Abstract
We used ChIP-Seq to map ERα-binding sites and to profile changes in RNA polymerase II (RNAPII) occupancy in MCF-7 cells in response to estradiol (E2), tamoxifen or fulvestrant. We identify 10 205 high confidence ERα-binding sites in response to E2 of which 68% contain an estrogen response element (ERE) and only 7% contain a FOXA1 motif. Remarkably, 596 genes change significantly in RNAPII occupancy (59% up and 41% down) already after 1 h of E2 exposure. Although promoter proximal enrichment of RNAPII (PPEP) occurs frequently in MCF-7 cells (17%), it is only observed on a minority of E2-regulated genes (4%). Tamoxifen and fulvestrant partially reduce ERα DNA binding and prevent RNAPII loading on the promoter and coding body on E2-upregulated genes. Both ligands act differently on E2-downregulated genes: tamoxifen acts as an agonist thus downregulating these genes, whereas fulvestrant antagonizes E2-induced repression and often increases RNAPII occupancy. Furthermore, our data identify genes preferentially regulated by tamoxifen but not by E2 or fulvestrant. Thus (partial) antagonist loaded ERα acts mechanistically different on E2-activated and E2-repressed genes.
Keywords: ChIP-Seq, estrogen receptor α, RNA polymerase II, SERMs
Introduction
Estradiol (E2) is a key regulator in the growth and differentiation of many target tissues and is involved in the development and progression of breast cancer (Anderson, 2002). Its genomic activity is to a large extent mediated by the estrogen receptor α (ERα; NR3A1), a member of the nuclear receptor super family. ERα regulates expression of target genes classically by binding directly to its cognate sequence, the estrogen response element (ERE). ERα binds to its cognate-binding sites as homodimer, recruits co-factors and activates or represses transcription in response to E2 (Shang et al, 2000). Alternatively, nonclassical regulation involves protein–protein interactions with other DNA-binding proteins such as Sp1, AP-1 and NF-κB. Identification of the ERα target gene network regulated by agonist and/or antagonist treatment is essential to understand the role of ERα in normal physiological processes and in cancer.
Several gene expression profiling studies in MCF-7 cells identified E2-responsive genes in the range of 100–1500 (Charpentier et al, 2000; Coser et al, 2003; Frasor et al, 2003; Rae et al, 2005; Carroll et al, 2006; Kininis et al, 2007; Kwon et al, 2007; Lin et al, 2007; Stender et al, 2007), whereas large scale ERα ChIP profiling showed that ERα interacts with many thousands genomic regions (Carroll et al, 2006; Kininis et al, 2007; Lin et al, 2007). This discordance is in part due to the fact that mRNA levels do not necessarily reflect gene activity because it is subject to degradation and regulation, and that likely not all ERα-binding sites are active under all conditions. Genome-wide profiling of RNA polymerase II (RNAPII) occupancy, however, does provide a much more direct readout and, thus, could yield insights beyond what is typically obtained by mRNA expression profiling.
Recent studies have shown that the promoters of a large number of genes are preloaded with RNAPII with minimal occupancy over the coding body, a phenomenon referred to as pausing or promoter proximal enrichment of RNAPII (PPEP). Collectively, these studies suggest that control of elongation rather than or in addition to transcription initiation has an important function in the activation of these genes, particular for genes rapidly responding to the developmental and cell signalling cues (Muse et al, 2007; Zeitlinger et al, 2007; Core et al, 2008).
Selective estrogen receptor modulators (SERMs) are (partial) E2 antagonists used for the treatment and prevention of breast cancer. One of the most widely used is tamoxifen, which has mixed agonistic/antagonistic properties and tissue-specific effects. Tamoxifen resistance develops ultimately in advanced breast cancer and is of major clinical significance (Ali and Coombes, 2002). SERMs induce an alternative conformation of the ERα ligand-binding domain that results in the recruitment of different co-factors and repression of transcription instead of activation. Fulvestrant (ICI 182 780) is a full antagonist that increases protein turnover and results in the degradation of ERα. Fulvestrant is used for the treatment of advanced breast cancer and tamoxifen-resistant tumours (Howell, 2006). The effect of SERMs on ERα binding and subsequent RNAPII recruitment has not been studied at a genome-wide level.
In this study, we used massive parallel sequencing of immunoprecipitated DNA fragments to identify ERα-interaction sites and RNAPII occupancy in response to E2 or the (partial) antagonists tamoxifen and fulvestrant. Combining ERα-binding site and RNAPII occupancy allowed us to measure the consequence of E2 treatment on RNAPII occupancy, that is ongoing transcription. RNAPII analyses also allowed us to assess whether PPEP is a general phenomenon in rapid E2 response. We identified a large number of ERα-interaction sites and a much smaller number of direct target genes, and show that tamoxifen and fulvestrant alter but not abolish ERα binding, and have differential effects on E2-upregulated and E2-downregulated genes. E2-mediated activation is antagonized by both compounds, whereas at E2-downregulated genes, tamoxifen shows agonistic behaviour in contrast to fulvestrant, which antagonizes E2-mediated repression.
Results
Identification of ER-interaction sites
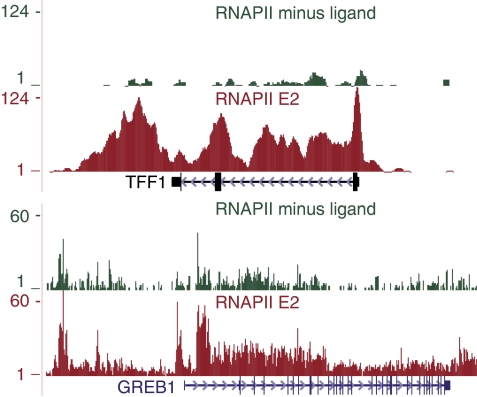
ChIP followed by deep sequencing was performed using the MCF-7 breast cancer cell line, which was hormone starved for 48 h and subsequently mock-treated (minus ligand) or stimulated for 1 h with 10 nM E2. The numbers of sequenced and mapped tags are shown in Supplementary Table SI. Classical ERα target genes, for example TFF1 and GREB1, showed vast enrichment of tags over a narrow range in their promoter and enhancer regions in the E2 dataset as compared with minus ligand (Figure 1A). Overlapping tags were joined into peaks and the number of tags per peak (peak scores) was counted. The frequency distribution of peak scores shows a wide range in the E2 dataset going up to nearly a 1000 tags/peak (Supplementary Figure S1). In the absence of ligand, the majority of peaks are found in the bins with lower peak scores; two high peak score bins are observed. Because substantial ERα binding is not expected in the absence of ligand, we visually inspected these genomic regions and observed local high tag densities over large areas reminiscent of copy number variation (CNV). Indeed, the outlier regions coincide with CNV as determined by arrayCGH data (Shadeo and Lam, 2006) and includes the amplified in breast cancer-1 gene (AIB1 or NCOA3) on chromosome 20. Regions with high CNV obviously compromise peak calling and were, therefore, corrected for prior to peak calling. Using an FDR of <1 × 10−4, we identified 10 205 ERα-interaction sites. ChIP-qPCR on three independent biological replicas validates the binding of ERα to randomly selected sites (20/20) (Supplementary Figure S2). The majority of the binding sites (41%) are located in introns and only a small percentage (7%) in promoter regions (Figure 1B), in good agreement with published data (Carroll et al, 2006; Lin et al, 2007).
