Abstract
The yeast filamentous growth (FG) MAP kinase (MAPK) pathway is activated under poor nutritional conditions. We found that the FG-specific Kss1 MAPK is activated by a combination of an O-glycosylation defect caused by disruption of the gene encoding the protein O-mannosyltransferase Pmt4, and an N-glycosylation defect induced by tunicamycin. The O-glycosylated membrane proteins Msb2 and Opy2 are both essential for activating the FG MAPK pathway, but only defective glycosylation of Msb2 activates the FG MAPK pathway. Although the osmoregulatory HOG (high osmolarity glycerol) MAPK pathway and the FG MAPK pathway share almost the entire upstream signalling machinery, osmostress activates only the HOG-specific Hog1 MAPK. Conversely, we now show that glycosylation defects activate only Kss1, while activated Kss1 and the Ptp2 tyrosine phosphatase inhibit Hog1. In the absence of Kss1 or Ptp2, however, glycosylation defects activate Hog1. When Hog1 is activated by glycosylation defects in ptp2 mutant, Kss1 activation is suppressed by Hog1. Thus, the reciprocal inhibitory loop between Kss1 and Hog1 allows only one or the other of these MAPKs to be stably activated under various stress conditions.
Keywords: filamentous growth, glycosylation, MAP kinase, signal transduction, yeast
Introduction
Under optimal growth conditions, the budding yeast Saccharomyces cerevisiae grows as round-shaped, single cells. On nitrogen starvation or in the presence of poor carbon sources, however, diploid cells of certain strains (such as Σ1278 h) initiate a fungus-like filamentous growth (FG) (Gimeno et al, 1992). Haploid cells also undergo a related phenomenon whereby the elongated cells invade solid media (Roberts and Fink, 1994). These responses, which we will refer to collectively as FG, are considered as an adaptive mechanism that allows normally sessile yeast to forage for scarce nutrients or to evade harmful waste products. An FG response requires at least three distinct protein kinases: the cAMP-dependent protein kinase Tpk2, the 5′-AMP-dependent protein kinase Snf1, and the MAP kinase (MAPK) Kss1 (Chen and Thorner, 2007). This article focuses on the signalling pathway that activates Kss1, namely, the FG MAPK pathway, whose core is a three-kinase cascade composed of the Ste11 MAPKKK, the Ste7 MAPKK and the Kss1 MAPK (Roberts and Fink, 1994).
Another MAPK cascade, termed the high osmolarity glycerol (HOG) MAPK pathway, is activated by, and required for adaptation to, an increased external osmolarity (Brewster et al, 1993; Hohmann, 2002). The HOG MAPK pathway is composed of two functionally redundant upstream signalling branches that regulate the common Hog1 MAPK (Maeda et al, 1994, 1995). Of these branches, the SHO1 branch is closely related to the FG MAPK pathway. The SHO1 branch is initiated by an interplay between the membrane anchor protein Sho1 and the putative osmosensor membrane proteins Msb2 and Hkr1, leading to activation of Ste11, that is, the same MAPKKK that is involved in the FG MAPK pathway (Maeda et al, 1995; Tatebayashi et al, 2006, 2007). In the HOG pathway, however, activated Ste11 activates the Pbs2 MAPKK, which then activates the Hog1 MAPK (Posas and Saito, 1997) (see Figure 7A). Activated Hog1 governs a series of adaptive responses to high osmolarity, including temporarily arrest of the cell-cycle progression, readjustment of the transcription and translation patterns, and synthesis and intracellular retention of the compatible osmolyte glycerol (Bilsland-Marchesan et al, 2000; Teige et al, 2001; Hohmann, 2002; Escote et al, 2004; O'Rourke and Herskowitz, 2004).
Although both the FG and the HOG MAPK pathways involve Ste11, each controls completely different, and mutually incompatible responses. Naturally, osmostress normally activates only the Hog1 MAPK (Posas and Saito, 1997). However, osmostress can activate the FG MAPK pathway in mutant cells, such as pbs2Δ or hog1Δ, in which the Hog1 MAPK cannot be activated (O'Rourke and Herskowitz, 1998; Davenport et al, 1999). In other words, activated Hog1 inhibits undesirable cross-talk activation of the FG MAPK pathway by osmostress in normal cells. In the pathogenic Candida albicans yeast, its Hog1 homolog inhibits, or raises the activation threshold of, the filamentation-inducing Cek1 MAPK (the Candida homolog of Kss1) by various natural inducers, thereby affecting its virulence (Eisman et al, 2006). Thus, inhibition of Kss1 by Hog1 is a conserved feature of the yeast MAPK pathways.
It is, however, unknown whether or how an inhibition of cross-talk in the reverse orientation occurs, that is, does activation of the FG pathway suppress the HOG pathway? Using a novel approach based on the use of glycosylation defective mutants, we show here that activation of the FG-specific Kss1 MAPK in fact suppresses a concomitant activation of the HOG-specific Hog1 MAPK. When activation of Kss1 is blocked by mutations, such as ste7Δ or kss1Δ, the Hog1 MAPK is activated by glycosylation defects. Kss1 inhibits Hog1 indirectly through the Ptp2 protein phosphatase. Thus, the FG MAPK pathway and the HOG MAPK pathway suppress each other so that only the relevant pathway is activated by a specific stimulus.
Results
A combination of O- and N-glycosylation defects induces FUS1-lacZ expression
To investigate the possible cross-regulation between the FG and the HOG pathways, a rapid and reliable method of activating the FG MAPK cascade is necessary. The commonly used method, namely the nutritional limitation, is inadequate, as it takes days to effect significant FG responses. A more promising approach is to use mutants that are defective in protein glycosylation. It has been shown that mutants of genes that are required for general glycosylation, such as OCH1, PMI40 and DPM1, constitutively activate an FG-like response (Lee and Elion, 1999; Cullen et al, 2000). However, because mutants of these genes are severely defective in protein glycosylation, they are either very sick or nonviable. It was also reported that the inhibitor of N-glycosylation tunicamycin moderately activated the FG MAPK pathway in rich media after 16 h (Cullen et al, 2000). As we will describe in this article, however, tunicamycin alone could not appreciably activate the FG MAPK pathway, at least under our experimental conditions. We, therefore, examined the possibility that a combination of separate, and more limited, defects in O-glycosylation and N-glycosylation might induce the FG response more robustly.
We found that the commonly used reporters for the FG response, such as FRE-lacZ and FG(TyA)-lacZ, were unsuitable for our purposes because they had high baseline expression levels without stimulation and relatively moderate increases on stimulation (Mosch et al, 1996; Madhani and Fink, 1997; Davenport et al, 1999; Cullen et al, 2004). We, therefore, used the FUS1-lacZ reporter, which had much lower baseline expression levels and larger increases on stimulation (Cullen et al, 2000). Furthermore, all strains used in this study carried the ssk2Δ ssk22Δ double mutation unless otherwise specified, to eliminate any potential influence from the SLN1 branch of the HOG pathway.
