Abstract
AMPA and kainate receptors mediate fast synaptic transmission. AMPA receptor ligand-binding domains form dimers, which are key functional units controlling ion-channel activation and desensitization. Dimer stability is inversely related to the rate and extent of desensitization. Kainate and AMPA receptors share common structural elements, but functional measurements suggest that subunit assembly and gating differs between these subtypes. To investigate this, we constructed a library of GluR6 kainate receptor mutants and directly measured changes in kainate receptor dimer stability by analytical ultracentrifugation, which, combined with electrophysiological experiments, revealed an inverse correlation between dimer stability and the rate of desensitization. We solved crystal structures for a series of five GluR6 mutants, to understand the molecular mechanisms for dimer stabilization. We demonstrate that the desensitized state of kainate receptors acts as a deep energy well offsetting the stabilizing effects of dimer interface mutants, and that the deactivation of kainate receptor responses is dominated by entry into desensitized states. Our results show how neurotransmitter receptors with similar structures and gating mechanisms can exhibit strikingly different functional properties.
Keywords: analytical ultracentrifugation, crystallography, desensitization, glutamate receptors, synaptic transmission
Introduction
Ionotropic glutamate receptors (iGluRs) mediate excitatory synaptic transmission by coupling the free energy of agonist binding to the opening and desensitization of a transmembrane ion channel (Gouaux, 2004; Mayer, 2006; Hansen et al, 2007). Central to the function of the 18 iGluR genes that form the AMPA, kainate and NMDA receptor families of ligand-gated ion channels is a structural unit formed by a dimer assembly of the ligand-binding domains (Armstrong and Gouaux, 2000; Mayer et al, 2001; Furukawa et al, 2005; Nanao et al, 2005). This dimer undergoes conformational changes driven by agonist-binding energy, producing transitions between the resting, conducting, and desensitized states of the ion channel. The rates of transitions between these states, which are finely tuned at individual excitatory synapses to allow information processing in the brain over a wide spectral bandwidth (Geiger et al, 1995), are controlled by intermolecular contacts in the dimer assembly (Sun et al, 2002; Horning and Mayer, 2004). An understanding of this process, which is much less well understood in kainate and NMDA receptors, is a key requirement for resolving the role of individual iGluR subtypes in synaptic plasticity.
Following release of neurotransmitter by the presynaptic cell, receptors in the postsynaptic membrane bind glutamate in a two-domain clamshell-shaped structure (LBD) formed by S1 and S2 polypeptide segments (Stern-Bach et al, 1994; Kuusinen et al, 1995). Structural studies on AMPA receptor GluR2 S1S2 have shown that activation occurs on glutamate binding through rotation of domain 2 and closure of the clamshell. A critical inter-subunit interface, formed between individual protomers in a dimer pair through the rear surface of domain 1, allows domain closure to place a torque on the membrane-embedded ion channel, causing it to open (Armstrong and Gouaux, 2000). A major advance in our understanding of AMPA receptor gating was the serendipitous discovery, by construction of chimeric receptors, that the GluR3 L485Y mutation blocks desensitization (Stern-Bach et al, 1998). Structural analysis for the equivalent GluR2 L483Y mutant revealed that the tyrosine side chain stabilizes dimer assembly by forming a cation–π interaction with a lysine side chain in the dimer partner subunit (Sun et al, 2002). This finding has profoundly influenced our understanding of AMPA receptor gating, and led to a model in which the domain 1 interface is under strain in the glutamate-bound state. During desensitization the interface ruptures, allowing the channel to close even with the agonist bound (Armstrong et al, 2006).
Paradoxically, the genes for GluR5–GluR7 subtype kainate receptors encode an aromatic amino acid at the position equivalent to GluR2 L483Y, but desensitize rapidly and nearly completely in response to glutamate (Schiffer et al, 1997; Swanson et al, 1997). Functional analysis of the LBD dimer interface and recent crystal structures of kainate receptor dimer assemblies, which bear striking structural similarity to their AMPA receptor counterparts, have not resolved this puzzle. It is to be noted that numerous differences exist between the two receptor families in their sensitivities to allosteric ligands such as cyclothiazide and concanavalin A (Partin et al, 1993; Wong and Mayer, 1993; Yamada and Tang, 1993), and to external ions that modulate desensitization (Bowie, 2002; Bowie and Lange, 2002; Plested et al, 2008). An approach taken by several groups has been to rebuild the dimer interface of kainate receptors by introducing AMPA receptor residues, in an attempt to recreate the non-desensitizing GluR2 L483Y phenotype (Fleck et al, 2003; Zhang et al, 2006; Weston et al, 2006b). However, with the exception of disulphide cross-links, which perturb the dimer interface, render glutamate a partial agonist, and disrupt trafficking to the plasma membrane, all attempts to generate non-desensitizing kainate receptor mutants through rational design have ultimately fallen short, having greater effects on the rate rather than the extent of desensitization (Swanson et al, 1997; Stern-Bach et al, 1998; Fleck et al, 2003; Yelshansky et al, 2004; Priel et al, 2006; Zhang et al, 2006; Weston et al, 2006b).
Here we explore the reason for this and test two plausible mechanisms that have important consequences for understanding kainate receptor function. The first is that the kainate receptor desensitized state acts as a deep energy well, competing with strengthening of the dimer assembly obtained by introduction of AMPA receptor residues. To address this, we measured rates of onset and recovery from desensitization using electrophysiological assays for a family of dimer interface mutants. An alternative mechanism would be that the dimer interface of kainate receptors is intrinsically less stable than that of AMPA receptors. These two possibilities represent extremes, which are not mutually exclusive. As the driving force for desensitization arises from strain imposed on LBD dimers by the ion channel, this complicates comparisons between AMPA and kainate receptor mutants based solely on electrophysiological studies. To avoid this, we directly measure the Kd for dimer formation by isolated LBDs for GluR6 in the absence of the ion channel using analytical ultracentrifugation (AUC). To test whether mutants in the dimer interface sense different local environments in the two receptors, we solved a library of five high-resolution crystal structures for GluR6 mutants, together with the structure for the wild-type GluR6 dimer crystallized under identical conditions. Our results indicate that even following extensive engineering, the stability of kainate receptor dimers is at most one half of that of their AMPA counterparts, and that even if it were possible to generate dimers as stable as those for GluR2 L483Y, these would be insufficient to block kainate receptor desensitization because of the deep energy well of the desensitized state. We propose that this intrinsic difference in dimer stability contributed to the evolution of subtype-specific allosteric regulators, for example, the recently described Na+- and Cl−-binding sites in kainate receptors (Plested and Mayer, 2007; Plested et al, 2008).
Results
Desensitization is regulated by clusters of dimer interface residues
Analysis of dimer crystal structures in the protein data bank for GluR2, GluR5, and GluR6 reveals that, although they share a similar dimer structure, conserved clusters of residues that contribute to the dimer interface differ in the two receptor families (Figure 1). Structure-based sequence alignments reveal that in addition to earlier identified amino-acid differences in α-helices D, F, and J, which flank the GluR2 L483Y tyrosine mutant that blocks AMPA receptor desensitization (Figure 1A), helix B harbors conserved Lys and His side chains in AMPA receptors, which are absent in kainate receptors. Mutation of these residues to their AMPA counterparts would be expected to alter the electrostatic environment in the dimer interface, and, together with earlier reported mutants of residues in helices D, F, and J, facilitate intermolecular interactions with the native (GluR6 Y490) tyrosine side chain in kainate receptors (Zhang et al, 2006; Weston et al, 2006b). To examine the influence of these residues on dimer stability, we created a library of 10 GluR6 mutants, which, using different combinations, progressively switch the sequence of seven amino acids in helices B, D, F, and J to the sequence found in GluR2. The mutants were T441K, I442H, K494E, K665R, I749L, Q753K, and E757Q; for brevity we use dashes to indicate the wild-type GluR6 sequence for mutants in which a limited number of amino acids was changed; for example, the GluR6 K665R mutant is designated - - -R- - -. These mutants were then assayed for changes in gating using full-length GluR6 (Figure 2), and for changes in dimer stability using analytical ultracentrifugation for the isolated LBDs (Figure 3).
Figure 1.
