on T cell development (28). However, removal of E2A on Id3 knockout background effectively prevented hyper production of γδ T cells induced by Id3 knockout (Fig.8). Therefore, the enhanced γδ T cell development observed in Id3 knockout mice is dependent on E2A activity in developing T cells.
Figure 8. Analysis of γδ T cells from Id3−/− E2Aflox/flox LckCretg mice.
The relative ratio of αβ and γδ T cells in thymus, spleen, bone marrow, and blood of wild type, Id3−/−, and Id3−/−E2Aflox/flox LckCretg mice were analyzed by dual staining. Plots were gated on live lymphocytes excluding B cells. Results were representative of two independent groups of mice from the Id3−/− E2Aflox/flox LckCretg breeding.
DISCUSSION
We report here that deletion of Id3 promotes γδ T cell development. This function of Id3 appears to be independent of its inhibitory role in αβ lineage development. Several models are considered here to explain the possible effect of Id3 knockout on αβ vs. γδ lineage development. First, Id3 deficiency may promote γδ T lineage development by exclusively acting on a subset of γδ T cell progenitors independent of αβ lineage development. The high frequency of TCRβ rearrangement observed in the γδ T cells of Id3−/− mice opposes this idea and strongly suggests that these γδ T cells arise from common precursors which are permissive for TCRβ rearrangement.
Second, loss of Id3 may promote γδ T cell development by blocking the preTCR signals and diverting these post β-rearranging cells to the γδ lineage. Upregulation of Id3 by preTCR is thought to be critical for blocking E-protein activity and releasing DN3 cells from the cell cycle checkpoint (15). A lineage switch at this developmental point will result in the production of γδ T cells which have already acquired a functional TCRβ chain. For example, deletion of the preTα gene resulted in enhanced production of γδ T cells which express ic-TCRβ (25). However, the frequency of ic-TCRβ expression among γδ T cells in Id3 knockout mice was slightly decreased rather than increased. Therefore, a block in pre-TCR signaling can not account for the dramatic increase of γδ T cells in Id3−/− mice. Most γδ T cells in Id3−/− mice are apparently derived from a developmental stage lacking a functional TCRβ chain. This conclusion was further supported by the fact that Id3 knockout enhances γδ T cell development even in the absence of a functional TCRβ gene.
We propose that Id3 deficiency promotes γδ T cell development by extending the window of TCRγ gene rearrangement among DN3 cells which have completed V-DJβ rearrangement but failed to produce a pre-TCR. These cells normally would die due to the lack of β selection. In the absence of Id3, E2A promotes further TCRγ rearrangements, which provide additional opportunity for these cells to acquire a functional γδTCR and progress to the γδ lineage. This interpretation is consistent with the observation that mature γδ T cells but not mature αβ T cells exhibit enhanced rearrangements of TCRγ genes. Based on this idea, we hypothesized that a block in TCRβ gene expression on Id3−/− background will make more precursor cells available for γδ T cell development. We tested our Id3 mutation on the background of a TCRβ mutant allele, which lacks both the Cβ1 and Cβ2 coding regions and thus is incapable of producing a functional TCRβ chain. If the majority of γδ T cells in Id3−/− mice come from DN3 cells, which have failed to produce a functional TCRβ chain, approximately 44% (2/3 × 2/3) DN3 progenitors have the opportunity to give rise to γδ T cells. When one allele of TCRβ renders non-functional, the maximal frequency for DN3 cells to contribute to the γδ lineage should be increased to 66% (2/3). Indeed, blocking TCRβ expression from one TCRβ allele resulted in a further increase in numbers of γδ T cells on the Id3−/− background. Deletion of Id3 on TCRβ−/− background also resulted in a substantial increase in numbers of γδ T cells, although the number of γδ T cells was lower than that on TCRβ+/− or TCRβ+/+ background. The less potent effect on γδ T cell development from TCRβ−/− background than from the TCRβ+/− background is most likely due to a requirement for DP cells in modulating γδ T cell development (29). Furthermore, a complete block of αβ lineage development also perturbs the overall architecture of the thymic environment which may adversely affect γδ lineage development. Although multiple factors may contribute to γδ T cell development after removal of both Id3 and TCRβ genes, the Id3−/− TCRβ−/− mice provide an informative model to further dissect the role of Id3 in regulating γδ lineage development in the absence of competition from αβ lineage development.