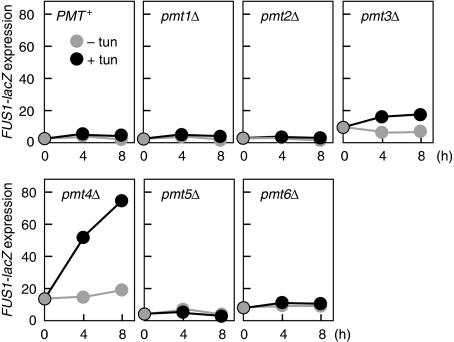
To inhibit O-glycosylation, we disrupted the genes that encode a family of protein O-mannosyltransferases (Pmt) (Gentzsch and Tanner, 1996; Girrbach and Strahl, 2003). Disruption of any of the six PMT genes (PMT1–PMT6), individually, had no significant effect on expression of FUS1-lacZ (Figure 1). Inhibition of N-glycosylation by tunicamycin, in wild-type cells, also did not induce FUS1-lacZ expression. In clear contrast, tunicamycin treatment of pmt4Δ mutant cells strongly induced FUS1-lacZ expression (Figure 1). Tunicamycin treatment of other pmt single mutants did not induce FUS1-lacZ expression. Furthermore, disruption of one or two additional PMT genes in the pmt4Δ background (e.g. pmt3/4/5Δ or pmt4/5/6Δ) did not have any stronger effect than pmt4Δ alone (Supplementary Figure S1). Thus, FUS1-lacZ expression is induced when N-glycosylation and a specific type of O-glycosylation is inhibited.
Figure 1.
Induction of the FUS1-lacZ reporter gene by glycosylation defects. The indicated yeast mutant cells carrying the pFUS1-lacZ plasmid were grown exponentially and were treated with (+tun) or without (−tun) tunicamycin (final conc. 25 μg/ml) for the indicated times. Activity of β-galactosidase in cell extracts was normalized using cell densities and expressed as Miller units (Miller, 1972). The strains used, and their full genotypes, are listed in Supplementary Tables S1 and S2, respectively.
Induction of FUS1-lacZ expression by glycosylation defects requires signalling elements of the FG pathway
We next examined whether induction of FUS1-lacZ expression by glycosylation defects is due to the activation of the FG pathway, because the FUS1-lacZ reporter can be induced also by the mating MAPK pathway. To determine which pathway is activated by the glycosylation defects, we disrupted, in the pmt4Δ background, genes that are specific for, or shared between, the mating and FG pathways. The results of an extensive survey are summarized in Figure 2A, which is a compilation of data from several different experiments. Although the mean value of the positive control strain (pmt4Δ) was somewhat variable from experiment to experiment (the range is between 34.3 and 61.9), only a typical control result is shown to save space. Thus, the exact values may not be directly comparable, but the following general conclusion is statistically valid.
Figure 2.
Signalling elements required for induction of FUS1-lacZ expression by glycosylation defects. (A) Induction of the FUS1-lacZ reporter gene by glycosylation defects. The indicated yeast mutant cells carrying the pFUS1-lacZ plasmid were grown exponentially, treated with (+tun) or without (−tun) tunicamycin for 8 h, and assayed for β-galactosidase activity as in Figure 1. (B) The indicated mutants were assayed as in (A). (C) Exponentially growing cells of the indicated genotypes were treated with 25 μg/ml tunicamycin for the indicated times. Cell extracts were probed for phosphorylated (activated) forms of Fus3 (P-Fus3), Kss1 (P-Kss1) and Mpk1 (P-Mpk1) by immunoblotting using anti-phospho-p44/42 antibody.
From these data, two components in FUS1-lacZ expression are discernible: the basal component seen without tunicamycin treatment, and the tunicamycin-induced component. On the basis of the differential effect of the mutants on these two components, we could classify the mutants into four groups. The first group (ste4Δ and ste5Δ) abolished only the basal component without affecting the induced component. The second group, composed of ste20Δ, ste50Δ, ste11Δ, ste7Δ, ste12Δ and the fus3Δ kss1Δ double mutation, abolished both the basal and induced components. The third group, composed of msb2Δ, opy2Δ and ptp2Δ, affected only the induced component. Finally, the fourth group (tec1Δ, fus1Δ, sho1Δ, hkr1Δ, hog1Δ, yps1Δ and ptp3Δ) affected neither the basal nor the induced expression levels. Genes in groups 1 and 2, whose disruption abolishes the basal components, are known to be involved in the mating pathway. On the other hand, genes in the groups 2 and 3, whose disruption abolishes the tunicamycin-induced components, are known to be required for FG response, with the notable exception of PTP2. Any role of the Ptp2 protein tyrosine phosphatase (PTP) in the FG MAPK pathway has not been known or suspected. Later in this article, we will explain why inactivation of the PTP2 gene suppresses FUS1-lacZ expression. Tec1 is required for the full FG response but is dispensable for induction of the FUS1-lacZ reporter (Madhani and Fink, 1997). Thus, in general, there is a good concordance between the genes that are required for the FUS1-lacZ expression induced by glycosylation defects and those that have been shown to be involved in the FG MAPK pathway.
Glycosylation defects induce characteristic FG responses
We examined whether glycosylation defects do indeed induce the FG response. A hallmark of the FG response is activation of the Kss1 MAPK, as opposed to the preferential activation of the Fus3 MAPK by the mating pathway (Chen and Thorner, 2007). As shown in Figure 2B, disruption of KSS1 completely abolished tunicamycin-induced FUS1-lacZ expression, whereas disruption of FUS3 only moderately reduced it, indicating that the Kss1 MAPK is mainly responsible for FUS1-lacZ expression by glycosylation defects. We then examined whether glycosylation defects activate Kss1 and/or Fus3 by probing for activation-associated phosphorylation of these kinases. The combination of a pmt4Δ mutation and tunicamycin treatment activated Kss1 but not Fus3 (Figure 2C). Disruption of STE7, which encodes the direct activator of Kss1, completely inhibited Kss1 phosphorylation, whereas disruption of STE4, which encodes an upstream signalling element in the mating pathway, had no effect. The preferential activation of Kss1 by glycosylation defects is thus consistent with activation of the FG pathway.
Tunicamycin also activates the cell-wall integrity Mpk1 MAPK pathway (Bonilla and Cunningham, 2003; Cohen et al, 2008). Phosphorylation of Mpk1, however, occurs very slowly after tunicamycin treatment and is independent of Kss1/Fus3 (Figure 2C, upper panel). As there was no apparent connection between Mpk1 and the FUS1-lacZ response under study, we did not analyse Mpk1 any further.
We also examined whether glycosylation defects induce FG. As tunicamycin inhibits cell growth, however, it cannot be used to induce FG. Therefore, as an alternative to tunicamycin, we used 2-butanol that is known to accelerate FG in liquid culture (Lorenz et al, 2000; Andersson et al, 2004). When the parental (PMT4+) yeast strain was grown in SC medium containing 1% 2-butanol, no FG was observed over a 16-h period (Supplementary Figure S2). In clear contrast, pmt4Δ mutant cells became elongated and formed filaments. The pmt4Δ msb2Δ double mutant cells did not form any filaments. Thus, we conclude that O-glycosylation defects caused by pmt4Δ significantly lower the threshold of other stimuli for induction of the FG response.