Conserved clusters of residues differ in the dimer interface of AMPA and kainate receptors. (A) Amino-acid sequence alignment for AMPA and kainate receptor gene families; dimer interface residues exchanged between GluR2 and GluR6 are indicated by Δ; additional residues that play key roles in the effects of individual mutations are indicated by *; cylinders above the alignment indicate location of α-helices B, D, F, and J in GluR6 crystal structures; + indicates the L/Y switch in helix D. (B) Ribbon diagram for wild-type GluR6 shows the location of the critical Tyr490 side chain, surrounded by residues exchanged between GluR2 and GluR6, drawn as gold- and cyan-colored CPK spheres for the pair of subunits in a dimer assembly (a stereo view is shown in Supplementary Figure 1).
Figure 2.
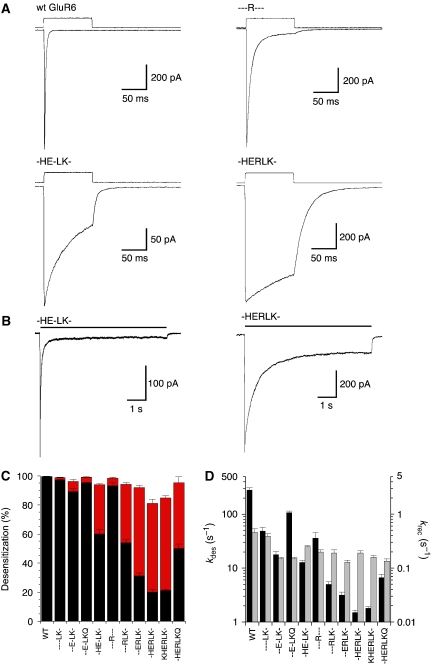
Kinetic analysis for a library of GluR6 dimer interface mutants. (A) Responses of outside-out patches to 100-ms applications of 10 mM glutamate are shown for wild type and three mutants. The rate of desensitization (kdes) is fastest for wild type, ∼200-fold slower for -HERLK-, whereas - - -R- - - and -HE-LK- produce intermediate kinetics. (B) Responses to 7-s applications of glutamate show the extent of desensitization for -HE-LK- and -HERLK-, highlighting the impact of the K665R mutation in the -HE-LK- background. (C) The extent of desensitization measured at 100 ms (black) and 7 s (red). (D) Rate of onset of desensitization (black bar) and recovery (grey bar), for wild-type GluR6 and the library of 10 dimer interface mutants; error bars indicate mean±s.e.m.
Figure 3.
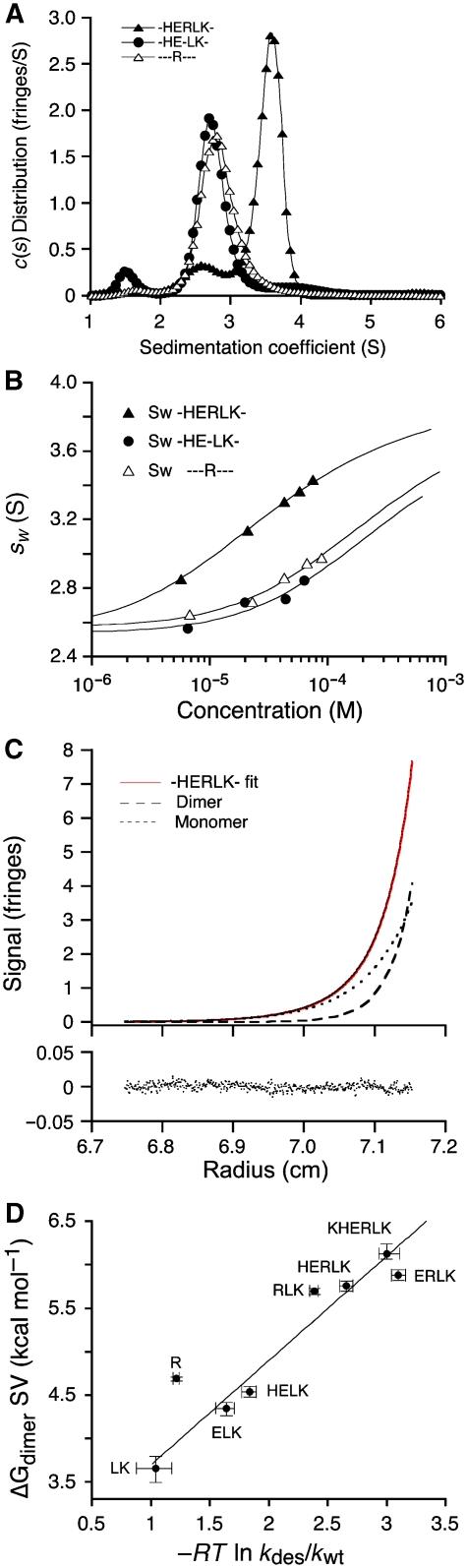
Dimer formation for GluR6 LBDs measured by analytical ultracentrifugation. (A) c(s) distributions from sedimentation velocity (SV) runs for -HERLK- (1.7 mg ml−1), -HE-LK- (1.9 mg ml−1) and - - -R- - - (2 mg ml−1); concentrations reported in parentheses are derived from peak integration (see also Supplementary Figure 2). Peak positions reflect sedimentation of rapidly interconverting monomer–dimer systems. The ∼3.6 S peak for -HERLK- reflects stronger association of this mutant compared with the 2.8 and 2.9 S peaks for -HE-LK- and - - -R- - -, respectively. (B) Dependence of the weight-average sedimentation coefficient sw on loading concentration for -HERLK-, -HE-LK- and - - -R- - - (symbols), and best fits with a binding isotherm for a monomer–dimer equilibrium (solid lines) with Kd values of 41.2 μM (1σ confidence interval 37–45 μM), 416 μM (1σ 370–460), 321 μM (1σ 307–333) for -HERLK-, -HE-LK- and - - -R- - -, respectively. (C) Representative sedimentation equilibrium profile for -HERLK- derived from a global analysis of data at a range of loading concentrations and rotor speeds, using a monomer–dimer model with a Kd of 102 μM (1σ 85–135). The black line indicates the model used to fit the data, red dots indicate experimental measurements, and dashed lines represent the best-fit populations of monomer and dimer; residuals of the fit are shown below the graph. (D) The free energy change for dimerization ΔGdimer plotted against the free energy change for onset of desensitization relative to wild type (−RT ln kdes/kwt) for each mutant (symbol and error bars). The dotted line shows the best fit from total least-squares optimization, suggesting a linear relationship in which stabilization of the dimer assembly slows the rate of onset of desensitization. A full colour version of this figure is available at The EMBO Journal online.
Successively introducing combinations of AMPA receptor residues into the wild-type GluR6 produced progressively slower rates of onset of desensitization (kdes) with rank-order responses to 10 mM glutamate of the wild-type GluR6 284±27 s−1, > - - - -LK- 48.9±8.5 s−1, > - - -R- - - 36.2±9.4 s−1, >- -E-LK- 17.8±2.6 s−1, >-HE-LK- 12.8±1.2 s−1, > - -ERLK- 3.2±0.4 s−1, >-HERLK- 1.5±0.1 s−1 ≈ KHERLK- 1.8±0.1 s−1. The most effective combination of mutations, -HERLK-, slowed kdes 190-fold compared with wild-type GluR6, producing a strong attenuation of desensitization measured 100 ms after the start of glutamate application, from 99.4±0.2% for wild-type GluR6 to 20±2% for -HERLK- (Figure 2A); however, when glutamate application was increased to 7 s, the extent of desensitization for -HERLK- at equilibrium increased to 81±3% (Figure 2B). This is in striking contrast to the nearly complete block of desensitization produced by GluR2 L483Y. Our analysis suggests that in -HERLK- the mutations K665R near the base of helix F and the pair I749L/Q753K in helix J act independently of each other, as reversion of each site to the wild-type sequence led to a similar 8- to 10-fold increase in the extent (Figure 2C) and rate of desensitization (Figure 2D). The mutants tested had only small effects on the rate of recovery from desensitization (krec), measured using twin pulse applications of glutamate (Figure 2D, Table I), and krec varied <3-fold from the value of 0.46±0.08 s−1 for the wild-type GluR6. In contrast to the stabilizing effects observed in the above series, E773Q at the base of helix J was clearly destabilizing, as indicated by a six-fold faster kdes for - -E-LKQ 108±6.9 s−1 compared with - -E-LK- 17.8±2.6 s−1, and the five-fold faster kdes for -HERLKQ 6.7±1.0 s−1 compared with -HERLK- 1.5±0.2 s−1 (Figure 2D, Table I).