Figure 1.
Overview of ERα-interaction sites. (A) ERα-binding sites at the TFF1 and GREB1 loci. The maximum number of overlapping tags, that is peak height is shown. Clear ERα peaks are detected in the promoter and enhancer region of the TFF1 and GREB1 gene on E2 treatment, whereas residual binding is observed in the absence of ligand. ERα binding is strongly decreased although not completely abolished on treatment with tamoxifen or fulvestrant compared with E2. (B) Genomic location of ERα-interaction sites. The majority of sites (41%) are located within an intron or distal from a gene (23%); 7% is located in promoter regions. (C) Comparison of large-scale ChIP profiling data. Venn diagram of the overlap of ERα-binding sites as identified in this study or reported by Lin et al and Lupien et al. 3305 and 1089 of the ChIP-Seq interaction sites are overlapping with the Lupien et al and Lin et al analysis (57 and 88%, respectively). (D) Venn diagram of the overlap between ERα-binding sites induced on E2, tamoxifen and fulvestrant treatment. The E2 and tamoxifen profile overlap to a large extent, but also contain preferential binding sites. Fulvestrant-liganded ERα interacts with a small number of sites that largely overlap with those found on E2 or tamoxifen induction.
Next, we compared our 10 205 ERα-interaction sites with genome-wide profiles determined in MCF-7 cells using either a micro-array platform or ChIP-PET identifying 5782 sites and 1234 sites, respectively (Figure 1C) (Lin et al, 2007; Lupien et al, 2008). Our ChIP-Seq and the ChIP-chip datasets show a substantial overlap (57%). Including the less deeply sequenced ChIP-PET dataset, which showed a 88% overlap with the ChIP-Seq targets, we obtained 615 ERα-binding sites that are identified with all three platforms. Sites shared between all three datasets are likely to encompass high affinity sites. Indeed, our ERα-binding sites common to all three datasets had a higher average peak height (84) as compared with sites present only in our dataset (average 34). The good enrichment obtained with our ERα monoclonal antibody combined with the high accuracy, sensitivity and sequence depth achieved with the Illumina genome analyzer allowed for the identification of more transient and likely indirect interaction sites in addition to high affinity and direct DNA-binding sites. However, small variations in cells and culture conditions, that is biological variation and sample handling likely also account for some of the differences. In conclusion, our ERα-binding site analysis reveals a very large number of sites encompassing direct as well as indirect interactions sites.
(Partial) antagonists affect ER binding
The effect of tamoxifen and fulvestrant is hitherto only studied on a small number of genes. The prevailing view is that SERMs do facilitate DNA binding of ERα (Shang et al, 2000). We performed ChIP-Seq of ERα in MCF-7 cells treated with either tamoxifen or fulvestrant to assess whether this holds true on a genome-wide scale. A total of 8854 tamoxifen and 4284 fulvestrant-induced ERα sites are detected and representative examples of ERα binding to the TFF1 and GREB1 loci are shown (Figure 1A). Globally, the E2 and tamoxifen profiles overlap to a large extent (54%), whereas a smaller proportion of the binding sites are shared between E2 and the fulvestrant profile (33%) (Figure 1D). These data corroborate and extend the notion that ERα is able to bind to its regulatory regions in vivo when loaded with these (partial) antagonists. We note that binding is reduced or even abolished at some sites (Supplementary Figure S3). The altered DNA binding of ERα in response to fulvestrant is not due to receptor degradation (Pink and Jordan, 1996), because we did not observe a reduction of the protein levels during the 1h treatment applied in these experiments (data not shown). We conclude that ERα binding is affected, but not abolished when liganded with SERMs.
Motif analysis
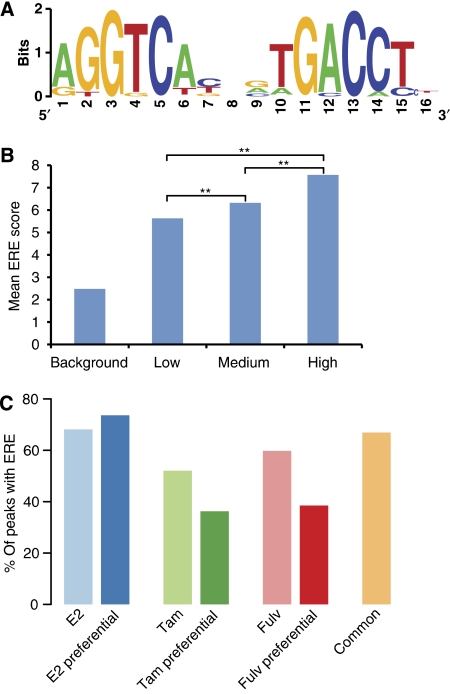
Next, we interrogated the sequence of the binding sites for overrepresentation of DNA motifs using the MDmodule program (Liu et al, 2002). The full ERE—palindromic arrangement of half sites with a 3bp spacer—turned out to be the by far most prevalent motif (Figure 2A). Using the weight matrix generated by MDmodule and an ERscan algorithm similar to that used earlier (Smeenk et al, 2008), we find that 68% of the ERα-interaction sites contain one or more ERE (motif score cut-off of 5, FPR of 15%). A clear positive correlation can be observed between peak height and the mean of the motif scores indicating that the ERα indeed binds most strongly to sites encompassing a consensus motif (Figure 2B). Next, we screened the ERα-interaction site sequences for the presence of other motifs. In line with published data (Carroll et al, 2006), we find significant enrichment of the Sp1, C/EBP and FOXA1 (HNF3α) motifs in addition to the ERE (Supplementary Table SII). Although the FOXA1 motif is statistically enriched in our dataset (P-value <0.0001; 400 bp window), the total number of potential FOXA1 sites in our data is low (748/10 205 or 7%). We separately examined the 3251 sites that do not encompass an ERE for the enrichment of transcription factor motifs. Among others, we find the FOXA1 motif in 308 peaks (9%). Given the apparent discordance between the Lupien and co-workers and our data, we also performed a peak calling using MACS to exclude any bias based on the peak detection algorithm (Zhang et al, 2008). MACS detects 7713 peaks, that is a 75% overlap with the 10 205 sites called by FindPeaks. Motif analysis showed that the FOXA1 motif is present in 6.6% of peaks called by MACS. To further rule out that this discordance is due to the use of different weight matrices and algorithms, we directly determined the overlap (400 bp window) between our ERα-interaction sites and the 12 904 reported FOXA1-binding sites and found an overlap of 15 and 13.6% with binding sites determined by FindPeaks and MACS, respectively. Taken together, our analysis reveals the statistical enrichment of a number of sequence motifs including the FOXA1 motif, but only a minor co-occurrence of ERα and FOXA1-interaction sites was detected.
Figure 2.