Opy2 and Msb2 are O-glycosylated by Pmt4
Next, we investigated the potential Pmt4 target(s) whose underglycosylation might activate the FG response. Pmt4 is unique among the protein O-mannosyltransferases in that it specifically glycosylates Ser/Thr-rich extracellular segments in membrane-attached proteins (Hutzler et al, 2007). This fact suggests that two membrane proteins required in the FG response, Opy2 and Msb2, are both potential substrates of Pmt4. Indeed, Opy2 has been shown to be glycosylated by Pmt4 (Hutzler et al, 2007).
Opy2 has two potential glycosylated Ser-rich (SR) regions, SR1 (aa 2–22) and SR2 (aa 75–84), in its extracellular domain (Figure 3A). The full-length, GFP-tagged Opy2 protein (Opy2-FL-GFP) isolated from PMT4+ cells was significantly larger than the same protein isolated from pmt4Δ cells (Figure 3B, compare lanes 1 and 5), indicating Pmt4-dependent glycosylation of Opy2. Deletion of the SR2 region did not reduce the Pmt4-dependent glycosylation (Figure 3B, lanes 3 and 7). In contrast, Opy2-ΔSR1-GFP had the same size in PMT4+ and pmt4Δ cells (Figure 3B, lanes 2 and 6), indicating that Pmt4 modifies only the SR1 region of Opy2.
Figure 3.
Glycosylation of Opy2 by Pmt4, and its role in the FG response. (A) Schematic diagrams of the Opy2 constructs used in (B–D). FL, full-length; SR, Ser-rich; TM, transmembrane segment. (B) C-terminally GFP-tagged Opy2 constructs were generated using a single-copy vector with the GAL1 promoter (p414GAL1). Expression of Opy2-FL-GFP, or the indicated deletion constructs, was induced by 2% galactose for 4 h in PMT4+ or pmt4Δ host cells. Total cell extracts were probed with an anti-GFP antibody by immunoblotting. The horizontal arrow indicates cleavage products. The estimated cleavage site is shown in (A) by an upright arrow. (C) The full-length OPY2 gene (with the OPY2 promoter) carried on a single-copy vector (pRS414) or its deletion constructs were individually expressed in pmt4Δ ste4Δ opy2Δ host cells that also carried the pFUS1-lacZ reporter plasmid. Cells were treated with (+tun) or without (−tun) tunicamycin (final conc. 25 μg/ml) for 8 h, before extracts were prepared for β-galactosidase assays. (D) GST-tagged Ste50 (GST-Ste50), or GST alone, was constitutively expressed using pFP122 or the vector p426TEG1, and expression of GFP-tagged Opy2-FL (Opy2-FL-GFP), or Opy2ΔC-GFP, was induced by 2% galactose for 4 h using the expression constructs based on the p414GAL1 vector, in pmt4Δ ste50Δ opy2Δ host cells. GST-Ste50 (or GST) was immunoprecipitated by glutathione-Sepharose beads (bottom panel), and co-precipitated Opy2-GFP protein was detected by immunoblotting (top panel). Expression levels of the Opy2-GFP proteins are shown in the middle panel.
Msb2 is also a glycosylated transmembrane protein (Cullen et al, 2004). Its extracellular domain contains an extended Ser/Thr rich (STR) region (aa 52–956) that is likely to be O-glycosylated (Hutzler et al, 2007) (see Figure 4A). The full-length HA-tagged Msb2 (Msb2-FL-HA) isolated from pmt4Δ cells migrated slightly faster in SDS–PAGE than the same molecule obtained from PMT4+ cells, indicating a contribution of Pmt4-dependent glycosylation. However, both molecules migrated at positions much higher than the calculated molecular weight of Msb2-FL-HA (ca. 150 kDa), reflecting the presence of Pmt4-independent glycosylation as well (Figure 4B). To clarify the roles of individual Pmt enzymes in Msb2 glycosylation, we used Msb2-ΔSTR1-HA and Msb2-ΔSTR2-HA for further analyses (see Figure 4A). The smaller size of these constructs allowed easier evaluation of the effect of pmt mutations. Both proteins, when expressed in pmt1Δ, pmt2Δ or pmt4Δ mutant cells, were smaller than when expressed in PMT+, pmt3Δ, pmt5Δ or pmt6Δ cells (Figure 4C and D), indicating that Pmt1, Pmt2 and Pmt4 contribute to Msb2 O-glycosylation. The fact that pmt1Δ and pmt2Δ reduced Msb2 glycosylation to a similar extent is consistent with the earlier observation that Pmt1 and Pmt2 form a functional heterodimer (Girrbach and Strahl, 2003). More important, the size reduction of Msb2 in pmt4Δ cells is quite different from that in pmt1Δ and pmt2Δ cells, clearly indicating that Pmt4 contributes to a distinct subset of Msb2 O-glycosylation. The Msb2-ΔSTR3-HA mutant protein, in which most of the STR region is absent, migrated closer to the calculated peptide molecular weight (ca. 50 kDa) (Figure 4E). Taken together, we conclude that Pmt4 plays a significant role in O-glycosylation of Msb2. Pmt1 and Pmt2 also glycosylate Msb2, but such glycosylation is either irrelevant or insufficient for the FG response.
Figure 4.
Pmt4 glycosylation of Msb2. (A) Schematic diagrams of the deletion constructs of Msb2 used in (B–F). Each construct contains two HA epitopes, one following amino acid position 48 and another at the C terminus. STR, Ser/Thr rich; HMH, Hkr1-Msb2 Homology domain; TM, transmembrane segment; FL, full-length. (B–E) Indicated Msb2-HA constructs, under the control of the GAL1 promoter in the single-copy vector p416GAL1, were induced by 2% galactose for 3 h in PMT+ or the indicated pmt mutant cells. Cell extracts were probed by immunoblotting with an anti-HA antibody. (F) Endo H treatment of Msb2. The Msb2-ΔSTR2-HA construct, under the control of the GAL1 promoter in the single-copy vector p416GAL1, was induced by 2% galactose for 3 h in PMT + cells and in the indicated pmt mutant cells. Cell extracts were prepared in Buffer N, immunoprecipitated with anti-HA antibody (3F10), treated with (+) or without (−) 0.4 U/ml endoglycosidase H (Endo H) at 37°C for 15 h, applied to SDS–PAGE, and probed with another anti-HA antibody (12CA5).
Msb2 has a number of potential N-glycosylation sites in addition to its numerous O-glycosylation sites. As defects in Pmt4-dependent O-glycosylation can interfere with the N-glycosylation status of the same protein (Ecker et al, 2003), we estimated the degree of Msb2 N-glycosylation by endoglycosidase H (Endo H) treatment. To this end, the Msb2-ΔSTR2-HA mutant protein was expressed in PMT+, pmt1Δ and pmt4Δ host cells, and the size of the protein before and after Endo H treatment was compared by SDS–PAGE. As shown in Figure 4F, Endo H treatment reduced the size of Msb2-ΔSTR2-HA to a similar extent irrespective of the host pmt mutation. Thus, at least in these cases, O-glycosylation defects did not substantially affect the status of Msb2 N-glycosylation.