Table 1.
Energetics of dimer dissociation and functional analysis of desensitization
| Mutant | Kd-SE (μM) | KdSV (μM) | ΔGdd SE (kcal mol−1) | ΔGdd SV (kcal mol−1) | kdes (s−1) | kdeact (s−1) | % des | krec (s−1) |
|---|---|---|---|---|---|---|---|---|
| KHERLK | 54.0 (52–58) | 24.8 (22–30) | 5.4 | 6.13 | 1.8±0.1 (11) | 51.2±4.2 (9) | 84.8±1.5 (11) | 0.16±0.01 (6) |
| -HERLK- | 102 (85–135) | 41.2 (37–45) | 5.11 | 5.88 | 1.5±0.1 (9) | 45.0±3.8 (7) | 81.0±3.0 (12) | 0.19±0.02 (5) |
| - -ERLK- | 90.5 (82–99) | 50.9 (46–56) | 5.13 | 5.76 | 3.2±0.4 (8) | 57.0±8.3 (5) | 92.0±1.5 (8) | 0.13±0.01 (4) |
| - - -RLK- | 97.4 (89–105) | 56.2 (53–60) | 5.09 | 5.70 | 5.0±0.6 (7) | 66.5±4.9 (7) | 94.2±1.0 (7) | 0.19±0.03 (6) |
| - - -R- - - | 995 (826–1250) | 321 (307–333) | 3.81 | 4.69 | 36.2±9.4 (6) | 88.7±11 (6) | 98.4±0.6 (6) | 0.20±0.02 (5) |
| -HE-LK- | 843 (814–882) | 416 (370–460) | 3.90 | 4.54 | 12.8±1.2 (6) | 152±22 (5) | 94.0±0.7 (7) | 0.25±0.01 (6) |
| - -E-LK- | 816 (776–861) | 569 (507–664) | 3.92 | 4.35 | 17.8±2.6 (6) | 297±48 (6) | 96.2±1.5 (6) | 0.15±0.01 (5) |
| - - - -LK- | 896 (741–1120) | 1880 (1480–2460) | 3.87 | 3.66 | 48.9±8.5 (5) | 397 ±87 (5) | 98.8±0.3 (5) | 0.39±0.05 (5) |
| -HERLKQ | 341 (320–363) | 233 (219–250) | 4.40 | 4.87 | 6.70±1.0 (10) | 59.1±6.6 (7) | 95.2±4.0 (10) | 0.14±0.01 (5) |
| - -E-LKQ | >6000 | ND | <2.6 | — | 108±6.9 (11) | 391±34 (9) | 99.0±0. 3 (11) | 0.22±0.02 (6) |
| Wt GluR6 | >8000 | ND | <2.8 | — | 284±27 (7) | 506±43 (6) | 99.2±0.4 (7) | 0.46±0.08 (6) |
| Kd-SE and Kd-SV are dimer dissociation constants determined from sedimentation equilibrium and velocity experiments, respectively; error estimates are reported as 1σ confidence intervals in parentheses and calculated as described in Materials and methods; ND signifies not determined. Kd's for - -E-LKQ and wild-type GluR6 are lower limits determined as described previously. ΔGdd SE and SV are the corresponding ΔG of dimer dissociation, calculated as ΔG=−RT ln Kd (R=1.987 cal(mol K)−1); T=277 or 298 K for SE and SV experiments, respectively). kdes, kdeact, % des, and krec are the rates of desensitization, deactivation, extent of desensitization, and rate of recovery from desensitization as measured by fast-solution exchanges; measurements are the mean±s.e.m., with the number of observations in parentheses. Kd's for - -E-LKQ and wild-type GluR6 are lower limits determined as described previously. KHERLK and Q mutations correspond to the following changes in full-length GluR6, respectively: T441K, I442H, K494E, K665R, I749L, Q753K, and E757Q. | ||||||||
Dimer mutants promote co-assembly of GluR6 ligand-binding domains
Sedimentation velocity (SV) and sedimentation equilibrium (SE) analytical ultracentrifugation experiments were performed for the isolated LBDs of a library of eight GluR6 mutants, designed on the basis of electrophysiological results. As reported earlier, for both wild-type GluR6 and - -E-LKQ, dimer formation is too weak to be determined by either approach (Weston et al, 2006b), and error analysis indicates a lower limit of 6–8 mM for the dimer Kd. In contrast, the dimerization of each of the eight mutants was sufficiently strong to be detected by both SV and SE, yielding Kd values with very similar rank order as predicted from electrophysiological analysis (Table I).
Examples of the SV approach, showing the c(s) sedimentation coefficient distribution at 20°C for -HERLK-, -HE-LK-, and - - -R- - -, at similar concentrations (Figure 3A), exhibit the expected behaviour for a monomer–dimer assembly in equilibrium (Schuck, 2000), with rapid interconversion on the sedimentation time-scale (Supplementary Figure 2). It is evident from visual inspection of overlaid c(s) distributions that -HERLK stabilizes dimer formation to a greater extent, judged by an increase in the sedimentation coefficient, relative to - - -R- - - and -HE-LK-, which behave similarly. The isotherm of weight-average sedimentation coefficients (sw) as a function of protein concentration followed the mass action law for a monomer–dimer equilibrium, yielding best-fit Kd values of 41.2, 321, and 416 μM for -HERLK-, - - -R- - -, and -HE-LK-, respectively (Figure 3B). In parallel, SE experiments at 4°C were conducted and global non-linear least squares fits of these datasets to a monomer–dimer model allowed independent determination of the dimerization Kd. The results from both analyses were in good agreement (Table I), with Kd values for SV ∼2-fold lower than those measured by SE (see Supplementary data). When the free energy of dimer dissociation is plotted against the rate constant for onset of desensitization measured by electrophysiological analysis (Figure 3D), an almost linear relationship emerges, indicating that dimer stability is a major determinant of desensitization for kainate receptors.
Dimer mutant crystal structures
To define the molecular mechanisms underlying changes in dimer stability and attenuation of desensitization by these mutants, a structural library of GluR6 dimers in their active conformation was determined for the glutamate complexes of -HERLK-, -HE-LK-, - - -RLK-, - - -R- - -, and wild type crystallized in the presence of a physiological concentration of NaCl, with data for Bragg spacings ranging from 1.5 to 1.32 Å (Table II). A key finding in all of the structures was that Na+ and Cl− were present in the allosteric ion-binding sites, as found earlier for GluR5 dimer crystal structures with the partial agonist kainate (Plested and Mayer, 2007; Plested et al, 2008). As these ions do not bind to AMPA receptors, this indicates that, despite the introduction of up to six GluR2 residues into GluR6, the dimer interface maintains unique properties characteristic of kainate receptors. The dimer interface buries a similar solvent accessible surface of 1164, 1160, 1151, 1121, and 1204 A2 per subunit for -HERLK-, -HE-LK-, - - -RLK-, - - -R- - -, and wild-type GluR6, respectively, and superposition of the dimer assemblies using domain 1 (D1) Cα coordinates gave r.m.s. deviations of <0.2 Å, indicating that despite profound differences in their electrophysiological and biophysical properties, these mutants have nearly identical structures to wild-type GluR6. However, because the crystals diffract to high resolution, we were able to detect local conformational differences in each structure that explain the mutational effects on dimer stability (Figure 4). The tyrosine side chains on the exposed face of helix D move upwards by 1.2 and 1.3 Å in the pair of subunits in -HERLK- compared with wild-type GluR6; this movement occurs without a change in the intermolecular distance between helices D and J measured using the Cα coordinates of Tyr490 and Ile749, and instead results from both a rotation of the aromatic ring and a change in the χ1 side-chain dihedral angle. This movement has a profound effect on cation–π interactions made by Tyr490, which is a key determinant of the high dimer stability in the GluR2 L483Y mutant (Sun et al, 2002). To analyze the strength of cation–π interactions (Gallivan and Dougherty, 1999), we used the program CAPTURE. In wild-type GluR6, there are intra-subunit cation–π interactions between Lys494 and Tyr490 in the same subunit (−1.31 and −2.34 kcal mol−1 for subunits A and B, respectively), which are replaced by new intermolecular cation–π interactions with the mutant Lys753 side chain in -HERLK- (−2.04 kcal mol−1) and -HE-LK- (−2.19 kcal mol−1).