The increased development of γδ T cells reported here is complementary to the earlier reports that E2A or HEB disruption led to reduced numbers of γδ T cells (27, 30). In addition, E-proteins have been implicated in regulation of TCRγ gene rearrangement. Ectopic expression of E47 and RAG1/2 were shown to be sufficient to activate V-Jγ recombination in a non-lymphoid human cell line (31). Thus, the lack of Id3 or impaired upregulation of Id3 after completion of TCRβ rearrangement may lead to persistent or increased E-protein activity, which could allow for enhanced or extra rounds of TCRγ rearrangement. This idea is supported by our finding that removal of E2A in developing thymocytes prevented any significant increase of γδ T cells induced by Id3 knockout. It is worth pointing out that the complete removal of E-proteins at the DN3 stage of T cell development has a differential impact on αβ vs. γδ T cell development. Removal of both E2A and HEB genes by double conditional knockout (32) or blocking E2A and HEB protein activities with a dominant negative form of HEB (33) all led to a near complete block in αβ lineage development but only a modest loss of γδ T cells. Because these experimental systems inactivate E-proteins at approximately the DN3 stage, it remains to be further determined whether E-proteins play any decisive role at the early stage of γδ lineage development prior to full activation and rearrangement of the TCRβ locus.
The Id3 mediated suppression of γδ T cell development may serve as an important mechanism to prevent the generation of T cells with inappropriate TCR and function. The enhanced production of Vγ1.1 positive cells and the appearance of Vγ3 and Vγ1.1 double positive cells in Id3−/− mice suggest a dysregulation at the TCRγ locus. γδ T cell development starts in fetal thymus and persists throughout postnatal life. Vγ3 expressing γδ T cells represent the first wave of γδ T cells that develop exclusively in fetal thymus. They migrate to the skin and stay there as dendritic epidermal T cells (DETC) for the rest of adult life (34). In contrast, Vγ1.1 and Vγ2 are exclusively used in late fetal and postnatal life and the resulting γδ T cells primarily stay and function in lymphoid tissues. However, Vγ1.1 but not Vγ2 usage is increased in Id3−/− mice. This explanation is consistent with the observation that ectopic expression of E2A and HEB led to activation of only a subset of human Vγ genes (31). Alternatively, Vγ1.1 may have a selective advantage over Vγ2 in pairing with certain TCRδ chains, resulting in under-representation of Vγ2 positive cells (20). Although the exact role of Id3 in regulating Vγ gene usage still remains to be resolved, our study of Id3 deficient mice clearly defines a unique window of T cell development during which γδ T cell development is suppressed by Id3. Further investigation of Id3 mediated pathways during this window of development should shed light on the mechanism that limits γδ T cell production in the postnatal thymus.
ACKNOWLEDGEMENTS
We thank Dr. Meifang Dai for establishing the 11 generation C57Bl/6 backcross of the Id3−/− strain, Dr. David Raulet for the generous gifts of Vγ specific antibodies, Dr. Juan-Carlo Zuniga-Pflucker for the gift of OP9Dl-1 stromal cells, Dr. Juan Carabana for primer designs in germline TCRVβ assay, Dr. Lindsay Cowell for advice on Vγ sequence analysis, and Dr. Michael Krangel and members of Zhuang laboratory for discussions and comments. We thank Lynn Martinek and Dr. Mike Cook of Duke Cell Sorter Facility for assistance in cell sorting. This work has been supported by the Ministry of Education, Science, and Culture of Japan (to I.U-H.), National Institute of Health (to Y.Z.), and Arthritis Foundation (to Y.Z.).
References
- 1.Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 2.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential notch dependency at the alphabeta and gammadelta T lineage bifurcation. Immunity. 2006;25:105–116. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman ES, Passoni L, Crompton T, Leu TM, Schatz DG, Koff A, Owen MJ, Hayday AC. Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- 4.Hayday AC, Pennington DJ. Key factors in the organized chaos of early T cell development. Nat Immunol. 2007;8:137–144. doi: 10.1038/ni1436. [DOI] [PubMed] [Google Scholar]
- 5.Kreslavsky T, Garbe AI, Krueger A, von Boehmer H. T cell receptor-instructed α β versus γ δ lineage commitment revealed by single-cell analysis. J Exp Med. 2008;205:1173–1186. doi: 10.1084/jem.20072425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khor B, Sleckman BP. Intra- and inter-allelic ordering of T cell receptor beta chain gene assembly. Eur J Immunol. 2005;35:964–970. doi: 10.1002/eji.200425806. [DOI] [PubMed] [Google Scholar]