In some cases, O-glycosylation is important for targeting proteins to the plasma membrane (Proszynski et al, 2004). Using YFP-fusion constructs, we examined whether removal of a substantial portion of the O-glycosylated STR region might affect the subcellular localization of Msb2. As shown in Supplementary Figure S3, both Msb2-FL-YFP and its deletion derivative Msb2-ΔSTR4-GFP are similarly localized on the plasma membrane, although a substantial amount of both constructs is also found in intracellular compartments, as observed earlier for Msb2-FL-YFP (Tatebayashi et al, 2007). Thus, removal of a substantial portion of the O-glycosylated STR region does not affect the cellular localization of the Msb2 protein.
Glycosylation defects of Msb2, but not of Opy2, activate the FG MAPK pathway
We then asked whether defective glycosylation of Opy2 or Msb2 induces the FG response. First, we examined the possibility that the glycosylated region in Opy2 is required for the FG response. However, an Opy2 mutant lacking the entire O-glycosylated domain (Opy2-ΔSR1) could fully support FUS1-lacZ expression (Figure 3C). In contrast, deletion of the cytoplasmic C-terminal region (aa 184–360) completely abolished FUS1-lacZ expression (Figure 3C). Although the full-length Opy2 protein binds to Ste50, Opy2-ΔC does not (Figure 3D). These results are consistent with the idea that the major function of Opy2 is to anchor Ste50 to the membrane, which is necessary for efficient signalling in the FG and HOG pathways (Wu et al, 2006; Tatebayashi et al, 2007). However, Opy2 glycosylation, or lack of it, may not be a factor in the FG response.
We then examined the possible role of the Msb2 glycosylation region in the FG response. When Msb2-ΔSTR4, in which a large segment of the glycosylated STR domain is deleted, was expressed in pmt4Δ host cells using the MSB2 own promoter, FUS1-lacZ was induced even in the absence of tunicamycin (Figure 5A and B). When less extensive deletion mutants, Msb2-ΔSTR7 and Msb2-ΔSTR8, were expressed, weaker induction of FUS1-lacZ was observed in the absence of tunicamycin. Expression of the least extensive deletion mutant, Msb2-ΔSTR5, did not induce FUS1-lacZ. Thus, there was a clear correlation between the extent of STR deletion and the extent of FUS1-lacZ induction in the absence of tunicamycin. All these Msb2 STR deletion mutants supported tunicamycin-induced FUS1-lacZ expression to similar extent. Thus, deletion of the glycosylated STR segment constitutively activated Msb2. Indeed, overexpression of Msb2-ΔSTR4 using an inducible GAL1 promoter activated the FG pathway even in glycosylation-proficient PMT4+ host cells, whereas expression of full-length Msb2 did not (Figure 5C). Thus, deletion of the STR region has a similar effect as glycosylation defects. In contrast, overexpression of glycosylation-defective Opy2-ΔSR1 did not activate the FG MAPK pathway at all, further supporting the earlier conclusion that glycosylation of Opy2 does not affect the FG MAPK pathway. Combining all the data, we conclude that deficiency of Pmt4-mediated O-glycosylation of Msb2 in the STR region is a part of signal that activates the FG MAPK pathway.
Figure 5.
Domains of Msb2 essential for the FG response. (A) Schematic diagrams of the Msb2 deletion constructs used in (B) and (C). STR, Ser/Thr rich; HMH, Hkr1-Msb2 Homology domain; TM, transmembrane segment; FL, full-length. (B) Induction of the FG reporter gene FUS1-lacZ by glycosylation defects. The full-length MSB2 gene, or its deletion constructs, carried on the single-copy vector pRS416 were individually expressed in pmt4Δ ste4Δ msb2Δ host cells in which the FUS1-lacZ reporter gene has been integrated. Cells were treated with (+tun) or without (−tun) tunicamycin (final conc. 25 μg/ml) for 8 h, before extracts were prepared for β-galactosidase assays. In HMH(Hkr1), the HMH domain of Msb2 was replaced by the homologous HMH domain from Hkr1. (C) Expression of the indicated Msb2 or Opy2 construct, placed under the GAL1 promoter, was induced by galactose for 2 h. Induction of the FG reporter gene FUS1-lacZ was assayed.
Msb2 has seven potential N-glycosylation sites that conform to the N-X-T/S motif (X being any aa other than P) in its extracellular domain (Asn-30, Asn-859, Asn-885, Asn-945, Asn-1049, Asn-1088 and Asn-1175). To test whether the effect of tunicamycin can be accounted for by defective Msb2 N-glycosylation, we generated a mutant termed Msb2-6N/A in which six of the potential N-glycosylation sites were mutated to Ala (Asn-30, which is outside the STR region, was left unmodified). Expression of the Msb2-6N/A construct in pmt4Δ host cells at the endogenous level (i.e. from the MSB2 promoter in a single-copy plasmid) did not induce FUS1-lacZ expression any better than wild-type Msb2, either in the absence or presence of tunicamycin (Supplementary Figure S4). Likewise, overexpression of Msb2-6N/A in pmt4Δ host cells using the GAL1 promoter did not induce FUS1-lacZ expression any better than wild-type Msb2 (Supplementary Figure S5). These results suggest that N-glycosylation defects in Msb2 alone are not sufficient to account for the effect of tunicamycin, and that there might be (an)other protein(s) whose N-glycosylation defects are involved in induction of FUS1-lacZ expression.
Activated Kss1 inhibits the Hog1 MAPK
With a reliable method to activate the FG pathway in hand, we investigated the interaction between the FG and the HOG MAPK pathways, using the HOG-specific reporter 8xCRE-lacZ (Tatebayashi et al, 2006). The pmt4Δ defect (in the absence of tunicamycin treatment) neither constitutively activated the HOG MAPK pathway in the absence of osmostress, nor inhibited the HOG response in the presence of osmostress (Supplementary Figure S6, open bars). We found, however, that tunicamycin treatment of pmt4Δ mutant cells, which activates the FG MAPK pathway, did not activate the HOG MAPK pathway (Figure 6A). In clear contrast, however, if activation of the Kss1 MAPK is blocked by ste7Δ, the HOG MAPK pathway was activated by tunicamycin treatment in the pmt4Δ cells (Figure 6A). Inactivation of both Kss1 and Fus3 (kss1Δ fus3Δ) also activated the HOG MAPK pathway by glycosylation inhibition (Figure 6A). Expression of the Kss1 MAPK (by a plasmid-carried KSS1) in the kss1Δ fus3Δ mutant completely inhibited the HOG pathway, whereas expression of the Fus3 MAPK only weakly inhibited it (Figure 6B). Disruption of either KSS1 or FUS3 separately gave essentially identical results (Supplementary Figure S7).
Figure 6.
