Abstract
In budding yeast Saccharomyces cerevisiae, telomere length maintenance involves a complicated network as more than 280 telomere maintenance genes have been identified in the nonessential gene deletion mutant set. As a supplement, we identified additional 29 telomere maintenance genes, which were previously taken as essential genes. In this study, we report a novel function of Sua5p in telomere replication. Epistasis analysis and telomere sequencing show that sua5Δ cells display progressively shortened telomeres at early passages, and Sua5 functions downstream telomerase recruitment. Further, biochemical, structural and genetic studies show that Sua5p specifically binds single-stranded telomeric (ssTG) DNA in vitro through a distinct DNA-binding region on its surface, and the DNA-binding ability is essential for its telomere function. Thus, Sua5p represents a novel ssTG DNA-binding protein and positively regulates the telomere length in vivo.
Keywords: DNA binding, OB-fold, Sua5, telomerase, telomere
Introduction
Telomeres are highly organized DNA–protein structures at the ends of linear eukaryotic chromosomes (McEachern et al, 2000). They not only help to distinguish chromosome ends from DNA double-strands breaks, but also provide a way to replicate chromosomes completely. The telomeric DNA consists of simple repetitive sequence with the G-rich strand extending in the 3′ direction to form a single-stranded tail (Vega et al, 2003) and is bound and protected by specific proteins (Smogorzewska and de Lange, 2004). The single-stranded telomeric (ssTG) G-rich DNA-binding proteins, including Oxytricha nova TEBPα/β, mammalian Pot1 and TPP1, fission yeast Pot1, budding yeast Cdc13p and Stn1p (Horvath et al, 1998; Baumann and Cech, 2001; Lei et al, 2003; Mitton-Fry et al, 2004; Gao et al, 2007; Wang et al, 2007), contain an OB (oligonucleotide/oligosaccharide binding)-fold, which is responsible for ssTG binding (Theobald et al, 2003). In addition to telomere protection (Nugent et al, 1996; Gao et al, 2007), the OB-fold ssTG-binding proteins also participate in telomerase recruitment (Evans and Lundblad, 1999) and/or activity regulation (Wang et al, 2007).
Telomere is elongated by the specialized reverse transcriptase telomerase (Blackburn, 1992). In budding yeast Saccharomyces cerevisiae, telomeres in each chromosome have approximately 350 bp heterogeneous TG1–3/C1–3A sequence, and telomerase is composed of at least four subunits, Est1p, Est2p, Est3p and TLC1 RNA (Lundblad and Szostak, 1989; Singer and Gottschling, 1994; Lendvay et al, 1996). Est2p, the catalytic subunit of telomerase (Counter et al, 1997), and TLC1, the RNA subunit (Singer and Gottschling, 1994), are required for the elongation activity of oligo primer in vitro, and comprise the catalytic core of telomerase (Counter et al, 1997; Lingner et al, 1997). Est1p interacts with Cdc13p to recruit and activate telomerase (Evans and Lundblad, 1999; Taggart et al, 2002). Est3p is an OB-fold protein (Lee et al, 2008; Young Yu et al, 2008) and its molecular function is not clear. Telomerase-dependent telomere elongation is mainly achieved through effective telomerase recruitment and activation (Smogorzewska and de Lange, 2004). Telomerase is recruited to telomere through Ku80p–TLC1 RNA interaction in G1 phase (Peterson et al, 2001; Stellwagen et al, 2003; Fisher et al, 2004), and synergistic action of ssDNA-binding proteins Cdc13p and RPA in late S phase (Evans and Lundblad, 1999; Taggart et al, 2002; Schramke et al, 2004). Telomere addition is inhibited by Pif1p, a 5′–3′ DNA helicase, which disassociates telomerase from telomeric DNA by unwinding RNA–DNA substrates (Zhou et al, 2000; Boule et al, 2005). Telomerase preferentially binds shorter telomeres to promote their elongation in a Tel1-dependent manner in vivo (Bianchi and Shore, 2007; Chang et al, 2007; Hector et al, 2007; Sabourin et al, 2007). Deregulation of telomerase protein/RNA levels or assembling will affect telomere homeostasis in an indirect manner. For example, deficiency of nonsense mRNA decay (NMD) pathway, which controls mRNA levels of EST1, EST2 and other telomere regulators, shortens telomere lengths (Dahlseid et al, 2003).