Table 2.
Data collection and refinement statistics
| Data set | WTR6 | - - -R- - - | - - -RLK- | -HE-LK- | -HERLK- | -HERLKQ |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | P21 | P21 | P21 | P21 | P21 | P21 |
| Unit cell a, b, c (Å) | 51.2, 114.2, 52.4 | 51.0, 113.5, 52.0 | 51.1, 113.5, 52.2 | 51.2, 114.2, 52.4 | 50.9, 113.7, 51.9 | 51.1, 113.8, 52.1 |
| a=γ, β | 90, 115.3 | 90, 115.2 | 90, 115.1 | 90, 115.3 | 90, 115.3 | 90, 115.3 |
| Number per a.u. | 2 | 2 | 2 | 2 | 2 | 2 |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.97930 |
| Resolution (Å)a | 30–1.38 (1.43) | 30–1.3 (1.35) | 30–1.5 (1.55) | 30–1.37 (1.42) | 35–1.32 (1.37) | 40–1.2 (1.24) |
| Unique observations | 102 842 | 129 531 | 83 975 | 109 022 | 124 266 | 149 021 |
| Mean redundancyb | 4.9 (4.8) | 6.3 (6.0) | 2.8 (2.4) | 7.3 (7.2) | 2.8 (2.6) | 4.6 (2.8) |
| Completeness (%)b | 94.2 (84.0) | 99.1 (91.4) | 97.9 (94.0) | 94.3 (89.3) | 94.7 (88.6) | 98.6 (86.5) |
| Rmergeb,c | 0.043 (0.315) | 0.039 (0.309) | 0.053 (0.347) | 0.038 (0.241) | 0.044 (0.369) | 0.064 (0.239) |
| I/σ(I)b | 15.0 (2.4) | 30.6 (3.4) | 17.6 (2.8) | 18.0 (2.9) | 24.0 (4.1) | 22.5 (3.4) |
| Refinement | ||||||
| Resolution (Å) | 28.71–1.38 | 29.26–1.30 | 29.53–1.50 | 28.69–1.37 | 34.5–1.32 | 35.9–1.24 |
| Protein atoms (AC)d | 4276 (223) | 4302 (227) | 4275 (271) | 4364 (275) | 4331 (246) | 4680 (560) |
| Glutamate atoms | 20 | 20 | 20 | 20 | 20 | 20 |
| Ions/Cl/Na atoms | 1/4 | 3/2 | 2/3 | 1/3 | 2/4 | 5/2 |
| Water atoms | 581 | 665 | 721 | 694 | 618 | 724 |
| Rwork/Rfree (%)e | 15.21/17.49 | 14.63/16.76 | 14.74/17.64 | 14.93/17.60 | 15.61/17.89 | 14.59/16.50 |
| r.m.s. deviations | ||||||
| Bond lengths (Å) | 0.012 | 0.016 | 0.010 | 0.010 | 0.010 | 0.017 |
| Bond angles° | 1.18 | 1.39 | 1.13 | 1.14 | 1.11 | 1.39 |
| Mean B-values (Å2) | ||||||
| Protein overall | 20.37 | 14.94 | 15.58 | 18.37 | 17.37 | 15.62 |
| MC/SCf | 17.08/23.5 | 12.46/17.30 | 12.85/18.17 | 15.45/21.11 | 14.89/19.74 | 13.01/17.88 |
| Glutamate | 12.65 | 7.13 | 7.45 | 9.50 | 8.91 | 7.61 |
| Cl/Na | 15.2/25.3 | 16.8/15.1 | 11.9/19.2 | 12.9/23.7 | 16.7/29.3 | 18.35/16.6 |
| Water | 32.3 | 26.6 | 28.4 | 31.2 | 31.2 | 27.6 |
| Ramachandran %g | 98.0/0 | 98.2/0 | 98.19/0 | 98.22/0 | 98.0/0 | 98.2/0 |
| aValues in parentheses indicate the low-resolution limit for the highest-resolution shell of data. | ||||||
| bValues in parentheses indicate statistics for the highest-resolution shell of data. | ||||||
| cRmerge==(Σ∣II–〈II〉∣)/ΣI∣II∣, where 〈II〉 is the mean II over symmetry-equivalent reflections. | ||||||
| dAlternate conformations. | ||||||
| eRwork=(Σ∣∣Fo∣–∣Fc∣∣)/Σ∣Fo∣, where Fo and Fc denote observed and calculated structure factors, respectively; 5% of the reflections were set aside for the calculation of the Rfree value. | ||||||
| fMain chain/side chain. | ||||||
| gPreferred/disallowed conformations. | ||||||
Figure 4.
High-resolution crystal structures for GluR6 LBD mutants. (A) Side chains drawn in stick representation are shown for wild-type GluR6, - - -R- - -, - - -RLK-, -HE-LK- and -HERLK- following least-squares superposition of dimer assemblies using D1 Cα coordinates; a stereo view with CPK spheres for -HERLK- is shown in Supplementary Figure 1. (B) σA-weighted 2mFo−DFc electron density maps contoured at 1σ for amino-acid side chains surrounding the native Tyr490 residue; for visualization side chains have been rotated from their orientation in the dimers. (C) Interactions between Tyr490 on one subunit with I749L and Q753K on helix J of the dimer partner for wild-type GluR6 (grey) and -HERLK- (gold). The van der Waals radii of Ile749 for wild-type GluR6 show a steric clash (top panel) with those for Tyr490 in the -HERLK- structure, which is eradicated on mutation to a Leu (bottom panel). (D) Interdomain interactions between helices B and J on one subunit and helices D and F on the other for HERLK and wild-type GluR6. Van der Waals interactions (distance <4 Å) are represented by a solid line connecting partner amino acids; hydrogen bonds by arrows pointing in the direction of the hydrogen bond acceptor; electrostatic (cation–π and salt-bridge) interactions by a thick line; water-mediated interactions with a dashed line; grey and gold indicate bonds for wild-type GluR6 and -HERLK-, respectively.
Electron density maps were of high quality for Tyr490 in all structures, whereas for Q753K, side-chain electron density, assessed by real space correlation coefficients and by visual inspection of omit maps, was best modelled by alternative conformations in several constructs (Figure 4B and Supplementary Figure 3). In -HERLK-, the replacement of Ile442 by the large positively charged imidazole ring of histidine biases the conformation of Lys753, increasing the probability for making a cation–π interaction with Tyr490. Indeed, in the library of constructs with the Q753K mutation, the side chain was best ordered in -HERLK- (Figure 4B). Estimation of the strength of cation–π interactions is highly sensitive to both the distance and orientation of the Lys side chain, and this likely explains why energetically significant interactions were observed in only one subunit of the dimer pair for -HERLK- and -HE-LK-, and were absent in both subunits for - - -RLK-. Superposition of the -HERLK-, - - -RLK-, -HE-LK-, and wild-type GluR6 structures reveals that replacement of Ile749 with a leucine, which has an unbranched side chain, leads to removal of a steric clash between the Ile749 Cβ atom in wild-type GluR6 and the conformation Tyr490 adopts in -HERLK- and related crystal structures (Figure 4C). This switch, which contributes to formation of a Leu749/Lys753 sandwich with Tyr490, is part of a complex network of contacts that regulate dimer affinity in the kainate receptor constructs analyzed in our study (Figure 4D).