Cross-inhibition of the Hog1 MAPK by the FG MAPK pathway. (A–C) Induction of the HOG reporter gene 8xCRE-lacZ by glycosylation defects. Yeast strains of the indicated genotypes, carrying the reporter plasmid, were stimulated with (+) or without (−) 25 μg/ml tunicamycin for 8 h, before cell extracts were prepared for β-galactosidase assay. [KSS1] and [FUS3] in (B) indicate that the strain contains a single-copy plasmid carrying the KSS1 or FUS3 gene with its own promoter. (D) Activation of the Hog1 MAPK by glycosylation defects. Exponentially growing cells of the indicated genotypes were treated with (+) or without (−) 25 μg/ml tunicamycin for 2 h. Cell extracts were probed for phosphorylated Hog1 (P-Hog1) and total Hog1 by immunoblotting. (E) Induction of the FUS1-lacZ reporter gene by glycosylation defects. The indicated yeast mutant cells carrying the pFUS1-lacZ plasmid were grown exponentially, treated with tunicamycin for 8 h (+tun) or untreated (−tun), and assayed for β-galactosidase.
These results suggest that activated Kss1 inhibits the HOG MAPK pathway. If so, then osmotic activation of Hog1 MAPK might be also inhibited in tunicamycin-treated pmt4Δ cells. Indeed, after 2 h incubation with tunicamycin, osmotic activation of the Hog1 MAPK in pmt4Δ cells was significantly weaker than that in tunicamycin-untreated pmt4Δ cells, or in tunicamycin-treated PMT4+ cells (Supplementary Figure S6). The inhibition was not complete, probably because osmotic stress activated the Hog1 pathway much more intensely than glycosylation defects did. Thus, we conclude that activation of Kss1 by glycosylation defects suppresses Hog1 activation.
Mutual inhibition between the Hog1 MAPK and the Kss1 MAPK
To elucidate how activation of Kss1 inhibited the HOG pathway, we first examined the possibility that activated Kss1 induced an inhibitor of the HOG pathway. Thus, we disrupted STE12 and TEC1, which are involved in Kss1-dependent gene expression (Chou et al, 2006), and FUS1, whose expression is induced by activated Kss1 and/or Fus3. As shown in Figure 6C, however, disruption of these genes did not activate the HOG pathway by glycosylation defect. Thus, the mechanism by which Kss1 inhibits the HOG pathway is not under the control of the Ste12 or Tec1 transcription factors. Although Fus1 has been shown to bind and inhibit Sho1 (Nelson et al, 2004), it does not seem to have a role in the inhibition of the HOG pathway by Kss1, which is consistent with the fact that Sho1 was dispensable for the tunicamycin-dependent activation of the FG MAPK pathway (see Figure 2A).
We then examined another possibility that is suggested by the fact that yeast MAPKs are negatively regulated by PTPs. In particular, Ptp2 preferentially inactivates Hog1, whereas Ptp3 primarily inactivates Kss1 and Fus3 (Jacoby et al, 1997; Wurgler-Murphy et al, 1997; Zhan and Guan, 1999). Therefore, we tested whether either Ptp2 or Ptp3 might be responsible for inhibition of the HOG MAPK pathway by activated Kss1. As seen in Figure 6C, tunicamycin-treatment induced the HOG-specific 8xCRE-lacZ reporter in pmt4Δ ptp2Δ mutant cells, but not in pmt4Δ ptp3Δ mutant cells. The expression level of the 8xCRE-lacZ reporter in pmt4Δ ptp2Δ treated with tunicamycin was about 30% of the reporter expression induced by osmostress (0.4 M NaCl) (compare Figure 6C with Supplementary Figure S6B). Furthermore, on tunicamycin treatment, phosphorylation of Hog1 was seen in pmt4Δ ptp2Δ mutant cells, but not in pmt4Δ cells (Figure 6D). The time-course of Hog1 phosphorylation in pmt4Δ ptp2Δ cells is comparable to that of Kss1 phosphorylation in pmt4Δ cells (compare Figure 2C with Supplementary Figure S8). Hog1 phosphorylation was also induced in PMT4+ ptp2Δ host cells (Supplementary Figure S8). In PMT4+ host cells, tunicamycin activates the Kss1 pathway only marginally, but perhaps the signal was sufficient to phosphorylate Hog1 in the absence of Ptp2. Thus, Ptp2 is required to prevent the Hog1 MAPK from being activated by the glycosylation defects, although we cannot yet conclude that Ptp2 is the direct target of Kss1.
This finding suggested a possible explanation for the earlier observation that ptp2Δ mutation inhibited Kss1 activation by glycosylation defects, namely that perhaps activated Hog1 MAPK, in its turn, might inhibit the Kss1 MAPK. Indeed, when the HOG1 gene was disrupted in pmt4Δ ptp2Δ mutant cells, induction of FUS1-lacZ expression by glycosylation defects was restored (Figure 6E). In contrast, disruption of the PTP3 gene in pmt4Δ ptp2Δ mutant cells did not restore FUS1-lacZ expression, indicating that Ptp3 is not responsible for the suppression of Kss1 by Hog1. We tested the possibility that Hog1 directly inhibits Kss1 by phosphorylation, by mutating the only MAPK substrate motif in Kss1, namely Thr244-Pro245. Activation of the Kss1-T244A mutant, however, was also inhibited by Hog1 (data not shown). Thus, it is yet unclear how activated Hog1 inhibits Kss1 activation.
Integrating all results, we conclude that glycosylation defects activate the Ste11 MAPKKK, which can activate, if uninhibited, both the Ste7-Kss1 and the Pbs2-Hog1 kinase cascades. However, only one kinase or the other is stably activated because of the reciprocal inhibitory loop between Hog1 and Kss1. Signal flows in several key mutants on glycosylation defects are interpreted schematically in Figure 7.
Figure 7.
Schematic diagrams of signal flow in the Kss1 and Hog1 MAPK pathways in three yeast mutants: (A) pmt4Δ, (B) pmt4Δ ptp2Δ, and (C) pmt4Δ ptp2Δ hog1Δ. Black arrows and T-shaped bars indicate, respectively, activation and inhibition. Gray arrows and bars represent lack of activation or inhibition. Crosses (X) signify disrupted genes. The precise molecular mechanisms by which Kss1 inhibits Hog1, or Hog1 inhibits Kss1, are currently unknown.
Discussion
In the first part of this report, we extended earlier observations that general glycosylation defects activate the FG response (Lee and Elion, 1999; Cullen et al, 2000) by showing that inhibition of N-glycosylation and a specific type of O-glycosylation, effected by a combination of tunicamycin treatment and pmt4Δ mutation, activates the FG MAPK pathway. In particular, we have identified a critical role for Pmt4-mediated O-glycosylation of the membrane protein Msb2 in FG pathway activation.