ERE motif and correlation with peak height. (A) The weight matrix of the highly overrepresented ERE motif. (B) Correlation of peak height with ERα motif score. The mean ERE motif score was determined using ERscan. ERα-interaction sites were binned according to peak height; random genomic regions were used as background. A clear positive correlation is obtained between the height of an ERα peak and the motif score. The mean of the motif scores in the three bins is significantly different as assessed by the Mann–Whitney test, with a P-value of <0.01, indicated by a double asterisk (C) Percentage of interaction sites containing an ERE. The different binding site profiles were searched for the presence of an ERE using ERscan. The ‘E2 preferential' group contains the highest percentage of ERE motifs as compared with the tamoxifen and fulvestrant preferential groups.
On treatment of MCF-7 cells with tamoxifen and fulvestrant, we observe many ‘SERM-specific' ERα-interaction sites (Figure 1D). The selective binding of the receptor in the presence of different ligands may be dictated by the sequence composition of the cis-acting element. Therefore, we assessed the presence of the ERE and other known transcription factor motifs in the different categories of compound-specific interaction sites as well as of sites common to all three compounds. We find that 74% of the ‘E2-specific' interaction sites contain an ERE, whereas 36 and 39% of the tamoxifen- and fulvestrant-specific sites contain an ERE (Figure 2C). Besides the ERE, no differentially enriched transcription factor motifs could be detected. In addition, we assessed the evolutionary conservation of the ERα-binding sites. The shared as well as the compound-specific groups are significantly more conserved compared with random regions (P-value <0.01) as shown in Supplementary Figure S4. This indicates that binding sites present in MCF-7 cells are conserved between species and play a general role in the regulation by ERα.
Ligand triggers rapid changes in RNAPII occupancy
Our and earlier genome-wide analyses have provided a wealth of ERα-binding sites. However, assigning the target genes has remained problematic because a large proportion of the ERα-binding sites are located at great distances from genes. To more directly identify genes responding to E2 treatment (1 h), we performed ChIP-Seq using an antibody against RNAPII and determined the log2 ratio of E2/minus ligand. RNAPII occupancy throughout the gene body provides a direct readout of transcriptional activity and thus yields insights beyond what is typically achieved by mRNA expression profiling. Classical ERα target genes, such as TFF1 and GREB1, show a clear increase in RNAPII occupancy over their gene body already after 1 h exposure to E2 (Figure 3). At a global scale, RNAPII occupancy over 596 genes significantly changes in response to E2 stimulation (mean±1.5 × s.d.), with 349 genes showing an increase and 247 genes showing a decrease in RNAPII occupancy. Comparing our E2-regulated genes with mRNA expression profiles (Kininis et al, 2007; Lin et al, 2007) revealed an overlap of 64 and 47 genes, respectively. When including genes that change less then two-fold but are significant (P-value <0.05) in the Kininis dataset, the overlap increased to 195 genes. Note that with our ChIP-Seq of RNAPII occupancy and the short E2 treatment (1 h), we will only or predominantly identify direct and immediate/early responding target genes, whereas in gene expression profiling at 3 or 8 h after E2 addition delayed/late responding and indirect targets may also have been identified.
Figure 3.
RNA polymerase II occupancy at ERα target genes. The RNAPII occupancy is depicted for the TFF1 (top panel) and GREB1 locus (lower panel) in response to solvent (green) or E2 (red).
Next, we examined E2-responsive genes for the presence of nearby ERα-interaction sites (within 50 kb). Of the 349 upregulated genes, 309 (89%) encompass 1226 ERα-interaction sites, that is 4 on average, whereas of the 247 downregulated genes, 116 (47%) encompass 192 ERα-interaction sites (1.5 on average). Besides that upregulated genes more frequently encompass ERα-binding sites than downregulated genes, the sites in upregulated genes more frequently contain an ERE that conforms better to the consensus ERE and displays a higher mean motif score (Supplementary Table SIII). Motif analysis shows that ERα-binding sites near up- and downregulated genes do not contain differentially enriched transcription factor motifs at statistically significant P-values. Gene Ontology (GO) analysis shows that E2-regulated genes are enriched for a diverse set of cellular processes and functions, including ovulation cycle process, female gonad development and female meiosis (Supplementary Table SIV).
In conclusion, we show that 596 genes change in RNAPII occupancy over the gene body in response to E2, of which 59% are upregulated and 41% are downregulated. A higher number of ERα-bindings sites are present near upregulated genes compared with downregulated genes and sites near upregulated genes conform better to the ERE consensus sequence than those of nearby downregulated genes.
Promoter proximal enrichment of RNAPII
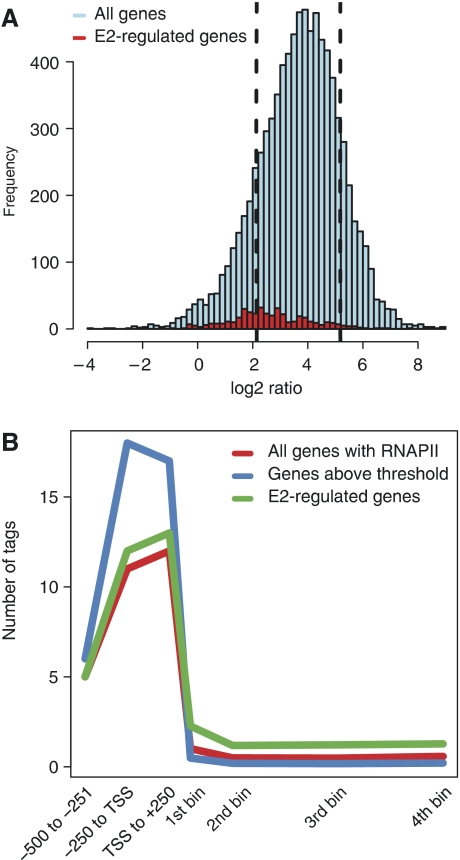
Recent genome-wide (ChIP-chip) studies have shown that a large fraction of the promoters of developmental and cell signalling genes as well as genes responding to external stimuli display PPEP or pausing of RNAPII, which is thought to facilitate rapid upregulation of transcription (Guenther et al, 2007; Muse et al, 2007; Zeitlinger et al, 2007). GRO-seq (global nuclear run-on-sequencing) revealed that up to 30% of genes display promoter proximal pausing (Core et al, 2008).
Nuclear hormone receptors such as ERα are regulators of rapid response par excellence and hence, it seemed likely that pausing of RNAPII might be involved in the fast regulation of immediate early E2-responsive genes. Therefore, we determined the number of tags in the promoter and body of genes; in the minus ligand dataset (i.e. before induction) of the 8465 genes that are significantly enriched for RNAPII, 1228 (15%) display PPEP (Figure 4A). RNAPII enrichment in promoter regions was validated on 6/6 genes using the 8WG16 antibody (Supplementary Figure S5). Furthermore, we validated PPEP using a number of phospho-specific (phosphoserine 2, 5 and 7) and an N-terminal RNAPII antibody (N-term). The transition of RNAPII from the initiation to the elongating form can be monitored by phosphorylation of specific serine residues in the CTD. Serine 5 is phosphorylated at the initiating phase of transcription, whereas serine 2 is a mark of productive RNAPII and occurs more in the 3′ end of a gene. Serine 7 phosphorylation is a mark for elongating RNAPII (Phatnani and Greenleaf, 2006; Chapman et al, 2007). The RNAPII phosphorylation status of three genes was assessed in the absence or presence of E2 using ChIP in combination with phospho-specific antibodies. The presence of phosphoserine 5 (and surprisingly 7) combined with the absence of phosphoserine 2 shows PPEP in the presence and absence of ligand (Supplementary Figure S6).