The K665R mutant forms a novel intermolecular salt bridge
Substitution of the native Lys665 side chain on helix F, with the arginine found in GluR2 (Priel et al, 2006; Zhang et al, 2006), has a profound impact on desensitization (Figure 2) and dimer stability measured by AUC (Figure 3). In crystal structures of the - - -R- - -, - - -RLK-, and -HERLK- mutants, electron density for the Arg side chain was unambiguous and revealed formation of an inter-subunit salt bridge with Glu756 at the base of helix J in the dimer partner subunit (Figure 5A). To form this intermolecular contact, the χ2 dihedral angle of the arginine side chain rotates by 110° in - - -R- - -, - - -RLK-, and -HERLK- compared with the conformation of the lysine side chain found in wild-type GluR6 and -HE-LK- (Figure 4A), and the χ3 dihedral angle of the Glu756 side chain rotates by 55°. Electron density for the native lysine side chain was well ordered for the wild-type GluR6 dimer but surprisingly, because of the change in the χ2 angle, Lys665 does not make a salt bridge with Glu756, and instead projects into a region filled by a network of water molecules in the K665R mutant structures, forming a water mediated hydrogen bond with Asp462 in the same subunit (Figure 5B). As expected for non-interacting sites, both the K665R salt bridge with Glu756, and the Leu749/Lys753 sandwich observed in -HERLK- are faithfully recapitulated in the - - -R- - - and -HE-LK- structures, respectively. It is noteworthy that K665R is on helix F in D2, distinguishing it from the rest of the dimer interface mutations, which reside on helices in D1.
Figure 5.
The K665R mutation forms an intermolecular salt bridge linking helix F with helix J. (A) Dimer assembly for -HERLK- showing side chains for K665R, Glu756 and surrounding water molecules with σA-weighted 2mFo−DFc electron density maps at 1.5σ; (B) dimer assembly for wild-type GluR6 illustrating formation of an intramolecular contact between Lys665 and Asp462 in the same subunit; note the 55° difference in χ3 for Glu756. Water molecules W1 and W2 are conserved in the two structures, but the native Lys665 side-chain projects into the second network of water molecules present in -HERLK-.
The rate of desensitization for wild-type GluR2, 126 s−1, is almost four-fold faster than for GluR6 K665R, suggesting that in AMPA receptors the native arginine side chain is unlikely to contribute to dimer stabilization. Consistent with this, compared with the GluR6 K665R mutant structures, analysis of GluR2 structures in the PDB reveals 128° and 106° flips, in opposite directions, about the side-chain χ1 and χ2 dihedral angles for residues equivalent to Arg665 and Glu756, respectively, increasing the distance between the Cz and Cδ atoms by 6 Å (Supplementary Figure 4). As a result of these large movements, in GluR2, Arg 665 is poised to form an intramolecular salt bridge with Asp661 located on same face of helix F, replacing the intermolecular salt bridge found in kainate receptor mutants. This competing intramolecular salt bridge cannot form in kainate receptors because of replacement of Glu657 in helix F by a threonine side chain in GluR5–GluR7 (Figure 1).
The base of helix J is destabilized by the E757Q mutant
The GluR6 E757Q mutant was designed based on the rationale that a glutamate at position 757 may provide a potential binding site for an alternative conformation of Lys753, preventing it from forming a cation–π interaction with Tyr 490. However, in earlier work the - -E-LKQ mutant desensitized with nearly wild-type kinetics (Weston et al, 2006b), whereas the - -E-LK- mutant showed an 11-fold slowing (Zhang et al, 2006), suggesting unexpectedly that the E757Q is destabilizing. In the present experiments, we establish that in both - -E-LK- and -HERLK- backgrounds, the E757Q mutation increased the extent and rate of onset of desensitization (Figure 6A and Table I), and performed AUC experiments to determine the effect on dimer stability. Although we had found earlier that - -E-LKQ fails to dimerize in solution (Weston et al, 2006b), dimer formation by - -E-LK- was easily measured in SV experiments, which gave a Kd of 569 μM. A similar disruption of dimer formation was observed for the -HERLKQ mutant, Kd 233 μM measured by SV, compared with the value of 41 μM for -HERLK- (Figure 6B). To investigate the underlying mechanism for this, -HERLKQ was crystallized under the same conditions used for other members of the structural library, with data to 1.2 Å (Table II). Differences between the structure of -HERLKQ and that of -HERLK- map to the C terminus of helix J in the vicinity of the E757Q mutation. Following superposition of D1 coordinates for both structures, the RMSD for Cα positions was only 0.1 Å, but increased to 0.6 Å for the region from Gln755 to Lys759. This RMSD increase results from a distortion in the last helical turn of helix J, which produces a 5° rotation and 1 Å translation of the base of helix J into the dimer interface (Figure 6C). An omit map of this region reveals strong electron density for both backbone and side-chain alternative conformations for Glu756, E757Q, and Gly758 in -HERLKQ (Figure 6D), indicating that the E757Q mutation produces a loss of conformational stability in this region, thus disrupting formation of intermolecular bonds between helix J and the partner subunit that stabilize dimerization. This destabilization also occurs in the wild-type GluR6 background, for which the E757Q mutant increased the rate of desensitization to 325±29 s−1 (n=6). Calculation of ΔG values from desensitization rate constants (Figure 3D) gave changes of 0.88, 0.83, and 0.91 kcal mol−1 for the effect of the E757Q mutation in the wild-type, - -E-LK-, and -HERLK- backgrounds, indicating that the effect is independent of that of the stabilizing mutations.
Figure 6.
The E757Q mutation destabilizes dimer assembly. (A) Responses to 100-ms applications of 10 mM glutamate for -HERLK- and -HERLKQ illustrating the five-fold faster rate of onset of desensitization due to E757Q substitution. (B) Sedimentation equilibrium scans for -HERLKQ at 1 mg ml−1 and 24 000 r.p.m. analyzed identically as for -HERLK- (Figure 3C); the dimerization Kd decreases ∼6-fold to 628 μM. (C) Base of helix J tilts by 5° in -HERLKQ; dimer assemblies for -HERLKQ (red) and -HERLK- (gold) were superimposed by least-squares using D1 Cα coordinates; the solid line shows a Cα trace illustrating that movement is limited to the base of helix J. (D) Residues at the base of helix J and in the linker leading to helix K are disordered in -HERLKQ. An Fo−Fc omit electron density map contoured at 3.5σ was calculated with residues Glu756–Lys759 omitted from the Fc calculation; stick representation for these residues show main chain alternative conformations for Glu756, E757Q and Gly758, and side-chain alternative conformations for Glu756, E757Q and Lys759.
Disruption of D1–D2 interdomain contacts further attenuates desensitization
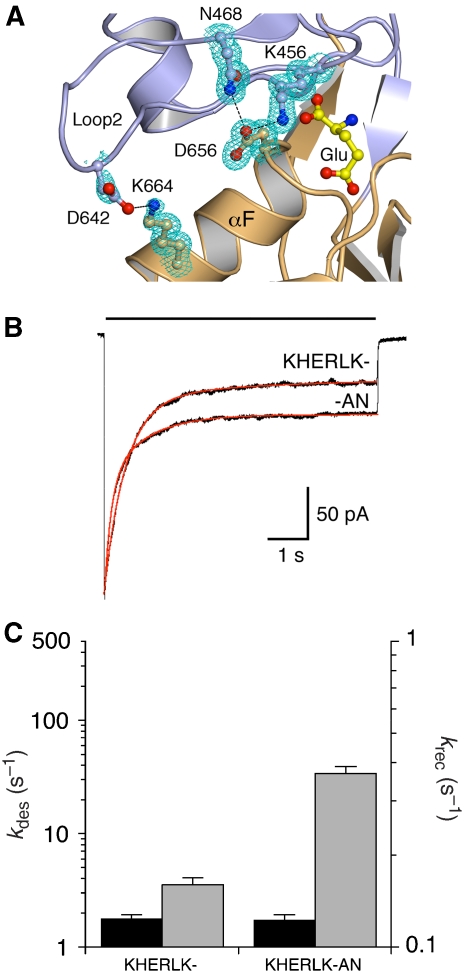
Does the desensitized state of kainate receptors act as a deep energy well that counteracts the effect of mutations that stabilize the dimer interface? To test this, we examined the effect of two mutations that reduce desensitized state stability (Weston et al, 2006a), D462A at the tip of loop2 in D1, and D656N near the N terminus of helix F in D2, referred to here as AN. The mechanism of action of such mutations, which speed the rate of recovery from desensitization for GluR6 by disrupting intramolecular contacts between domains 1 and 2 that stabilize the closed-cleft conformation of the LBD, is well established, and has been studied also in AMPA receptors (Robert et al, 2005; Zhang et al, 2008). The D462A and D656N mutants disrupt cross-domain salt-bridge contacts to Lys664 (D2) and Lys456 (D1), respectively, accelerating krec by 6- to 1.5-fold (Weston et al, 2006a). We confirmed that these interdomain interactions are preserved across all the dimer mutant structures (Figure 7A), and then tested the effect of the AN mutant on the extent of desensitization in the KHERLK- background, with the prediction that the AN mutation should increase steady-state currents by producing faster recovery from desensitization; both effects were observed. The equilibrium current increased 2.6-fold (Figure 7B), and krec increased 2.7-fold with no change in the rate of onset of desensitization (Figure 7C).