Although there are still major gaps in our knowledge, the specific role of Msb2 in the FG pathway became clearer. It is apparent that there is a considerable mechanistic similarity among the osmostress activation of the Hog1 MAPK (the HOG response), the activation of the Kss1 MAPK by osmostress in the absence of Pbs2 or Hog1 (cross-talk), and the authentic FG response. Thus, to understand the role of Msb2 in the FG response, it would be helpful to recall the known role of Msb2 in the HOG response as well as the cross-talk activation of Kss1 by osmostress (Tatebayashi et al, 2007). The SHO1 branch of the HOG pathway is initiated by an interplay between the essential HMH domain and the inhibitory STR domain in the Msb2 molecule. The extracellular HMH domain (residues 960–1113) is juxtaposed to the glycosylated STR domain and is required for activation of the HOG MAPK pathway as well as of the FG MAPK pathway (see Figure 5B). It is possible that the positively regulatory HMH domain is normally masked by the O-glycosylated STR domain, and that underglycosylation, or deletion of a large segment, of the STR domain unmasks the HMH domain. An analysis by deletion scanning has shown that the entire region between 979 and 1117 is required for activation of the HOG pathway (Tatebayashi et al, 2007), as well as for the cross-talk activation of the FG pathway by osmostress (KT and HS, unpublished data). It is therefore reasonable to assume that the HMH domain serves a similar positive role in the FG response. If so, activated Msb2 may interact with Sho1 to initiate an intracellular signal flow (Tatebayashi et al, 2007). As indicated in Figure 2A, however, Sho1 is not required for the glycosylation defect-dependent activation of the FG MAPK pathway. This is not contradictory, however, as we have shown that Msb2 can activate the Hog1 MAPK (and also the Kss1 MAPK by cross-talk) through two different mechanisms, or modes (Tatebayashi et al, 2007). The mode 1 mechanism entails the interaction between Msb2 and the ectodomain of Sho1. The mode 2 mechanism of the HOG pathway, however, involves only the Sho1 cytoplasmic domain, which is needed to activate the Pbs2 MAPKK. As the FG response does not involve Pbs2 at all, activation of the Kss1 MAPK by the mode 2 mechanism can proceed in the complete absence of Sho1.
We might then speculate that the Msb2 STR domain serves as a dual-purpose sensor for environmental conditions. Under hyperosmotic conditions, the physico-chemical changes in the oligosaccharide gel structure might unmask the essential HMH domain, whereas under poor nutritional conditions, underglycosylation of STR unmasks the HMH domain.
It was recently proposed that the aspartyl protease Yps1 activates the FG MAPK pathway by cleaving the extracellular domain of Msb2. Proteolytic degradation of overexpressed Msb2, as revealed by shedding of the Msb2 ectodomain, is nearly completely blocked in yps1Δ mutant (Vadaie et al, 2008). The primary cleavage site was mapped within the residues 1045–1145, which was termed the cleavage domain (CD). It was reported without data that yps1Δ mutant was only partially defective in activating the FG MAPK pathway (Vadaie et al, 2008), which is perhaps consistent with our result that yps1Δ mutation does not inhibit the induction of FUS1-lacZ by glycosylation defects (Figure 2A). Deleting the CD abrogated the Yps1-dependent Msb2 degradation, and completely inhibited the FG response. However, because deleting the CD also affects the essential HMH domain, it does not provide evidence that Yps1-dependent cleavage itself is essential for the FG response. Clearly, further investigations are required to elucidate the individual roles of, and possible interrelationship between, the Yps1-dependent cleavage and glycosylation defects in activating the Kss1 MAPK cascade.
The Pmt4-dependent O-glycosylation of the pathogenic yeast C. albicans has been shown to affect hyphal morphology, which is closely related to the FG of S. cerevisiae. Hyphal morphogenesis of a homozygous pmt4 mutant is defective under aerobic conditions, but enhanced in embedded or hypoxic conditions (Prill et al, 2005). In live mouse model system, pmt4 mutants are markedly less virulent than PMT4 counterparts (Prill et al, 2005). Thus, Pmt4-dependent glycosylation serves a complex role in environment-dependent signalling that is critical for pathogenicity of C. albicans. It is hoped that our current results shed light on the complex role of the Pmt4-dependent O-glycosylation in this important pathogen.
In the second part of this report, we investigated the interaction between the FG and the HOG MAPK pathways. In the budding yeast, three functionally distinct MAPK signalling pathways share the same Ste11 MAPKKK: the mating pheromone pathway, the FG MAPK pathway, and the SHO1 branch of the osmoregulatory HOG MAPK pathway (Chen and Thorner, 2007). Nevertheless, each pathway is only activated by cognate stimuli.
Mating pheromones activate the Ste11 MAPKKK, but Ste11 thus activated does not lead to any activation of the Hog1 MAPK (Posas and Saito, 1997). It is well established that the HOG pathway is insulated from the pheromone stimulation, because the Ste11-Ste7-Fus3 MAPK cascade of the mating pathway is tethered by the scaffold protein Ste5, which also interacts with the mating-specific upstream components (Elion, 2001). Direct inhibition of the Hog1 MAPK by activated Fus3 may also contribute to the lack of Hog1 activation by the mating pheromones (McClean et al, 2007). On mating pheromone stimulation, Kss1 is activated only transiently, whereas Fus3 is activated for the duration of the mating response (Sabbagh et al, 2001). The mechanism by which activated Fus3 inhibits Kss1 is unknown, but an involvement of a phosphatase has been suggested (Chen and Thorner, 2007). Stimulation of the FG pathway through the Msb2 sensor does not entail any Fus3 activation, probably because Fus3 is sequestered by Ste5 binding. Indeed, when binding of Fus3 to Ste5 is abrogated by nondocking mutation in Ste5 (Ste5ND), Fus3 behaves like Kss1 (Hao et al, 2008). Thus, there is perhaps no need for Kss1 to directly inhibit Fus3.
The relationship between the HOG and the FG MAPK pathways appears to be more subtle. These MAPK pathways use basically the same signalling machinery that includes the putative sensor Msb2, the G-protein Cdc42, the PAK-like kinase Ste20, the adaptor protein Ste50 and the Ste11 MAPKKK (Chen and Thorner, 2007). Nevertheless, osmostress does not activate the FG-specific Kss1 MAPK. The Kss1 MAPK is directly or indirectly inhibited by activated Hog1, because osmostress does potently activate the Kss1 MAPK in hog1Δ or pbs2Δ mutant strains (O'Rourke and Herskowitz, 1998; Davenport et al, 1999).
It has been difficult, however, to examine the reverse situation, namely whether activated FG MAPK pathway interferes with the HOG pathway or not, because no suitable way to activate the FG pathway was available for such studies. We found that the HOG MAPK pathway was indeed inhibited when the FG MAPK pathway was activated by glycosylation defects. Furthermore, if activation of the Kss1 MAPK was prevented, for example, by ste7Δ, then there was a robust activation of the Hog1 MAPK by glycosylation defects. We also found that inhibition of the Hog1 MAPK was mediated by the Ptp2. It is possible that activated Kss1 affects expression or activity of Ptp2, but the alternative explanation that Kss1 and Ptp2 both inhibit Hog1 independently of each other has not been excluded. Further experiments are required to clarify the detailed mechanism of inhibition of Hog1 by Kss1.
An important finding from this study is that activation of the FG MAPK pathway and that of the HOG MAPK pathway are mutually exclusive. When either one of these MAPK pathways is activated, the second pathway is suppressed, thereby achieving bistability of the system. However, a tantalizing question remains; that is, how is the correct MAPK pathway chosen for activation by a specific stimulus to begin with?
A clue to a possible solution might be in the different kinetic properties of activating stimuli. Activation of the FG pathway by accumulation of underglycosylated Msb2 molecules is relatively weak and gradually increases in strength. In contrast, activation of the Sho1 branch of the HOG pathway by a hyperosmolarity is rapider and stronger in intensity (Tatebayashi et al, 2007). Perhaps, both the FG and the HOG MAPK pathways are initially activated to limited extents, but, depending on the intensity and duration of the stimulus, cross-inhibition in one or the other direction eventually dominates, resulting in activation of only one pathway.