Figure 4.
Promoter proximal enrichment of RNAPII (PPEP). (A) Histogram of the RNAPII occupancy ratio at promoter versus gene body. The distribution of the promoter/gene body ratio of all genes containing RNAPII (light blue) and of E2-responsive genes (red). The dashed lines represent the mean±1 × s.d. The 1228 genes display paused RNAPII of which only 21 are E2-responsive genes. (B) RNAPII occupancy profile. Genes were divided into bins relative to the transcription start site; −500 to −251, −250 to TSS, TSS to +250 and the remaining gene body was divided into four equal bins. For each group of genes, the mean number of tags per bin is plotted. E2-regulated genes on average have less RNAPII at their promoter regions as compared with the mean of all genes. Genes above the set threshold (mean+1 × s.d.) have a higher RNAPII occupancy at their promoter as compared with all genes and E2-regulated genes.
Strikingly, only 21 of the 596 E2-regulated genes (4%) display PPEP. Moreover, the median RNAPII occupancy profile over E2-regulated genes in the promoter region and the coding body does not significantly deviate from that of all genes but is very significantly lower than the profile of PPEP genes (Figure 4B). Of the 64 E2-regulated genes shared between our and Kininis datasets, 8% display PPEP, a percent wise increase as compared with our entire data but still a minor fraction. (Kininis et al, 2007). Together, these results show that a large fraction of all genes, but only a very minor fraction of E2-regulated genes display PPEP in MCF-7 cells. Nevertheless, the majority of the 349 E2-upregulated genes do show a rapid and highly significant increase in RNAPII occupancy already at 1 h of E2 induction.
Finally, we determined the effect of ligand administration on the 21 E2-regulated genes that display PPEP. E2 induction changes the RNAPII occupancy ratio between promoter and gene body resulting in a loss of PPEP (10/21), whereas tamoxifen and fulvestrant treatment resulted in the abolishment of RNAPII occupancy (and thus PPEP) on 21/21 genes and 15/21 genes, respectively. The observation that (partial) antagonists induce a rapid loss of RNAPII on E2-responsive promoters indicates that at large these ligands prevent the recruitment to and/or stabilization of RNAPII at the promoter and, thus, preinitiation complex (PIC) formation rather than affecting the transition of RNAPII into the elongating form.
Overlapping as well as distinct groups of genes respond to (ant)agonists
Given that in the majority of the cases, the binding of ERα is not abolished on (partial) antagonist treatment, we determined the effect of tamoxifen and fulvestrant on the RNAPII occupancy. On tamoxifen administration, 719 genes change in RNAPII occupancy; the majority (695/719) is downregulated as compared with the minus ligand control. Strikingly, more genes change their RNAPII occupancy on (partial) antagonist as compared with E2 treatment (596), which has also been observed in expression profiling studies (Frasor et al, 2004). On fulvestrant treatment, 319 genes change in RNAPII occupancy as compared with the minus ligand control of which 230 are downregulated. A typical example is shown in Supplementary Figure S7. Note that in mock-treated cells, the RNAPII occupancy of many ERα-regulated genes was low, but in many cases clearly enriched for RNAPII, which is likely due to incomplete ligand depletion. Collectively, the total number of genes with altered RNAPII occupancy for all three ligands is 1256.
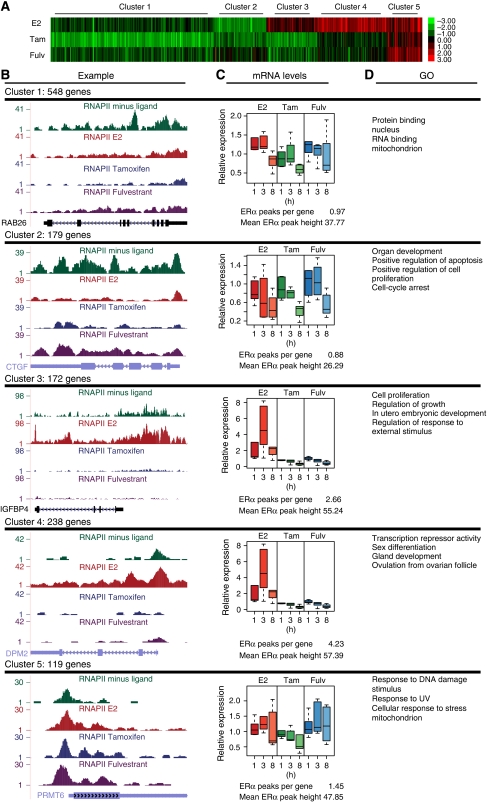
To classify genes based on their response to E2, tamoxifen and fulvestrant, we performed K-means clustering of the RNAPII ratios revealing five distinct clusters (Figure 5A and B). Using GO, we assessed whether the genes within the clusters are functionally related (Figure 5D). Supplementary Table SV shows an overview of the ERα-binding sites analysis per cluster. The changes in RNAPII occupancy in response to the various ligand treatments were validated by ChIP-qPCR on 7–8 randomly chosen examples from each cluster (Supplementary Figure S8). Next, we compared the changes in RNAPII occupancy of six genes from each cluster with the changes in the level of primary transcript and mRNA levels as measured by RT–qPCR at 0, 1, 3 and 8 h after treatment using intron-exon and exonic primer pairs, respectively. In particular, the changes in primary transcript levels (Supplementary Figure S9) and to a lesser extent of mRNA levels (Figure 5C; Supplementary Figure S10) of genes in clusters 2, 3 and 4 correlate very well to the changes in RNAPII occupancy in response to the various ligands as determined by ChIP-Seq.
Figure 5.
Cluster analysis based on changes in RNAPII occupancy. (A) The 1256 genes with changed RNAPII occupancy in response to the various ligands were clustered into five groups on the basis of the change in RNAPII occupancy relative to ‘no ligand' (ligand/no ligand ratio) using K-means clustering. (B) Representative examples of RNAPII occupancy in response to the various ligands. The number of genes in each cluster is indicated. (C) Boxplot presentation of the changes in mRNA levels triggered by ligand treatment for 1, 3 and 8 h of RT–qPCR analysis was performed on six randomly chosen genes out of each cluster using exonic primer pairs. Relative mRNA levels were normalized to that of RPS19. The number of ERα peaks and the mean ERα peak height is indicated. (D) Enriched GO categories in each cluster.