Figure 7.
Destabilization of ligand binding reduces desensitization. (A) Single subunit of -HERLK- showing glutamate bound in a cleft between D1 (blue) and D2 (orange). Interdomain salt bridge between Lys456 (D1) and Asp656 (D2) and also between Lys664 (D1) and Asp462 (D2) shown with σA-weighted 2mFo DFc electron density at 1σ. (B) Responses to 7-s applications of glutamate reveal an increase in steady-state response for KHERLK-AN compared with KHERLK-. (C) Bar plots showing the effect of K456A/D656N on the rate of desensitization (black) and recovery (grey) in KHERLK-.
Disruption of desensitization greatly slows deactivation
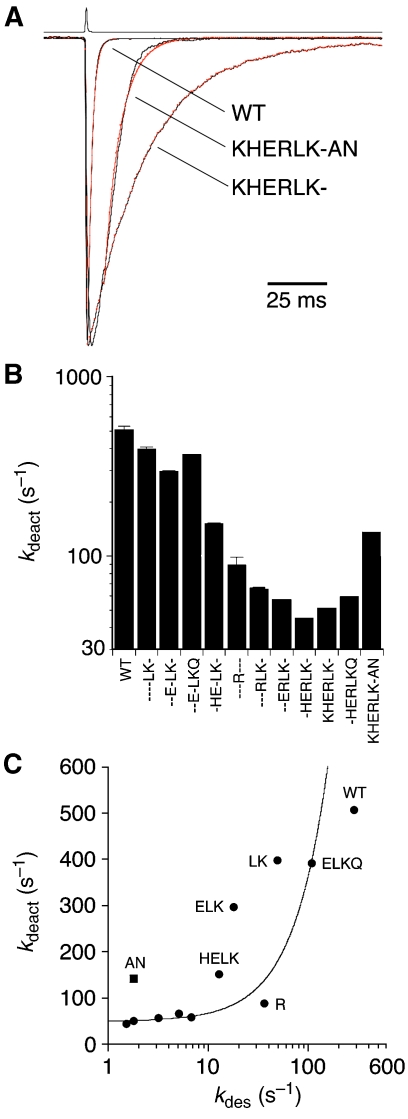
When glutamate is applied for brief durations to mimic the rapidly decaying concentration transient occurring during synaptic transmission, the rate of decay, termed deactivation (kdeac), is determined by a complex molecular sequence of events including bursts of channel opening and closing, unbinding of agonist, but also entry into desensitized states (Raman and Trussell, 1995). When we measured kdeac in response to 1-ms glutamate applications for wild-type GluR6 and our library of constructs, we observed a profound slowing of deactivation for mutants that attenuate desensitization (Figure 8). The KHERLK- mutant produced a 10-fold slowing of deactivation compared with responses for wild-type GluR6 (kdeac 51±4.2 versus 506±43 s−1). For KHERLK-AN, the construct with the least desensitization at equilibrium, kdeac was 142±11 s−1, three-fold faster than the parent construct, consistent with destabilization of the agonist-bound complex.
Figure 8.
Deactivation is greatly slowed in weakly desensitizing GluR6 mutants (A) The rate of deactivation for responses to 1-ms applications of glutamate is much slower for KHERLK- than for wild-type GluR6, 51 and 506 s−1, respectively, but increases for KHERLK-AN to 142 s−1. (B) Bar plots showing the deactivation rate for the library of dimer interface mutants. (C) The rate of deactivation saturates for mutants with stable dimer interfaces; data points for individual mutants are labelled except those for - - -RLK-, - -ERLK-, -HERLK-, KHERLK-, and -HERLKQ which cluster at 50 s−1; the data point for -KHERLK-AN lies above the trend for mutants with similar slow rates of desensitization.
The rates of deactivation varied 11-fold for the library of constructs studied (Figure 8B), but the rates of desensitization for the same constructs varied 190-fold. A plot of kdeact versus kdes reveals that in mutants with the slowest desensitization the rate of deactivation approaches an asymptote of 50 s−1 (Figure 8C). Responses for the AN mutant show faster deactivation, with no change in kdes, underscoring the distinct molecular processes controlling glutamate binding and dimer stability. We propose that the intrinsic rate for deactivation of wild-type GluR6 responses to glutamate is close to 50 s−1, and that the rapid onset of desensitization masks the true time course of deactivation. Thus, as a fortuitous consequence of blocking desensitization, we were also able to more accurately measure deactivation resulting from channel closing and unbinding of glutamate.
Discussion
The extreme functional diversity of excitatory synaptic currents masks the fact that AMPA, kainate, and NMDA receptors share a conserved structural fold. Although it has been proposed that AMPA and kainate receptors have different gating mechanisms (Bowie, 2002; Bowie and Lange, 2002), subsequent work suggests this might not be so (Zhang et al, 2006). Here we show that for kainate receptors there is an inverse correlation between the rate of onset of desensitization and stability of dimers formed by their LBDs, establishing that gating mechanisms and functional assembly of AMPA and kainate receptors as dimers of dimers are indeed conserved. Despite this, the generation of a non-desensitizing kainate receptor phenotype akin to GluR2 L483Y continues to be a formidable experimental challenge. Our results suggest that this is because of both a substantially more stable desensitized state in GluR6, as well as intrinsic differences in LBD dimer stability between the two receptors.
The molecular determinants of desensitization include (1) intermolecular interactions within the LBD domain that stabilize the D1 dimer interface; (2) intramolecular interactions between domains D1 and D2 that stabilize the agonist-bound closed cleft conformation; and (3) regions outside of the ligand-binding domain (Yelshansky et al, 2004). Direct measurement of LBD dimer stability in solution by AUC reveals that GluR6 -HERLK-, which rebuilds the local environment flanking the Tyr side chain in the non desensitizing GluR2 L483Y mutant, accounts for only 50% of the free energy of dimerization (5 versus 9.5 kcal mol−1). Notably, these cumulative mutations in the HERLK dimer interface act through stabilization of the active dimer assembly rather than by destabilization of the desensitized state since krec changes negligibly in stark contrast to an ∼200-fold decrease in kdes compared with wild-type GluR6. Second, the much greater stability of the desensitized state for kainate versus AMPA receptors, for which krec is >400-fold faster (Horning and Mayer, 2004), suggests that it acts as an energy well offsetting the stabilizing effects of dimer interface mutations. Thus, despite the striking 200-fold decrease in kdes observed for GluR6 -HERLK-, desensitization still reaches 81% at equilibrium because of the extreme stability of the desensitized state.
Although there is little knowledge of a bona fide structure of an iGluR desensitized state which would require structures of both the LBD and pore regions, its stability is proposed to arise from interactions holding the S1S2-binding domain in a closed-cleft conformation, together with interactions between juxta-membrane and transmembrane domains (Armstrong et al, 2006). In fact, the deep energy well of the kainate receptor desensitized state has been shown to result in part from unique interdomain contacts in the LBD closed-cleft conformation, absent in AMPA receptors, which contribute to higher stability on the glutamate-bound complex (Weston et al, 2006a). Consistent with this, removing some of these contacts in KHERLK-AN produces a further 2.5-fold attenuation in the extent of desensitization at equilibrium. Thus, it is evident that in kainate receptors, where the long agonist dwell time limits both kdeact and krec compared to AMPA receptors (Weston et al, 2006a), D1 dimer interface engineering alone is not sufficient to create a non-desensitizing receptor.