- 7.Capone M, Hockett RD, Jr, Zlotnik A. Kinetics of T cell receptor beta, gamma, and delta rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44(+)CD25(+) Pro-T thymocytes. Proc Natl Acad Sci U S A. 1998;95:12522–12527. doi: 10.1073/pnas.95.21.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livak F, Tourigny M, Schatz DG, Petrie HT. Characterization of TCR gene rearrangements during adult murine T cell development. J Immunol. 1999;162:2575–2580. [PubMed] [Google Scholar]
- 9.Hayes SM, Li L, Love PE. TCR signal strength influences alphabeta/gammadelta lineage fate. Immunity. 2005;22:583–593. doi: 10.1016/j.immuni.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, Kappes DJ, Wiest DL. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Mombaerts P, Clarke AR, Hooper ML, Tonegawa S. Creation of a large genomic deletion at the T-cell antigen receptor beta-subunit locus in mouse embryonic stem cells by gene targeting. Proc Natl Acad Sci U S A. 1991;88:3084–3087. doi: 10.1073/pnas.88.8.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- 13.Quong MW, Romanow WJ, Murre C. E protein function in lymphocyte development. Annu Rev Immunol. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- 14.Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- 15.Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–826. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Pan L, Sato S, Frederick JP, Sun XH, Zhuang Y. Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol Cell Biol. 1999;19:5969–5980. doi: 10.1128/mcb.19.9.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Sommers CL, Burshtyn DN, Stebbins CC, DeJarnette JB, Trible RP, Grinberg A, Tsay HC, Jacobs HM, Kessler CM, Long EO, Love PE, Samelson LE. Essential role of LAT in T cell development. Immunity. 1999;10:323–332. doi: 10.1016/s1074-7613(00)80032-1. [DOI] [PubMed] [Google Scholar]
- 19.Kuwata N, Igarashi H, Ohmura T, Aizawa S, Sakaguchi N. Cutting edge: absence of expression of RAG1 in peritoneal B-1 cells detected by knocking into RAG1 locus with green fluorescent protein gene. J Immunol. 1999;163:6355–6359. [PubMed] [Google Scholar]
- 20.Pereira P, Boucontet L. Rates of recombination and chain pair biases greatly influence the primary gammadelta TCR repertoire in the thymus of adult mice. J Immunol. 2004;173:3261–3270. doi: 10.4049/jimmunol.173.5.3261. [DOI] [PubMed] [Google Scholar]
- 21.Volpe JM, Cowell LG, Kepler TB. SoDA: implementation of a 3D alignment algorithm for inference of antigen receptor recombinations. Bioinformatics. 2006;22:438–444. doi: 10.1093/bioinformatics/btk004. [DOI] [PubMed] [Google Scholar]
- 22.Taghon TN, David ES, Zuniga-Pflucker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong N, Raulet DH. Development and selection of gammadelta T cells. Immunol Rev. 2007;215:15–31. doi: 10.1111/j.1600-065X.2006.00478.x. [DOI] [PubMed] [Google Scholar]
- 24.Kang J, Volkmann A, Raulet DH. Evidence that gammadelta versus alphabeta T cell fate determination is initiated independently of T cell receptor signaling. J Exp Med. 2001;193:689–698. doi: 10.1084/jem.193.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aifantis I, Azogui O, Feinberg J, Saint-Ruf C, Buer J, von Boehmer H. On the role of the pre-T cell receptor in alphabeta versus gammadelta T lineage commitment. Immunity. 1998;9:649–655. doi: 10.1016/s1074-7613(00)80662-7. [DOI] [PubMed] [Google Scholar]
- 26.Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. Regulation of T cell receptor beta gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity. 2007;27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Bain G, Romanow WJ, Albers K, Havran WL, Murre C. Positive and negative regulation of V(D)J recombination by the E2A proteins. J Exp Med. 1999;189:289–300. doi: 10.1084/jem.189.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan L, Hanrahan J, Li J, Hale LP, Zhuang Y. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J Immunol. 2002;168:3923–3932. doi: 10.4049/jimmunol.168.8.3923. [DOI] [PubMed] [Google Scholar]
- 29.Silva-Santos B, Pennington DJ, Hayday AC. Lymphotoxin-mediated regulation of gammadelta cell differentiation by alphabeta T cell progenitors. Science. 2005;307:925–928. doi: 10.1126/science.1103978. [DOI] [PubMed] [Google Scholar]
- 30.Barndt R, Dai MF, Zhuang Y. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during alpha beta thymopoiesis. J Immunol. 1999;163:3331–3343. [PubMed] [Google Scholar]
- 31.Ghosh JK, Romanow WJ, Murre C. Induction of a diverse T cell receptor gamma/delta repertoire by the helix-loop-helix proteins E2A and HEB in nonlymphoid cells. J Exp Med. 2001;193:769–776. doi: 10.1084/jem.193.6.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong N, Kang C, Raulet DH. Positive selection of dendritic epidermal gammadelta T cell precursors in the fetal thymus determines expression of skin-homing receptors. Immunity. 2004;21:121–131. doi: 10.1016/j.immuni.2004.06.008. [DOI] [PubMed] [Google Scholar]