Cluster 1 contains a large group of genes that were strongly downregulated only on tamoxifen treatment. In line with this, four of the six validated genes show diminishing primary transcripts levels, whereas the mRNA levels respond later and decrease after 8 h. On average, one peak per ERα-binding site is present in the vicinity of these genes. Among the genes in this cluster is the pro-apoptotic gene Bad whose overexpression inhibits estrogen-induced cell proliferation (Fernando et al, 2007). In addition, high Bad expression levels correlate with improved disease-free survival (Cannings et al, 2007). The cyclin A gene has a similar response to the different ligands and has prognostic value in early breast cancer. Overexpression of cyclin A in tamoxifen-treated tumours is significantly associated with poorer outcome (Michalides et al, 2002). Intriguingly, ERα peaks in the vicinity of genes in this cluster were slightly higher when liganded with tamoxifen as compared with E2-liganded ERα.
The second cluster contains strongly E2-downregulated genes that were derepressed by fulvestrant, but not or only to a limited extend by tamoxifen. Among these was cyclin G2, a negative regulator of the cell cycle that maintains cells in a quiescent state and is downregulated on E2 induction as described earlier (Stossi et al, 2006). Strikingly, we observed that tamoxifen acts as an agonist on cyclin G2, in contrast to Stossi et al who describe that tamoxifen antagonizes the E2-mediated downregulation. Another gene downregulated on E2 treatment was the pro-apoptotic Bak gene. It has been described earlier that Bak expression is downregulated on E2 treatment and reduction of Bak expression provides a growth advantage to cells (Leung et al, 1998). Bik, another pro-apoptotic gene was also present in cluster 2 and in good agreement with our data, it has been shown that Bik mRNA levels increase on estrogen starvation and antagonist treatment, whereas mRNA levels decrease on E2 induction (Hur et al, 2004). Interestingly, genes in this cluster display the lowest number of ERα-binding sites per gene (0.88 on average). Tamoxifen-liganded ERα bound on average with higher affinity to sites near cluster 2 genes compared with E2-liganded ERα. Interestingly, the primary transcripts of cluster 2 genes do not change or are even slightly elevated on tamoxifen or fulvestrant treatment, whereas the mRNA levels decrease likely due to regulation of mRNA stability. GO analysis showed that cluster 2 is enriched in genes involved in apoptosis and cell-cycle arrest.
Genes in the third cluster are upregulated by E2, which is strongly antagonized by tamoxifen and fulvestrant. In the absence of ligand, these genes were already expressed as revealed by the presence of RNAPII; on tamoxifen or fulvestrant treatment, the RNAPII occupancy level of these genes dropped below the minus ligand level. For example, transcription of the IGFBP4 gene is strongly boosted on E2 addition and decreased on tamoxifen treatment. IGFBP4 expression is used as a predictor of responsiveness to endocrine therapies (Walker et al, 2007). Another E2-upregulated gene in this cluster was the cell-cycle regulator cyclin D1. Overexpression of cyclin D1 is associated with better outcome for breast cancer patients, but its overexpression is also linked to tamoxifen resistance (Ishii et al, 2008). On average, genes in cluster 3 contain 2.7 ERα-binding sites. GO analysis revealed among others involvement in cell proliferation and insulin receptor signalling. The mRNA levels of genes in clusters 3 and 4 (see below) increase on E2 treatment, with the largest increase at 3 h of treatment and do not respond or are downregulated on tamoxifen or fulvestrant treatment.
Cluster 4 also contains E2-upregulated genes that are antagonized both by tamoxifen and fulvestrant, but genes in this cluster show no/minor RNAPII occupancy in the absence of ligand as opposed to cluster 3 genes. Interestingly, the nuclear receptor coactivator NCOA4 is upregulated on E2 treatment. NCOA4 can associate with ERα and has been reported to increase transcription of TFF1 (Lanzino et al, 2005). The anti-apoptotic Bcl-2 is present in this cluster. Bcl-2 has been shown to be upregulated on E2 induction and Bcl-2 expression is linked with better outcome in hormone or chemotherapy-treated patients (Nadler et al, 2008). Genes in the fourth cluster display on average the highest number of ERα-binding sites (4.2 on average per gene). In addition, ERα-binding sites found in the proximity of clusters 3 and 4 have a higher peak score (affinity) for the ERα as compared with the other clusters. Cluster 4 was enriched in the GO categories sex differentiation, ovulation from ovarian follicle and gland development.
Remarkably, cluster 5 contains genes upregulated by all three ligands, which is, however, not well reflected in the mRNA levels. Among the genes was HUS1, which is part of a cell-cycle checkpoint in the DNA damage response (Meyerkord et al, 2008). Genes in this cluster contained on average 1.4 ERα-binding site per gene. GO analysis showed enrichment for genes involved in stress response.
In conclusion, tamoxifen and fulvestrant have differential effects on RNAPII occupancy. The majority of the E2-upregulated genes are antagonized by both tamoxifen and fulvestrant, while tamoxifen displayed agonistic behaviour whereas fulvestrant antagonized E2-downregulation. Furthermore, tamoxifen specifically affects a rather large group of genes that are not affected by either E2 or tamoxifen.
Discussion
To unravel the target gene network of a transcription factor, identification of its interaction sites, cis-regulatory elements and target genes is essential. We have applied ChIP combined with massive parallel sequencing to identify ERα-interaction sites and to globally map changes in RNAPII occupancy in the presence and absence of three ligands. A small fraction of ERα-interaction sites is located in promoter regions, whereas the majority is found at large distances from annotated genes or in introns in line with earlier studies (Carroll et al, 2006; Lin et al, 2007). These distal sites most likely act as enhancers and interact with receptive promoters through looping to regulate gene expression, as has been described for some ERα target genes such as TFF1, GREB1 and bcl-2 (Carroll et al, 2005; Deschênes et al, 2007; Perillo et al, 2008). Our comprehensive analysis of the ERα-interaction site sequences showed that the majority of sites contain an ERE which is in agreement with other data (Lin et al, 2007). In line with observations that ERα physically and/or functionally interacts with other transcription factors, we reveal that among others the Sp1, C/EBP and FOXA1 motif are enriched (400 bp window). Lupien et al reported that 50–60% of ERα binding in MCF-7 cells co-occurred with FOXA1. Although statistically enriched, FOXA1 is only present in a small fraction of all our ERα-interaction sites. Use of the peak detection method used by Zhang and co-workers (MACS) did not significantly change the percentage of ERα-binding sites with a FOXA1 motif (Zhang et al, 2008). The intersection between the FOXA1-binding sites (Lupien et al, 2008) and our ERα profile (FindPeaks) reveals a 15% overlap. Our motif analysis and the limited overlap between both genomic binding sites indicates that the cooperativity between ERα and FOXA1 is under our condition much more restricted than reported earlier. The discordance is substantial and may be due to the use of different antibodies and platforms. More research is necessary to fully elucidate the role of FOXA1 in ERα transcriptional regulation.