Another clue to the problem is the observation that, even though Sho1 and Msb2 are involved in both the HOG and FG MAPK pathways, their participation in each pathway is not exactly identical. On the one hand, the Sho1 cytoplasmic SH3 domain, which binds the Pbs2 MAPKK, is essential for the HOG pathway, but is not required for the FG pathway (Maeda et al, 1995; Tatebayashi et al, 2007). On the other hand, the cytoplasmic region of Msb2, whose function is yet unknown, is essential for the FG pathway, but optional for the HOG pathway (Tatebayashi et al, 2007). It is possible that different stimuli might affect these cytoplasmic domains differently, so that the signal flow from activated Ste11 MAPKKK to its two potential targets, Pbs2 and Ste7, is properly steered. In this context, it will be interesting to investigate the potential role of Msb2 as a membrane anchorage protein specific to the FG MAPK pathway.
In conclusion, we showed the presence of a reciprocal inhibitory loop between the FG promoting Kss1 MAPK and the osmoregulatory Hog1 MAPK. This will serve as a basis for an integrated regulatory MAPK signalling network in yeast, as well as a model for similar networks in other organisms.
Materials and methods
Yeast strains
The yeast strains used, and their complete genotypes, are listed in Supplementary Tables S1 and S2, respectively.
Media and buffers
Yeast media YPD, YPGal, SC, SGal and CAD, and Buffers A and Z were defined earlier (Horie et al, 2008). Buffer N contains 50 mM Tris–HCl (pH 7.5), 15 mM EDTA, 15 mM EGTA, 2 mM dithiothreitol, 1% NP40, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 150 mM NaCl.
Plasmid vectors
The single-copy pRS41x series vectors and the multicopy pRS42x series vectors have been described (Sikorski and Boeke, 1991; Christianson et al, 1992). p414GAL1 (PGAL1 TRP1+ ARS) and p416GAL1 (PGAL1 URA3+ ARS) are galactose-inducible single-copy vectors (Mumberg et al, 1994). p424TEF (PTEF2 TRP1+ 2 μm) is a multicopy vector with the strong and constitutive TEF2 promoter (Mumberg et al, 1995). p426TEG1 (PTEF2-GST URA3+ 2 μm) is a multicopy vector that allows a constitutive expression of GST-fusion protein using the TEF2 promoter (Posas et al, 1996).
Expression plasmid constructs
The C-terminally GFP-tagged Opy2 constructs used in Figure 3B are based on the galactose-inducible single-copy vector p414GAL1. The Opy2 constructs used in Figure 3C are based on the single-copy vector pRS414 (TRP1+ ARS), and contains the OPY2 promoter. Opy2-ΔSR1 is Δ(2–22)/Ser27A, which eliminates all the Ser residues between the N terminus and Ser27. Opy2-ΔSR2 is Ser67A/Ser69A/Ser72A/Ser73A/Δ(75–84)/Ser86A/Ser91A, which eliminates all the Ser residues between Ser 67 and Ser 91 inclusive. Opy2-ΔC is Δ(184–360). pFP122 contains the full-length STE50 ORF fused to the C terminus of GST in the p426TEG1 vector (Posas et al, 1998). The HA-tagged Msb2 constructs used in Figure 4B–E (Msb2-FL HA and its ΔSTR1, ΔSTR2 and ΔSTR3 derivatives) are based on the galactose-inducible p416GAL1 vector, and contain two HA epitopes, one following amino acid position 48 and another at the C terminus. The Msb2 expression constructs used in Figure 5B are without epitope-tag, based on the single-copy vector pRS416 (URA3+ ARS), and expressed from the MSB2's own promoter. In the Msb2-HMH(Hkr1) mutant, the HMH domain of Msb2 (residues 961–1117) was replaced by the homologous HMH domain of Hkr1 (residues 1210–1427) (Tatebayashi et al, 2007). The Msb2 and Opy2 expression constructs used in Figure 5C are based on p414GAL1.
Reporter assays
Reporter assays using the FG reporter plasmid pFUS1-lacZ (pSB231) (CEN URA3+ FUS1-lacZ) and the HOG reporter plasmid pRS414-8xCRE-lacZ (CEN TRP1+ 8xCRE-lacZ), have been described (McCaffrey et al, 1987; Tatebayashi et al, 2003, 2006). All reporter assays were at least triplicated using independent cultures. Error bars represent standard deviations.
Microscopy
Microscopic images of yeast cells were captured using a Nikon TE2000-E inverted microscope equipped with a Roper Scientific CoolSnap HQ CCD camera.
Immunoblotting analyses
Immunoblotting analyses were carried out essentially as described earlier (Takekawa and Saito, 1998; Murakami et al, 2008). Buffer N was used to prepare cell extracts containing HA-tagged Msb2 constructs. Images were digitally captured by LAS-1000 Plus (Fujifilm) equipped with a CCD camera. The following antibodies were used as primary antibodies in immunoblotting: the anti-Hog1 goat antibody yC-20 (Santa Cruz), anti-GST monoclonal antibody (mAb) B-14 (Santa Cruz), anti-GFP mAb B-2, anti-HA mAb 12CA5 (Roche), anti-phospho-p38 antibody (Cell Signalling) for phosph-Hog1, and anti-phospho-p44/42 antibody (Cell Signaling) for phospho-Kss1, phospho-Fus3 and phospho-Mpk1.
In vivo binding assay
Exponentially growing cells in CARaf were adjusted to 2% galactose and cultured for an additional 4 h. Cell extracts were prepared in buffer A using glass beads, essentially as described earlier (Tatebayashi et al, 2003). A 200 μg aliquot of protein extract was incubated with 50 μl of glutathione-Sepharose beads for 3 h at 4°C. Beads were washed three times in buffer A, resuspended in SDS loading buffer, boiled and separated by SDS–PAGE. Proteins were detected by immunoblotting.
Other methods
Other methods including standard genetic procedures are as described earlier (Rose et al, 1990; Tatebayashi et al, 2003; Horie et al, 2008; Murakami et al, 2008).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr K Tanaka for yeast strains and the Kss1-T244A mutant plasmid, and Dr P O'Grady for critical reading of the paper. This work was supported in part by several grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (to HS and KT), a grant from the Salt Science Research Foundation (No. 0715) (to KT), and a grant from the Asahi Glass Foundation (to KT). HYY was supported by scholarships from the Interchange Association, Japan (IAJ) and the Hirose International Scholarship Foundation, and KY was supported by MEXT through the Program for Improvement of the Research Environment for Young Researchers.
Conflict of interest The authors declare that they have no conflict of interest.