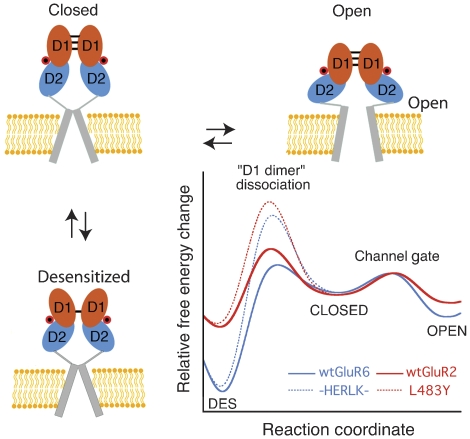
Illustrated in Figure 9 is a model that emerges from our data for AMPA and kainate receptor activation and desensitization, processes linked through the dimer interface. For both wild-type receptors, the energy barrier for desensitization is higher than for activation and therefore the receptor activates faster than it desensitizes, but as the desensitized state is more stable than the activated receptor, most of the receptors become desensitized during a prolonged incubation with agonist (see Table I). According to our model, relative to AMPA receptors, kainate receptors decrease the free energy of the open state, deepen the energy well for the desensitized state, and decrease the barrier for entry into the desensitized state. The net result is that to overcome the energetic sink of the GluR6 desensitized state many more compensatory interactions are required to stabilize the active-state dimer versus in GluR2; using ΔG=−RT ln(kdes/krec) and for simplicity considering only the influence of the ∼400-fold slower value of krec for kainate receptors gives 3.6 kcal mol−1 for the extra ‘compensatory' energy required. In both receptor subtypes the stability of the active dimer assembly is generated by a complex network of intermolecular and solvent-mediated contacts that are distributed over an area of ∼1000 Å2. In principle, it should be possible to identify additional sites for which mutations could be introduced to achieve a more stable GluR6 dimer, but the complex and unpredictable nature of interactions across the dimer interface revealed by our crystallographic experiments highlight how difficult it is to improve dimer packing by rational design. This is illustrated by the K665R mutant where the arginine side chain adopts a strikingly different conformation compared to the AMPA receptor donor, forming an inter-subunit salt bridge absent in AMPA receptors (Supplementary Figure 3).
Figure 9.
Conserved model for kainate and AMPA receptor gating. State diagram showing how agonist binding energy is used to open the ion channel (activation) or to rearrange the dimer interface (desensitization). D1 and D2 of the LBD domain are shown in cyan and orange, respectively, whereas the transmembrane segments comprising the ion-pore are shown as a grey cylinder. Each subunit binds agonist (red circle) and undergoes transitions between agonist-bound closed, open, and desensitized states. Hypothetical plots are constructed for an AMPA and kainate receptor comparing the free-energy changes that occur during gating. Estimates for ΔGc->o, the ΔG of the closed to open transition, ΔGdes, the ΔG of desensitization, and ΔGdd, the activation energy barrier to rupture the D1 dimer, were estimated using −RT ln 1/kdeact, −RT ln kdes/krec, and −RT ln kdes, respectively. Approximate values for kdes, krec, and kdeact used for wt GluR2 are 126, 102, and 175 s−1, respectively, and kdes for L483Y is 6 s−1 (Sun et al, 2002; Horning and Mayer, 2004); for wt GluR6 and HERLK values used are as reported in Table I. Note that kdeact measured for L483Y is ∼4-fold faster than HERLK.
Other examples of how small sequence differences within iGluR LBDs function to diversify receptor properties are well established. Alternative splicing of AMPA receptor flip and flop exons in helix J alters desensitization kinetics (Penn et al, 2008), as does the Arg to Gly switch encoded by mRNA editing of nucleic acids preceding the flip and flop exons (Sommer et al, 1990; Penn et al, 2008). Another example is the Met/Lys switch preceding helix J that renders AMPA receptors insensitive to cation modulation (Wong et al, 2007; Plested et al, 2008). Together with the results reported here, a picture emerges in which distributed small changes in sequence can be exploited to evolve diverse functional properties within a degenerate fold. We argue that nature has explored these regions of sequence space and chosen many of the possible solutions to building a two-domain clamshell structure, generating the discrete properties of AMPA and kainate receptors.
The presence of multimeric assemblies in a large number of channels suggests that dynamic oligomerization of LBDs may prove to be a general mechanism underlying ligand gating. In several cases, oligomeric conversions occur on the cytoplasmic side of the membrane, where binding of intracellular ligands to LBD structures acts as gating switches. Regulators of K+ conductance (RCK) domains, for example, ubiquitous at the intracellular carboxy terminus of many Ca2+-activated K+ channels, are α–β proteins with a Rossman-fold similar to bacterial periplasmic proteins and iGluR LBDs. The crystal structure of the MthK K+ channel from an archaebacterium shows how eight RCK domains form a gating ring leading to channel activation, where Ca2+ binds in the base of a cleft between RCK domains near a flexible hinge (Jiang et al, 2001, 2002), much like what we see in iGluR LBDs. Furthermore, recent functional and biochemical analyses suggest that the four functional states of MthK (closed, open, desensitized, and inactivated) correspond to distinct oligomeric states of its RCK domains (Kuo et al, 2007). Another reported example is in hyperpolarization-activated cation (HCN) channels, where intracellular cAMP binding to the C-terminal LBD promotes a transition from a dimer-of-dimers into a four-fold symmetric gating ring important for channel activation (Zhou et al, 2004). Rearrangements in LBD oligomerization in a diverse family of cation channels, inclusive of iGluRs, thus appears to provide a simple solution to a seemingly complicated function: driving a molecular machine through the states of its gating cycle.
Materials and methods
Molecular biology and protein purification
GluR6 point mutations were made in various combinations by overlap PCR and the amplified region confirmed by sequencing. Wild-type and mutant GluR6 S1S2 ligand-binding domain proteins were overexpressed in Escherichia coli and purified using methods described before (Mayer, 2005).
Physiology
Wild-type GluR6(Q) and mutant glutamate receptors were expressed in HEK-293 cells for outside-out patch recording with rapid solution exchange as described earlier (Weston et al, 2006b). Solution exchange time (20–80% to peak), estimated from open tip responses at the completion of a recording, ranged from 0.1 to 0.4 ms. Patch pipettes had a resistance of 3–5 MΩ and were filled with solution containing (in mM) 150 CSF, 20 HEPES, 10 NaCl, and 10 EGTA, pH 7.3. Extracellular medium contained (in mM) 140 NaCl, 2.4 KCl, 10 HEPES, 10 glucose, 4 MgCl2, and 4 CaCl2, pH 7.3, 305 mOsm. Currents were filtered at 2 kHz and digitized at 10 kHz. Further details are described in Supplementary data.
Analytical ultracentrifugation
Sedimentation velocity and equilibrium experiments were performed in a Beckman/Coulter XLI analytical ultracentrifuge using absorbance and Rayleigh interference optics. An additional protein purification step with an XK 16/60 Superdex 200 gel-filtration column in buffer containing (in mM) 10 HEPES pH 7.4, 165 NaCl, 1 EDTA, and 2 L-glutamic acid was used before experiments. SV experiments were conducted at 50 000 r.p.m. and 20°C using interference detection and double-sector cells loaded at 4, 3, 2, 1, 0.3, and 0.1 mg ml−1 typically. For SE experiments, data were collected at 4°C at 1, 0.3 and 0.1 mg ml−1 and 12 000, 18 000, and 24 000 r.p.m. by both absorbance detection at 280 and 250 nm and interference detection using sapphire windows and ‘aged' cell components. SV and SE data were analyzed using the SEDFIT and SEDPHAT programs (Schuck, 2000, 2003); further details of analysis methods and error statistics are provided in the Supplementary data.
Crystallography
Crystals of -HERLK- were grown in hanging drops at 8–12 mg ml−1 in buffer containing (in mM) 150 NaCl, 5 L-glutamic acid, 1 EDTA, and 10 HEPES, pH 7.4, mixed at a 1:1 volume ratio with reservoir solution containing 12–15% isopropanol, 15–18% PEG 4 K, and 100 mM sodium citrate pH 5.6. Microseeding techniques were used to obtain diffraction quality crystals for the other constructs. Crystals were cryoprotected by serial transfers to mother liquor with progressively higher concentrations of PEG 4000 (30% final concentration) and flash frozen in liquid nitrogen. Diffraction data were collected at the APS ID22 beamline using a MAR300 CCD detector and processed using HKL2000 (Otwinowski and Minor, 2001). The HERLK- structure was solved by molecular replacement with Phaser (McCoy et al, 2007), using a wild-type GluR6 monomer (PDB 1S7Y) with alanine substituted for the mutated residues as a search model. The other structures were solved by difference Fourier techniques. Model building was done using COOT (Emsley and Cowtan, 2004) and refinement using REFMAC5 (Winn et al, 2001) and PHENIX (Adams et al, 2002). Further details of crystallographic methods are provided in the Supplementary data.