We showed at a genome-wide scale that tamoxifen and fulvestrant affect but do not abolish binding of ERα to chromatin. Our analysis revealed an extensive overlap between ERα-interaction profiles in the presence of the different ligands but also detect interaction sites that show ligand-specific ERα binding. Additionally, our analysis showed that a higher percentage of sites in the ‘E2-specific' group conform to the consensus ERE as compared with the tamoxifen- and fulvestrant-specific group. One explanation of this ligand-dependent preference is that the receptors interact or cooperate with other transcription factors and that this cooperativity with other transcription factors may in part be determined by conformational changes in the DNA-binding domain instigated by E2, tamoxifen and fulvestrant binding. However, our motif analysis does not lend support to this hypothesis and hence, the ligand-specific binding is likely dependent on other factors. The local chromatin landscape and histone modifications likely play a decisive role as has been described for GR (John et al, 2008). Furthermore, it has been shown that E2 but not fulvestrant-loaded ERα recruits SWI/SNF, an ATP-dependent chromatin remodeller complex, to its binding sites resulting in chromatin remodelling and histone acetylation (Belandia et al, 2002). Likely the correct chromatin structure necessary for stable and recurring ERα–chromatin interactions is not induced by (partial) antagonists and as a consequence, ERα binds weaker or not at all.
The dispersed and often distal localization of ERα-binding sites complicates the assignment of ERα-interaction sites to E2-responsive genes. We identify for the first time genes responding to E2 induction by using ChIP-Seq profiling of RNAPII occupancy in the absence and presence of the various compounds. Of the 596 genes changing in RNAPII occupancy on E2 induction around half are upregulated and half are downregulated in keeping with micro-array expression profiling (Kininis et al, 2007; Lin et al, 2007). The limited overlap between our E2-responsive genes and mRNA expression profiles maybe due to differences in the MCF-7 sublines used, the different induction times and inherent differences between analyses based on RNAPII occupancy and on steady state mRNA levels (Frasor et al, 2004).
The majority of E2-activated genes are associated with one or more ERα-binding sites, indicating that ERα complexes bound to multiple interaction sites probably cooperate to regulate expression of target genes as observed earlier, for example for TCF4 and MYC (Bieda et al, 2006; Hatzis et al, 2008). Nevertheless, the requirement of multiple ERα-binding sites per E2-regulated gene alone cannot account for the striking discordance between the number of E2-regulated genes (596) and ERα-binding sites (10 205). Most likely other factors besides ERα binding such as promoter accessibility, the local chromatin structure, epigenetic state and the presence of specific co-factors are necessary for transcriptional regulation to occur.
We also analysed the RNAPII occupancy profiles to assess PPEP. Several recent studies have reported that between 12 and 30% of genes display PPEP. RNAPII profiling in Drosophila showed that pausing is occurring predominantly on genes responding to stimuli as well as developmental genes. It is postulated that the presence of RNAPII on the promoter allows rapid upregulation of these genes (Kim et al, 2005; Guenther et al, 2007; Muse et al, 2007; Zeitlinger et al, 2007) as observed for the heat shock genes (Petesch and Lis, 2008). We show that 15% of all active genes in the minus ligand data show PPEP in MCF-7 cells in keeping with other studies (Kim et al, 2005; Guenther et al, 2007; Muse et al, 2007; Zeitlinger et al, 2007; Core et al, 2008; Kininis et al, 2009). In our dataset, however, the genes displaying PPEP are not significantly enriched for developmental genes or genes responding to stimuli, but for a broad spectrum of GO classes. This may be due to the developmental or differentiated state of the cells under investigation (Supplementary Table SVI). We used the MCF-7 breast cancer cell line in contrast to the Drosophila embryos or the embryonal Drosophila S2 cell line (derived from late stage embryos) used by Muse et al and Zeitlinger et al. In differentiated cells, other genes may be poised for activation as compared with embryos/embryonic cell line. E2-responsive genes are responding to external stimuli par excellence and hence likely candidates for using PPEP as a way of regulation. However, only a small fraction of genes that display PPEP are also E2 responsive, indicating that PPEP is not a general mechanism of rapid E2-instigated gene regulation, although it may play a role on a limited number of E2-regulated genes. E2-responsive genes are, thus, predominantly regulated by RNAPII recruitment and not at the elongation level.
RNAPII pausing was recently reported to play a role in the regulation of a number of E2-regulated genes (Kininis et al, 2007, 2009). The authors assessed RNAPII pausing by performing ChIP-chip for RNAPII using a promoter array in addition to gene expression profiling to determine E2-responsive genes. They reported that 59% of E2-responsive genes are preloaded with RNAPII (Kininis et al, 2009). In contrast, our data indicates a minor role of pausing or PPEP in E2-mediated regulation. It seems likely that the choice of analysed E2-responsive genes that is selected either on the basis of mRNA profiling (Kininis et al, 2007) and on E2-induced RNAPII occupancy at very early times may have affected the respective seemingly opposing conclusions. As an example, the Myc and SIAH2 gene described by Kininis et al do not fulfill our criteria for being E2 responsive and displaying PPEP (Supplementary Figure S11). In our genome-wide RNAPII ChIP-Seq analysis, the Myc gene does not exceed our threshold for E2 responsiveness (mean±1.5 × s.d. of log2 ratio E2/minus ligand). PPEP is observed in the proximity of the Myc promoter as defined by RefSeq, but the peak for RNAPII is not near the start site as defined by Ensembl and hence drops out as an E2-responsive gene displaying PPEP. SIAH2 is clearly E2 responsive, but does not fulfill the PPEP threshold (mean±1 × s.d. of the log2 ratio of RNAPII occupancy over promoter/gene body). Further research will be necessary to resolve the discordance and to elucidate the possible role of RNAPII pausing in E2-mediated regulation.
Tamoxifen stimulation abolished RNAPII occupancy over the promoter and coding regions of all 21 E2-regulated genes displaying PPEP while fulvestrant results in the abolishment of PPEP on 15/21 genes. Tamoxifen- or fulvestrant-loaded receptor is able to prevent the recruitment of RNAPII and hence PPEP is not established. Therefore, we conclude that both tamoxifen and to a lesser extent fulvestrant inhibit RNAPII recruitment, and/or destabilization of the RNAPII/PIC complex. We show that the SERMs tamoxifen and fulvestrant have a profound effect on the RNAPII occupancy of many genes. Using clustering methods, we could divide E2-, tamoxifen- and fulvestrant-regulated genes into five distinct clusters based on their RNAPII ratio. The primary transcript levels for the clusters 2, 3 and 4 correspond very well to the RNAPII ChIP data validating our approach. After tamoxifen treatment, the primary transcript levels of genes randomly selected from cluster 2 do not change significantly while fulvestrant treatment increases the primary transcript levels. In contrast, mRNA levels show a different pattern for both tamoxifen and fulvestrant and are diminished indicating that mRNA stability plays a role in ERα-mediated regulation of this gene cluster again validating our RNAPII occupancy approach. Strikingly, cluster 1 contains a group of genes that show no or only minor downregulation on E2 induction but are strongly repressed by tamoxifen and not or to a lesser extent by fulvestrant. The presence of many ERα-binding sites in the vicinity of these genes indicates that the tamoxifen-mediated repression may indeed be directly mediated through ERα. Tamoxifen preferential gene regulation has been shown on mRNA levels (Frasor et al, 2006). Cluster 2 contains genes that are most strongly downregulated by E2 and on which tamoxifen acts as a weak agonist; in contrast, fulvestrant displays full antagonistic behaviour. Using reporter assays it has been shown that tamoxifen antagonizes E2-induced upregulation but acts as an agonist at E2-downregulated genes (Ramkumar and Adler, 1995). One possible mechanism could be that on E2-downregulated genes, ERα recruits nuclear receptor corepressors and associated HDAC's that subsequently deacetylates the chromatin and prevent recruitment of RNAPII and the basal transcriptional machinery (Stossi et al, 2006). It seems likely that tamoxifen prevents the recruitment and loading of co-activator complexes on E2-upregulated genes, as seen in cluster 3 and 4, but on E2-downregulated genes tamoxifen does not prevent the recruitment of repressing complexes or is not able to recruit activating complexes, as seen in cluster 2. Strikingly, fulvestrant does antagonize both E2-upregulated as well as E2-downregulated genes. A small group of genes is upregulated by stimulation with all three ligands. These genes are enriched for among others response to stress and DNA damage, indicating that ligand stimulation results in the activation of stress responses.
