EMBO J 28, 1442–1452 (2009); published online 20 May 2009
EMBO J 28, 1453–1465 (2009); published online 20 May 2009
The highly orchestrated movements of chromosomes during mitosis depend on the formation of stable connections between microtubules and kinetochores. How kinetochores generate these linkages to harness the forces produced by dynamic microtubule plus-ends remains unknown. Three recent studies make significant progress on this front, by identifying a third component of the kinetochore-associated Ska (spindle and kinetochore associated) complex and demonstrating that the complex is required to generate stable kinetochore–microtubule attachments during mitosis in human cells.
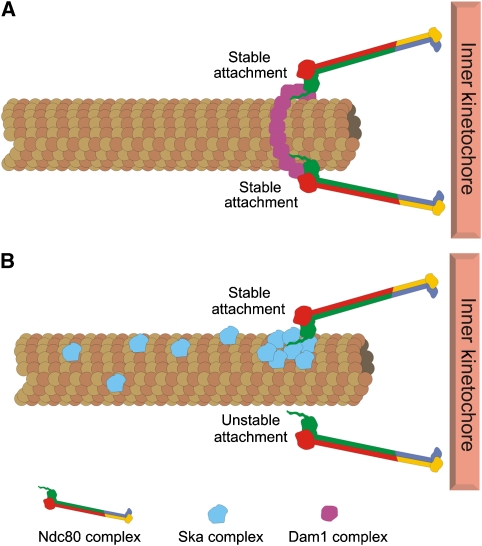
Chromosome alignment and segregation in mitosis requires strong yet flexible connections between spindle microtubules and kinetochores. Major puzzles are the molecular details of how kinetochores generate and maintain attachments to microtubule plus-ends that are continually gaining and losing tubulin dimers. In budding yeast, the Dam1 complex, which localizes along mitotic spindle microtubules, is required to form stable kinetochore–microtubule attachments (Jones et al, 2001). The Dam1 complex has been proposed to generate these attachments by partnering with the kinetochore-localized Ndc80 complex (Shang et al, 2003) (Figure 1). Dam1 complexes form rings and non-ring oligomers on microtubules in vitro, both of which are able to track with depolymerizing microtubule plus-ends (reviewed in Gardner and Odde, 2008), implying that these complexes may be competent to link microtubules to kinetochores in cells. Despite thorough searches, however, no Dam1 homologous have been identified in higher eukaryotes. The Ndc80 complex (which is present in all eukaryotes) may independently form these linkages, as this complex is essential for kinetochore–microtubule attachments in cells (reviewed in Ciferri et al, 2007) and similar to Dam1, can bind to and track with dynamic microtubule plus-ends (Powers et al, 2009). However, a host of additional proteins localize to the kinetochore–microtubule interface in mitotic cells, and it is likely that other components are needed to form the functional linkages required for chromosome bi-orientation. Three recent papers, two appearing in this issue of The EMBO Journal, illustrate this point: the Nigg, Buchholz and Cheeseman groups all identify a novel component of the Ska complex and demonstrate that the complex is required to form stable kinetochore–microtubule attachments in human cells (Gaitanos et al, 2009; Theis et al, 2009; Welburn et al, 2009).
Figure 1.
Models for kinetochore–microtubule attachment mediated through Dam1 and Ska complexes. (A) Dam1 ring complexes have been suggested to form kinetochore–microtubule interactions in budding yeast through Ndc80 complex interaction. (B) The Ska complex may have analogous functions to the Dam1 complex in higher eukaryotes. Ska components, bound along the lattice of spindle microtubules may serve to increase the stability of connections between Ndc80 complexes and microtubules, either directly or through an unknown linker protein.
The Ska complex was originally identified in a proteomic survey of the human mitotic spindle apparatus and included the proteins Ska1 and Ska2 (Hanisch et al, 2006), both of which localized along spindle microtubules and to kinetochores. Depletion of Ska1 or Ska2 resulted in a metaphase delay, but only minor defects in chromosome congression were observed. The Ndc80 complex remained bound to kinetochores after Ska1 or Ska2 depletion, and kinetochore–microtubule attachments were largely unperturbed. In the light of these findings, the Ska complex was not considered an essential factor in generating kinetochore–microtubule attachments. In this issue of The EMBO Journal, Gaitanos et al and Theis et al report the identification of a third component of the Ska1 complex, C13orf3, which they refer to as Ska3. Similar to Ska1 and Ska2, Ska3 localized to the outer kinetochore and along spindle microtubules, and its depletion from human cells resulted in mitotic arrest with mostly aligned chromosomes. Gaitanos et al further demonstrated that a more complete depletion of the Ska complex from HeLa cells, achieved by targeting multiple Ska complex components through RNAi, resulted in severe chromosome alignment defects, stemming from an inability to form stable kinetochore–microtubule attachments. As shown for depletion of Ska1 or Ska2 alone, depletion of the entire complex did not significantly reduce Ndc80 protein levels at kinetochores. These results suggest that the Ndc80 complex is not sufficient to generate kinetochore–microtubule attachments in eukaryotic cells, and the Ska complex joins the ranks of essential players in this process.