The genome-wide screenings in the nonessential gene deletion set have revealed a telomere maintenance network of about 280 nonessential genes (Askree et al, 2004; Gatbonton et al, 2006). The telomere maintenance genes are very diverse in their functions, including DNA and RNA metabolism, chromatin modification and vacuolar traffic-related genes (Askree et al, 2004; Gatbonton et al, 2006). In a genome-wide characterization of essential genes, we unexpectedly identified additional 64 nonessential genes, which were previously considered as essential genes in genome-wide deletion project (Giaever et al, 2002). As a supplement, we examined the telomere lengths of these mutants. Here, we document the screening result and describe a new telomere maintenance gene, SUA5. SUA5 (suppressor of upstream ATG) was first identified in a genetic screening of suppressor for translation initiation defect in the leader region of the CYC1 gene (Na et al, 1992). Eight SUA genes were identified in the screening: SUA7 and SUA8 encode homologs of transcription factor TFIIB and RNA polymerase II largest subunit B220, respectively, which are required for transcription start site selection (Pinto et al, 1992, 1994). SUA1 and SUA6, which latterly were renamed as NMD2 and UPF3, are important factors in NMD pathway (Cui et al, 1995). SUA5 encodes a 46-kDa protein, which has a conserved yrdC domain at the N terminus and a SUA5 domain at the C terminus, and its biological function is still unclear.
Our genetic, biochemical and structural analysis revealed that Sua5p is a conserved non-OB-fold ssTG DNA-binding protein. It positively regulates telomere elongation through affecting telomerase activity, and this positive regulatory function requires its DNA-binding ability.
Results
Deletion of SUA5 causes telomere progressively shortening
In a genome-wide characterization of essential genes in S. cerevisiae (FL Meng and JQ Zhou, in preparation), we found that 64 nonessential genes were mis-classified as essential genes in the Saccharomyces genome deletion project (Giaever et al, 2002) (representative tetrad dissection result is shown in Supplementary Figure 1). As a supplement to the screenings in nonessential gene deletion set, we examined the telomere lengths, and identified 29 additional telomere maintenance genes (Supplementary Table II). The mutant cells were streaked on YPD plate for five times after sporulation. Interestingly, three deletion mutants exhibited progressively shortened telomeres during passages (Figure 1A): MTR10 has been reported to be required for the normal accumulation of mature TLC1 RNA and its proper nuclear localization (Ferrezuelo et al, 2002); KAE1 encodes a putative peptidase, which is one component of telomere regulator KEOPS complex (Downey et al, 2006); SUA5 has not been reported to affect telomeres.
Figure 1.
SUA5 deletion causes telomere shortening. (A) Telomere blot shows telomere shortening in early passages (within 165 generations after sporulation) of the MTR10, KAE1 and SUA5 deletion strains. The ‘1st' and ‘5th' indicate the first- and fifth-restreaked cells, respectively. The genomic DNA was digested with XhoI, and hybridized with a telomeric TG probe. (B) The sua5Δ cells were restreaked on YPD plate for 15 times (about 400 generations after sporulation), and the restreaked times are labelled on the top of the panel. The genomic DNA was digested with PstI. PstI and XhoI digestions (panel A) of the wild-type yeast genomic DNA result in approximately 0.8 and approximately 1.3 kb terminal telomere signals, respectively. PstI digestion reveals subtle telomere length changes. (C) A CEN plasmid-borne SUA5 progressively restores the telomere lengths in sua5Δ cells. The numbers indicate the restreaked times. (D) Telomere blot analysis of the genetic interactions of SUA5 and some well-characterized telomere maintenance genes. The isogenic strains are labelled on the top of the panel. The cells were restreaked on YPD plate for three times after sporulation.
The progressively shortened telomere phenotype is reminiscent of the ever shorter telomere (EST) phenotype in telomerase-deficient cells (Lundblad and Szostak, 1989). The sua5Δ cells were continuously passaged for approximately 400 generations after sporulation. Telomeres were progressively shortened during early passages (approximately 150 generations after sporulation), and then kept at a short-and-stable state in the successive passages (Figure 1B, PstI digested). The short telomeres could be gradually restored to wild-type length when SUA5 was re-introduced into sua5Δ cells (Figure 1C), indicating that the short telomeres resulted from the SUA5 deletion rather than other mutations. SUA5 deletion in YPH499 background displayed a similar progressively shortened telomere phenotype (data not shown).
To investigate how Sua5 affects telomere replication, we examined the genetic interaction of Sua5 with some well-characterized telomere regulators. SUA5 deletion further shortened telomeres in yku70Δ, yku80Δ, rad50Δ, rad9Δ, dot1Δ and tel1Δ cells (Figure 1D), suggesting that Sua5p plays an independent positive regulatory role in telomere length control. SUA5 deletion cells express low levels of CYC1, and harbour dysfunctional mitochondria, that is, respiratory incompetent (ρ−) (Na et al, 1992; Supplementary Figure 2A). However, telomere length did not show significant change in cyc1Δ cells (Supplementary Figure 2B; Askree et al, 2004; Gatbonton et al, 2006), and loss of mitochondrial DNA did not affect telomere length (Supplementary Figure 2B). In addition, consistent with the earlier report (Cui et al, 1995), SUA5 is not involved in NMD pathway (Supplementary Figure 2C), and SUA5 deletion further shortened telomeres of nmd2Δ and upf3Δ cells (Figure 1D). These results indicated that telomere shortening in sua5Δ is not caused by either the dysfunctional mitochondria or the defective NMD pathway.