On SERM treatment, RNAPII is not significantly enriched over the coding body of most if not all E2-upregulated genes compared with background, whereas these same genes display clear RNAPII occupancy in the absence of ligand. On E2-downregulated genes, E2-loaded ERα represses transcription probably by interfering with PIC formation and/or by preventing its assembly as well as by recruiting corepressor complexes and histone deacetylases. On binding of a SERM-loaded receptor, transcription is not downregulated and the PIC remains present. Thus, tamoxifen- or fulvestrant-loaded ERα acts mechanistically different on E2-upregulated and downregulated genes. This differential behaviour at up- and downregulated genes might relate to the direct or indirect protein-mediated binding of ERα. Regulation through tethering to other transcription factors such as NF-κB, AP-1 and Sp1 might alter the properties of SERM-loaded receptor.
In conclusion, our study provides novel and important insight into the regulation of the ERα target gene network and serves as a resource for the further elucidation of ERα-regulated transcription. Pausing of RNAPII occurs frequently in MCF-7, but only at a very small number of ERα target genes. We provide compelling evidence that tamoxifen and fulvestrant behave differently; tamoxifen acts as an agonist at E2-downregulated genes, whereas fulvestrant antagonizes both E2-upregulated and downregulated genes. Strikingly, tamoxifen regulates a rather large number of genes that are not or much less responsive to E2 or fulvestrant. Furthermore, these genes might play a role in tamoxifen resistance.
Materials and methods
ChIP
MCF-7 cells were cultured in DMEM with 10% FCS at 37°C. Cells were maintained in DMEM w/o phenol red and 5% charcoal stripped FCS for 48 h before induction. MCF-7 cells were mock treated or with 10 nM 17β-E2, 1 μM 4-OH tamoxifen or 100 nM Fulvestrant for 1 h. Chromatin was harvested and ChIP and qPCR was performed as described (Denissov et al, 2007). ChIP was performed using one of the following antibodies: ERα (Diagenode AC-066-100), RNAPII (Diagenode AC-055-100), Ser5-P RNAPII (Abcam, ab24759), RNAPII N-terminus (Santa Cruz, sc-899 X), Ser2-P RNAPII and Ser7-P RNAPII, both a kind gift from Dirk Eick.
Illumina high throughput sequencing
Sample preparation was performed as described by the manufacturer. The 32 bp tags were mapped to the human genome HG18 using the eland program allowing one mismatch. The 32bp sequence reads were directionally extended to 133 bp, corresponding to the length of the original fragments used for sequencing. For each base pair in the genome, the number of overlapping sequence reads was determined, averaged over a 10bp window and visualized in the UCSC genome browser (http://genome.ucsc.edu).
CNV correction and peak detection
The distribution of tags in the minus ligand dataset was determined by counting the number of tags in a 500kb window. The mean and standard deviation of all windows was determined and a threshold was set at mean+3 × s.d. Windows above the threshold were corrected by uniformly removing tags such that the number of tags was equal to that of the mean of all windows. All ERα datasets were corrected; subsequently enriched regions were identified with FindPeaks using a FDR cut-off of <1 × 10−4, subpeaks 0.9, triangles distribution and duplicate filter (Fejes et al, 2008). To allow for direct comparison, datasets were uniformly equalized relatively to the sample with the lower number of tags.
Motif search
For all sequence analysis, the highest point in a peak (FindPeaks) was used and extended on both sides with either 200 or 1500 bp. MDmodule was used with varying window sizes (Liu et al, 2002). Sequence logos were constructed using Weblogo (http://weblogo.berkeley.edu/). To search for known motifs, the MATCH software and the Transfac 11.1 database were used (Matys et al, 2003). Random genomic sequences were used as background. Enrichment was calculated using Benjamin-Hochberg multiple testing correction. Motifs with a P-value of <0.001 and a score above 2 were called enriched. The presence of ERE was determined in a 400bp window using ERscan, an adapted version of p53scan (Smeenk et al, 2008).
Conservation
ERα-binding sites were extended from the highest point in a peak with 100 bp on both sides. For each binding site, the mean PhastCons28 conservation score was calculated and per group the mean of all sites was determined. A Mann–Whitney test was performed to determine whether the scores were significantly higher compared with random sequences.
GO
Enrichment of GO categories was determined with the Ontologizer software, using Topology-Elim, Benjamini-Hochberg and a cut-off of 0.01.
RNAPII analysis
To quantitate the change in RNAPII occupancy, the datasets were uniformly equalized by removing tags relatively to the sample with the lower tag count. For each gene, the number of tags in the gene body was counted and the log2 ratio between ligand and minus ligand control was calculated. A threshold was set at mean±1.5 × s.d. of the log2 ratio. Very low expressed genes were removed if under all conditions the number of tags was <5 or the length in bp/tag count was <100. Clustering was performed using the K-means algorithm with cosine distance using the ARMADA program (Chatziioannou et al, submitted for publication). The heatmap was created using Java TreeView.
PPEP
For each gene in the Ensembl 47 gene annotation, the number of tags was determined in the promoter (−250 to +500 bp with respect to the transcription start site) and gene body (+500 bp to gene end). Genes with 8 or more tags in the promoter region were selected and the number of tags per 750 bp in the gene body was determined. Subsequently, the log2 ratio of the number of tags in the promoter and in the gene body was calculated. A threshold was set at mean+1 × s.d. and genes above this threshold were defined as displaying PPEP.
mRNA and primary transcript expression analysis
RNA was harvested after 0, 1, 3 or 8 h ligand treatment using the Qiagen RNeasy kit including on column DNase treatment. RNA was reverse transcribed to cDNA using Invitrogen superscript III. Exon-intron and exonic primer pairs were used for primary transcript and mRNA analysis. Expression levels were normalized to RSP19.
Accession codes
Sequence and processed data has been submitted to National Center for Biotechnology Gene Expression Omnibus with GEO accession number GSE14664.
Supplementary Material
Supplementary Tables
Supplementary Figures
Acknowledgments
We thank Dr B Alako, K Françoijs and P Moulos for helpful bioinformatic assistance and Dr L Altucci, Dr G Veenstra, R Akkers and L Smeenk for helpful discussions and critical reading of the paper. We thank Dirk Eick for his generous gift of Ser2-P RNAPII and Ser7-P RNAPII antibodies. Funding for this work was provided by the Radboud University Nijmegen Medical Centre and Radboud University Science Faculty. NBIS and EPIgenetic TReatment Of Neoplastic disease (EPITRON) (LSH-2004-2.2.0-2 to M.A.vD.); X-TRA-NET (LSHG-CT-2005-018882 to M.A.vD.). HEROIC (High-throughput Epigenetic Regulatory Organization In Chromatin). The authors declare that they have no conflict of interest.