References
- Andersson J, Simpson DM, Qi M, Wang Y, Elion EA (2004) Differential input by Ste5 scaffold and Msg5 phosphatase route a MAPK cascade to multiple outcomes. EMBO J 23: 2564–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsland-Marchesan E, Ariño J, Saito H, Sunnerhagen P, Posas F (2000) Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol Cell Biol 20: 3887–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla M, Cunningham KW (2003) Mitogen-activated protein kinase stimulation of Ca2+ signaling is required for survival of endoplasmic reticulum stress in yeast. Mol Biol Cell 14: 4296–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC (1993) An osmosensing signal transduction pathway in yeast. Science 259: 1760–1763 [DOI] [PubMed] [Google Scholar]
- Chen RE, Thorner J (2007) Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1773: 1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Lane S, Liu H (2006) Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol Cell Biol 26: 4794–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122 [DOI] [PubMed] [Google Scholar]
- Cohen TJ, Mallory MJ, Strich R, Yao T-P (2008) Hos2p/Set3p deacetylase complex signals secretory stress through the Mpk1p cell integrity pathway. Eukaryot Cell 7: 1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sabbagh W Jr, Graham E, Irick MM, van Olden EK, Neal C, Delrow J, Bardwell L, Sprague GF Jr (2004) A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev 18: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Schultz J, Horecka J, Stevenson BJ, Jigami Y, Sprague GF (2000) Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics 155: 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport KD, Williams KE, Ullmann BD, Gustin MC (1999) Activation of the Saccharomyces cerevisiae filamentation/invasion pathways by osmotic stress in high-osmolarity glycogen pathway mutants. Genetics 153: 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker M, Mrsa V, Hagen I, Deutzmann R, Strahl S, Tanner W (2003) O-mannosylation precedes and potentially controls the N-glycosylation of a yeast cell wall glycoprotein. EMBO Rep 4: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisman B, Alonso-Monge R, Román E, Arana D, Nombela C, Pla J (2006) The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell 5: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA (2001) The Ste5p scaffold. J Cell Sci 114: 3967–3978 [DOI] [PubMed] [Google Scholar]
- Escote X, Zapater M, Clotet J, Posas F (2004) Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nature Cell Biol 6: 997–1002 [DOI] [PubMed] [Google Scholar]
- Gentzsch M, Tanner W (1996) The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J 15: 5752–5759 [PMC free article] [PubMed] [Google Scholar]
- Gimeno CM, Ljungdahl PO, Styles CA, Fink GR (1992) Unipolar cell divisions in the yeast Saccharomyces lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090 [DOI] [PubMed] [Google Scholar]
- Girrbach V, Strahl S (2003) Members of the evolutionarily conserved PMT family of protein O-mannosyl transferases form distinct protein complexes among themselves. J Biol Chem 278: 12554–12562 [DOI] [PubMed] [Google Scholar]
- Hao N, Nayak S, Behar M, Shanks RH, Nagiec MJ, Errede B, Hasty J, Elston TC, Dohlman HG (2008) Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell 30: 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66: 300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Tatebayashi K, Yamada R, Saito H (2008) Phosphorylated Ssk1 prevents unphosphorylated Ssk1 from activating the Ssk2 MAP kinase kinase kinase in the yeast HOG osmoregulatory pathway. Mol Cell Biol 28: 5172–5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutzler J, Schmid M, Bernard T, Henrissat B, Strahl S (2007) Membrane association is a determinant for substrate recognition by PMT4 protein O-mannosyltransferases. Proc Natl Acad Sci USA 104: 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby T, Flanagan H, Faykin A, Seto AG, Mattison C, Ota I (1997) Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J Biol Chem 272: 17749–17755 [DOI] [PubMed] [Google Scholar]
- Lee BN, Elion EA (1999) The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc Natl Acad Sci USA 96: 12679–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Cutler NS, Heitman J (2000) Chracterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol Biol Cell 11: 183–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Fink GR (1997) Combinatorial control required for the specificity of yeast MAPK signaling. Science 275: 1314–1317 [DOI] [PubMed] [Google Scholar]
- Maeda T, Takekawa M, Saito H (1995) Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269: 554–558 [DOI] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369: 242–245 [DOI] [PubMed] [Google Scholar]
- McCaffrey G, Clay FJ, Kelsay K, Sprague GF (1987) Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol 7: 2680–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean MN, Mody A, Broach JR, Ramanathan S (2007) Cross-talk and decision making in MAP kinase pathways. Nat Genet 39: 409–414 [DOI] [PubMed] [Google Scholar]
- Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Mosch H-U, Roberts RL, Fink GR (1996) Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 93: 5352–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucl Acids Res 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic background. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Tatebayashi K, Saito H (2008) Two adjacent docking sites in the yeast Hog1 Mitogen-activated protein (MAP) kinase differentially interact with the Pbs2 MAP kinase kinase and the Ptp2 protein tyrosine phosphatase. Mol Cell Biol 28: 2481–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B, Parsons AB, Evangelista M, Schaefer K, Kennedy K, Ritchie S, Petryshen TL, Boone C (2004) Fus1p interacts with components of the Hog1p mitogen-activated protein kinase and Cdc42p morphogenesis signaling pathways to control cell fusion during yeast mating. Genetics 166: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Herskowitz I (1998) The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev 12: 2874–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Herskowitz I (2004) Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression anaylsis. Mol Biol Cell 15: 532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Saito H (1997) Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: Scaffold role of Pbs2p MAPKK. Science 276: 1702–1705 [DOI] [PubMed] [Google Scholar]
- Posas F, Witten EA, Saito H (1998) Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol Cell Biol 18: 5788–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H (1996) Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 ‘two-component' osmosensor. Cell 86: 865–875 [DOI] [PubMed] [Google Scholar]
- Prill SK-H, Klinkert B, Timpel C, Gale CA, Schröppel K, Ernst JF (2005) PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol Microbiol 55: 546–560 [DOI] [PubMed] [Google Scholar]
- Proszynski TJ, Simons K, Bagnat M (2004) O-glycosylation as a sorting determinant for cell surface delivery in yeast. Mol Biol Cell 15: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, Fink GR (1994) Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev 8: 2974–2985 [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P (1990) Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sabbagh W, Flatauer LJ, Bardwell AJ, Bardwell L (2001) Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol Cell 8: 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD (1991) In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol 194: 302–318 [DOI] [PubMed] [Google Scholar]
- Takekawa M, Saito H (1998) A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell 95: 521–530 [DOI] [PubMed] [Google Scholar]
- Tatebayashi K, Takekawa M, Saito H (2003) A docking site determining specificity of Pbs2 MAPKK for Ssk2/Ssk22 MAPKKKs in the yeast HOG pathway. EMBO J 22: 3624–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K, Tanaka K, Yang H-Y, Yamamoto K, Matsushita Y, Tomida T, Imai M, Saito H (2007) Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J 26: 3521–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K, Yamamoto K, Tanaka K, Tomida T, Maruoka T, Kasukawa E, Saito H (2006) Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J 25: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Reiser V, Ruis H, Ammerer G (2001) Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc Natl Acad Sci USA 98: 5625–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ (2008) Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol 181: 1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Jansen G, Zhang J, Thomas DY, Whiteway M (2006) Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev 20: 734–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Maeda T, Witten EA, Saito H (1997) Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol 17: 1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X-L, Guan K-L (1999) A specific protein–protein interaction accounts for the in vivo substrate selectivity of Ptp3 towards the Fus3 MAP kinase. Genes Dev 13: 2811–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information