Protein data bank accession codes
Atomic coordinates and structure factors have been deposited with the PDB; accession codes are 3G3F, 3G3G, 3G3H, 3G3I, 3G3J, 3G3K.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Information
Acknowledgments
We thank Carla Glasser for preparing cDNAs; Andrea Balbo for technical assistance with AUC experiments; the NINDS DNA sequencing facility; Drs S Heinemann and P Seeburg for the gift of wild-type cDNAs; and Dr Andrew Plested for discussion. Synchrotron diffraction data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory. Use of APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This work was supported by the intramural research programs of NICHD, NIH, DHHS (MLM) and NIBIB, NIH, DHHS (PS); RO1 NS 050655 (CR); and the Helen Hay Whitney Foundation (CC).
Author contributions CC performed biochemistry and structural biology; MCW and CR performed electrophysiological experiments; AUC was performed by CC and PS; MLM assisted with data collection, structure solution, and analysis.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E (2000) Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28: 165–181 [DOI] [PubMed] [Google Scholar]
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E (2006) Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell 127: 85–97 [DOI] [PubMed] [Google Scholar]
- Bowie D (2002) External anions and cations distinguish between AMPA and kainate receptor gating mechanisms. J Physiol 539: 725–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D, Lange GD (2002) Functional stoichiometry of glutamate receptor desensitization. J Neurosci 22: 3392–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fleck MW, Cornell E, Mah SJ (2003) Amino-acid residues involved in glutamate receptor 6 kainate receptor gating and desensitization. J Neurosci 23: 1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E (2005) Subunit arrangement and function in NMDA receptors. Nature 438: 185–192 [DOI] [PubMed] [Google Scholar]
- Gallivan JP, Dougherty DA (1999) Cation–pi interactions in structural biology. Proc Natl Acad Sci USA 96: 9459–9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger JRP, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H (1995) Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principle neurons and interneurons in rat CNS. Neuron 15: 193–204 [DOI] [PubMed] [Google Scholar]
- Gouaux E (2004) Structure and function of AMPA receptors. J Physiol 554: 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Yuan H, Traynelis SF (2007) Structural aspects of AMPA receptor activation, desensitization and deactivation. Curr Opin Neurobiol 17: 281–288 [DOI] [PubMed] [Google Scholar]
- Horning MS, Mayer ML (2004) Regulation of AMPA receptor gating by ligand binding core dimers. Neuron 41: 379–388 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R (2002) Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417: 515–522 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Pico A, Cadene M, Chait BT, MacKinnon R (2001) Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel. Neuron 29: 593–601 [DOI] [PubMed] [Google Scholar]
- Kuo MM, Baker KA, Wong L, Choe S (2007) Dynamic oligomeric conversions of the cytoplasmic RCK domains mediate MthK potassium channel activity. Proc Natl Acad Sci USA 104: 2151–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusinen A, Arvola M, Keinanen K (1995) Molecular dissection of the agonist binding site of an AMPA receptor. Embo J 14: 6327–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML (2005) Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron 45: 539–552 [DOI] [PubMed] [Google Scholar]
- Mayer ML (2006) Glutamate receptors at atomic resolution. Nature 440: 456–462 [DOI] [PubMed] [Google Scholar]
- Mayer ML, Olson R, Gouaux E (2001) Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state. J Mol Biol 311: 815–836 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanao MH, Green T, Stern-Bach Y, Heinemann SF, Choe S (2005) Structure of the kainate receptor subunit GluR6 agonist-binding domain complexed with domoic acid. Proc Natl Acad Sci USA 102: 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (2001) Denzo and Scalepack. International Tables for Crystallography 1F: 226–235 [Google Scholar]
- Partin KM, Patneau DK, Winters CA, Mayer ML, Buonanno A (1993) Selective modulation of desensitization at AMPA versus kainate receptors by cyclothiazide and concanavlin A. Neuron 11: 1069–1082 [DOI] [PubMed] [Google Scholar]
- Penn AC, Williams SR, Greger IH (2008) Gating motions underlie AMPA receptor secretion from the endoplasmic reticulum. EMBO J 27: 3056–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plested AJ, Mayer ML (2007) Structure and mechanism of kainate receptor modulation by anions. Neuron 53: 829–841 [DOI] [PubMed] [Google Scholar]
- Plested AJ, Vijayan R, Biggin PC, Mayer ML (2008) Molecular basis of kainate receptor modulation by sodium. Neuron 58: 720–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel A, Selak S, Lerma J, Stern-Bach Y (2006) Block of kainate receptor desensitization uncovers a key trafficking checkpoint. Neuron 52: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Raman IM, Trussell LO (1995) The mechanism of alpha-amino-3-hydroxy-5-methyl-4-isoxazoleproprionate receptor desensitization after removal of glutamate. Biophys J 68: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A, Armstrong N, Gouaux JE, Howe JR (2005) AMPA receptor binding cleft mutations that alter affinity, efficacy, and recovery from desensitization. J Neurosci 25: 3752–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer HH, Swanson GT, Heinemann SF (1997) Rat GluR7 and a carboxy-terminal splice variant, GluR7b, are functional kainate receptor subunits with a low sensitivity to glutamate. Neuron 19: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Schuck P (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78: 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P (2003) On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem 320: 104–124 [DOI] [PubMed] [Google Scholar]
- Sommer B, Keinänen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Köhler M, Takagi T, Sakmann B, Seeburg PH (1990) Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science 249: 1580–1585 [DOI] [PubMed] [Google Scholar]
- Stern-Bach Y, Bettler B, Hartley M, Sheppard PO, O'Hara PJ, Heinemann SF (1994) Agonist-selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid binding proteins. Neuron 13: 1345–1357 [DOI] [PubMed] [Google Scholar]
- Stern-Bach Y, Russo S, Neuman M, Rosenmund C (1998) A point mutation in the glutamate binding site blocks desensitization of AMPA receptors. Neuron 21: 907–918 [DOI] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E (2002) Mechanism of glutamate receptor desensitization. Nature 417: 245–253 [DOI] [PubMed] [Google Scholar]
- Swanson GT, Gereau RW, Green T, Heinemann SF (1997) Identification of amino acid residues that control functional behavior in GluR5 and GluR6 kainate receptors. Neuron 19: 913–926 [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov CN (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57: 122–133 [DOI] [PubMed] [Google Scholar]
- Weston MC, Gertler C, Mayer ML, Rosenmund C (2006a) Interdomain interactions in AMPA and kainate receptors regulate affinity for glutamate. J Neurosci 26: 7650–7658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML (2006b) Conformational restriction blocks glutamate receptor desensitization. Nat Struct Mol Biol 13: 1120–1127 [DOI] [PubMed] [Google Scholar]
- Wong AY, MacLean DM, Bowie D (2007) Na+/Cl− dipole couples agonist binding to kainate receptor activation. J Neurosci 27: 6800–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LA, Mayer ML (1993) Differential modulation by cyclothiazide and concanavalin A of desensitization at native alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid- and kainate-preferring glutamate receptors. Mol Pharmacol 44: 504–510 [PubMed] [Google Scholar]
- Yamada K, Tang CM (1993) Benzothiadiazines inhibit rapid glutamate receptor desensitization and enhance glutamatergic synaptic currents. J Neurosci 13: 3904–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshansky MV, Sobolevsky AI, Jatzke C, Wollmuth LP (2004) Block of AMPA receptor desensitization by a point mutation outside the ligand-binding domain. J Neurosci 24: 4728–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Cho Y, Lolis E, Howe JR (2008) Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating. J Neurosci 28: 932–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nayeem N, Nanao MH, Green T (2006) Interface interactions modulating desensitization of the kainate-selective ionotropic glutamate receptor subunit GluR6. J Neurosci 26: 10033–10042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Olivier NB, Yao H, Young EC, Siegelbaum SA (2004) A conserved tripeptide in CNG and HCN channels regulates ligand gating by controlling C-terminal oligomerization. Neuron 44: 823–834 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Information