These findings are corroborated by Welburn et al (2009), who also report the identification of the third Ska complex component (referred to as Rama1 by this group). In line with the Theis et al and Gaitanos et al results, depletion of Rama1 from HeLa cells resulted in chromosome alignment defects that varied in severity, attributed again to variable Ska complex depletion. Biochemical characterization of the reconstituted Ska complex by this group suggested the presence of two copies of each subunit (Ska1/Ska2/Rama1) in the intact complex, which has a native molecular weight of ∼184 kDa. In the presence of polymerized microtubules, purified Ska complexes directly bound and formed oligomeric structures along the microtubule lattice. This binding was cooperative, with Ska1 serving as the major direct microtubule-binding component and Ska3 governing cooperativity. The authors went on to show that Ska complex-coated beads tracked depolymerizing microtubule plus-ends in a processive manner. On the basis of these findings, the authors suggest that Ska complex oligomers may directly link dynamic microtubule plus-ends to kinetochores in cells (Figure 1).
An intriguing possibility put forward by both Welburn et al and Gaitanos et al is that the Ska complex may be functionally homologous to the Dam1 complex (Figure 1). Findings from the three highlighted studies suggest that this may be the case. First, the Ska and Dam1 complexes have similar localization patterns in vivo: they both localize to spindle microtubules and to kinetochores, and for both complexes, kinetochore association is dependent on the presence of microtubules and the Ndc80 complex (Janke et al, 2002; Hanisch et al, 2006; Gaitanos et al, 2009; Welburn et al, 2009). Second, the Ska complex can bind microtubules and couple bead movement to depolymerizing microtubule ends (Welburn et al, 2009), similar to the Dam1 complex. Finally, Ska3 is phosphorylated in mitosis (Gaitanos et al, 2009; Theis et al, 2009), and phosphorylation is dependent, in part, on Aurora B kinase (Theis et al, 2009). Interestingly, Dam1 phosphorylation by the Ipl1 Aurora kinase in yeast is proposed to regulate kinetochore–microtubule binding, perhaps through the Ndc80 complex (Cheeseman et al, 2002; Shang et al, 2003). Given these similarities, it is possible that even though Ska1 components do not resemble Dam1 complex proteins at the primary sequence level, they may carry out similar functions in cells. Although it is exciting to speculate that the Ska complex may mediate kinetochore–microtubule attachment in concert with the Ndc80 complex, there is no evidence thus far of physical interaction between the two complexes (Gaitanos et al, 2009). Clearly, determining if and how the Ska complex interacts with the Ndc80 complex will be key in determining how functional kinetochore–microtubule attachments are made in mitosis.
In addition to providing evidence that the Ska complex functions to generate kinetochore–microtubule attachments, Theis et al suggest that it may have an additional function during mitosis. These authors demonstrated that chromosomes in cells depleted of Ska3 exhibited premature loss of sister chromatid cohesion, similar to cells depleted of the protein shugoshin, a known protector of centromeric cohesion (Watanabe, 2005). Here, depletion of shugoshin resulted in a reduction in Ska3 protein levels, suggesting a functional connection between the two proteins, even though a physical interaction between Ska3 and shugoshin could not be detected. Such a link between outer kinetochore proteins and shugoshin is not unprecedented, as depletion of Bub1 also results in the loss of sister chromatid cohesion, due to a defect in the centromere targeting of shugoshin (Boyarchuk et al, 2007). Further investigation will be needed to determine how the Ska complex and sister chromatid cohesion are functionally linked.
References
- Boyarchuk Y, Salic A, Dasso M, Arnaoutov A (2007) Bub1 is essential for assembly of the functional inner centromere. J Cell Biol 176: 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR III, Chan CS, Drubin DG, Barnes G (2002) Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163–172 [DOI] [PubMed] [Google Scholar]
- Ciferri C, Musacchio A, Petrovic A (2007) The Ndc80 complex: Hub of kinetochore activity. FEBS Lett 581: 2862–2869 [DOI] [PubMed] [Google Scholar]
- Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA (2009) Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J 28: 1442–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Odde DJ (2008) Dam1 complexes go it alone on disassembling microtubules. Nat Cell Biol 10: 379–381 [DOI] [PubMed] [Google Scholar]
- Hanisch A, Silljé HH, Nigg EA (2006) Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J 25: 5504–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortíz J, Tanaka TU, Lechner J, Schiebel E (2002) Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J 21: 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, He X, Giddings TH, Winey M (2001) Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc Natl Acad Sci USA 98: 13675–13680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL (2009) The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 136: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, Barnes G (2003) Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell 14: 3342–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis M, Slabicki M, Junqueira M, Paszkowski-Rogacz M, Sontheimer J, Kittler R, Heninger A-K, Glatter T, Kruusmaa K, Poser I, Hyman AA, Pisabarro MT, Gstaiger M, Aebersold R, Shevchenko A, Buchholz F (2009) Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division EMBO J 28: 1453–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y (2005) Shugoshin: guardian spirit at the centromere. Curr Opin Cell Biol 17: 590–595 [DOI] [PubMed] [Google Scholar]
- Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR III, Cheeseman IM (2009) The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell 16: 374–385 [DOI] [PMC free article] [PubMed] [Google Scholar]