References
- Ali S, Coombes RC (2002) Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer 2: 101–112 [DOI] [PubMed] [Google Scholar]
- Anderson E (2002) The role of oestrogen and progesterone receptors in human mammary development and tumorigenesis. Breast Cancer Res 4: 197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belandia B, Orford RL, Hurst HC, Parker MG (2002) Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J 21: 4094–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieda M, Xu X, Singer MA, Green R, Farnham PJ (2006) Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res 16: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannings E, Kirkegaard T, Tovey SM, Dunne B, Cooke TG, Bartlett JM (2007) Bad expression predicts outcome in patients treated with tamoxifen. Breast Cancer Res Treat 102: 173–179 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M (2006) Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 [DOI] [PubMed] [Google Scholar]
- Charpentier AH, Bednarek AK, Daniel RL, Hawkins KA, Laflin KJ, Gaddis S, MacLeod MC, Aldaz CM (2000) Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer Res 60: 5977–5983 [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coser KR, Chesnes J, Hur J, Ray S, Isselbacher KJ, Shioda T (2003) Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc Natl Acad Sci USA 100: 13994–13999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H (2007) Identification of novel functional TBP-binding sites and general factor repertoires. EMBO J 26: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschênes J, Bourdeau V, White JH, Mader S (2007) Regulation of GREB1 transcription by estrogen receptor alpha through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J Biol Chem 282: 17335–17339 [DOI] [PubMed] [Google Scholar]
- Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, Jones SJ (2008) FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics 24: 1729–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando R, Foster JS, Bible A, Ström A, Pestell RG, Rao M, Saxton A, Baek SJ, Yamaguchi K, Donnell R, Cekanova M, Wimalasena J (2007) Breast cancer cell proliferation is inhibited by BAD: regulation of cyclin D1. J Biol Chem 282: 28864–28873 [DOI] [PubMed] [Google Scholar]
- Frasor J, Chang EC, Komm B, Lin CY, Vega VB, Liu ET, Miller LD, Smeds J, Bergh J, Katzenellenbogen BS (2006) Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res 66: 7334–7340 [DOI] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS (2003) Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144: 4562–4574 [DOI] [PubMed] [Google Scholar]
- Frasor J, Stossi F, Danes JM, Komm B, Lyttle CR, Katzenellenbogen BS (2004) Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res 64: 1522–1533 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE, Welboren W, Versteeg R, Cuppen E, van de Wetering M, Clevers H, Stunnenberg HG (2008) Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol 28: 2732–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A (2006) Pure oestrogen antagonists for the treatment of advanced breast cancer. Endocr Relat Cancer 13: 689–706 [DOI] [PubMed] [Google Scholar]
- Hur J, Chesnes J, Coser KR, Lee RS, Geck P, Isselbacher KJ, Shioda T (2004) The Bik BH3-only protein is induced in estrogen-starved and antiestrogen-exposed breast cancer cells and provokes apoptosis. Proc Natl Acad Sci USA 101: 2351–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, Waxman S, Germain D (2008) Tamoxifen stimulates the growth of cyclin D1-overexpressing breast cancer cells by promoting the activation of signal transducer and activator of transcription 3. Cancer Res 68: 852–860 [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL (2008) Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell 29: 611–624 [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B (2005) A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL (2007) Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol 27: 5090–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kininis M, Isaacs GD, Core LJ, Hah N, Kraus WL (2009) Post-recruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol Cell Biol 29: 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, Fan JB, Duan L, Glass CK, Rosenfeld MG, Fu XD (2007) Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci USA 104: 4852–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzino M, De Amicis F, McPhaul MJ, Marsico S, Panno ML, Andò S (2005) Endogenous coactivator ARA70 interacts with estrogen receptor alpha (ERalpha) and modulates the functional ERalpha/androgen receptor interplay in MCF-7 cells. J Biol Chem 280: 20421–20430 [DOI] [PubMed] [Google Scholar]
- Leung LK, Do L, Wang TT (1998) Regulation of death promoter Bak expression by cell density and 17 beta-estradiol in MCF-7 cells. Cancer Lett 124: 47–52 [DOI] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y et al. (2007) Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet 3: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS (2002) An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol 20: 835–839 [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M (2008) FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Münch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerkord CL, Takahashi Y, Araya R, Takada N, Weiss RS, Wang HG (2008) Loss of Hus1 sensitizes cells to etoposide-induced apoptosis by regulating BH3-only proteins. Oncogene 27: 7248–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R, van Tinteren H, Balkenende A, Vermorken JB, Benraadt J, Huldij J, van Diest P (2002) Cyclin A is a prognostic indicator in early stage breast cancer with and without tamoxifen treatment. Br J Cancer 86: 402–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K (2007) RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler Y, Camp RL, Giltnane JM, Moeder C, Rimm DL, Kluger HM, Kluger Y (2008) Expression patterns and prognostic value of Bag-1 and Bcl-2 in breast cancer. Breast Cancer Res 10: R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV (2008) DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319: 202–206 [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT (2008) Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL (2006) Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20: 2922–2936 [DOI] [PubMed] [Google Scholar]
- Pink JJ, Jordan VC (1996) Models of estrogen receptor regulation by estrogens and antiestrogens in breast cancer cell lines. Cancer Res 56: 2321–2330 [PubMed] [Google Scholar]
- Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME (2005) GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat 92: 141–149 [DOI] [PubMed] [Google Scholar]
- Ramkumar T, Adler S (1995) Differential positive and negative transcriptional regulation by tamoxifen. Endocrinology 136: 536–542 [DOI] [PubMed] [Google Scholar]
- Shadeo A, Lam WL (2006) Comprehensive copy number profiles of breast cancer cell model genomes. Breast Cancer Res 8: R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103: 843–852 [DOI] [PubMed] [Google Scholar]
- Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, Denissov S, Stunnenberg HG, Lohrum M (2008) Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res 36: 3639–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender JD, Frasor J, Komm B, Chang KC, Kraus WL, Katzenellenbogen BS (2007) Estrogen-regulated gene networks in human breast cancer cells: involvement of E2F1 in the regulation of cell proliferation. Mol Endocrinol 21: 2112–2123 [DOI] [PubMed] [Google Scholar]
- Stossi F, Likhite VS, Katzenellenbogen JA, Katzenellenbogen BS (2006) Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J Biol Chem 281: 16272–16278 [DOI] [PubMed] [Google Scholar]
- Walker G, MacLeod K, Williams AR, Cameron DA, Smyth JF, Langdon SP (2007) Insulin-like growth factor binding proteins IGFBP3, IGFBP4, and IGFBP5 predict endocrine responsiveness in patients with ovarian cancer. Clin Cancer Res 13: 1438–1444 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA (2007) RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables
Supplementary Figures