Telomeric DNA in sua5Δ cells is not efficiently elongated
Telomere length is maintained by a dynamic process of lengthening and shortening. The balance between DNA loss and gain ensures telomeres with a constant length, and also defines an average point of divergence (APD) as yeast telomerase adds new variable sequences to telomere ends (Forstemann et al, 2000). In the proximal region of the divergence point telomeres have a stable sequence, whereas the distal telomere sequences are divergent (Forstemann et al, 2000). To find out the defect(s) of telomere replication caused by Sua5 deletion, Telomere I-L (TEL01L) was cloned and sequenced by Telomere PCR (Forstemann et al, 2000; Lee et al, 2007). The PCR-amplified TEL01L showed a decreased size during the early passages of sua5Δ cells (Figure 2A; Supplementary Figure 3A), which is consistent with the overall telomere length changes shown by telomere blot (Figure 1B). The APD in wild type and the first-restreaked sua5Δ cells was 161 nucleotides (nt) and 194 nt into the telomeric tract, respectively (Figure 2B), indicating that the depletion of Sua5p has not enhanced the nucleolytic degradation of telomeric DNA. The temperature sensitivity and quantification of telomeric single-stranded DNA also suggested that Sua5p is not involved in telomere end protection (Supplementary Figure 4A and B). Additionally, depletion of exonuclease Exo1p has no effect on the telomere length of sua5Δ cells (Supplementary Figure 4C). Therefore, Sua5p is not a telomere capping factor.
Figure 2.
Telomere sequencing reveals the defects of telomere replication in sua5Δ cells. (A) The analysis of Telomere I-L in wild type, first and fifth restreaked sua5Δ cells. Each column represents one sequenced telomere. The result of a representative clone of sua5Δ strain is shown here. (B) Statistics of the telomere sequencing results. The average point of divergence (APD) describes the average length of nondiverging telomere sequence, whereas the average length of divergence (ALD) stands for the average length of diverging telomere sequence. The results were summarized from two independent experiments shown in Supplementary Figure 3B. (C) Longer telomeres are not efficiently elongated in sua5Δ. The average lengths of the telomere extensions are determined for nondivergent telomeres that are longer or shorter than 170 nt in length. Telomeres shorter than 100 nt are excluded from the analysis because of the reported high recombination rate for short telomeres at TEL01L. The P-values associated with two-tailed unpaired t tests are shown. (D) Length of telomere extension as function of telomere length. The lengths of diverging regions are plotted as a function of the nondivergent sequence, which represents the original telomere sequence. (E) Frequency of telomere extension as function of telomere length. Telomere sequences are ordered according to the nondivergent sequence length and pooled into subgroups containing 10 telomeres each. The frequency of elongation in each subgroup is calculated and plotted as a function of telomere length.
The average length of divergence (ALD) sequences added by telomerase is 101 nt in wild-type cells, whereas the ALDs decreased to 16 and 27 nt in the first- and fifth-restreaked sua5Δ cells, respectively, indicating that telomerase activity is greatly reduced in sua5Δ cells. To analyse the relationship of telomere elongation and original length in sua5Δ cells, we counted the average nucleotide addition length per telomere for the telomeres that are longer and shorter than 170 nt, respectively. Telomeres that are shorter than 100 nt were excluded from the analysis, as the reported high recombination rate for short telomeres at chromosome I-L (Teixeira et al, 2004). The average lengths of nucleotides added at the longer telomeres in sua5Δ cells are significantly decreased when the telomere addition length as function of telomere size was plotted (Figure 2C and D). The sequencing result showed that many long telomeres are not elongated. Ten telomeres with close length of nondivergent sequence were grouped into subgroups, and the frequencies of telomere extension as function of telomere size was plotted (Figure 2E). The frequency of elongation in wild-type cells did not correlate significantly with telomere size after several cell-cycles replication (Figure 2E). This result is different from the telomere addition in one cell cycle revealed by the STEX method (Teixeira et al, 2004). In the first-restreaked sua5Δ cells, the elongation frequency negatively correlated with the telomere length (Figure 2E), and 93% of the telomeres that are shorter than 125 nt were elongated (Figure 2B), but only 23% of the telomeres that are longer than 150 nt were elongated (Figure 2B). Taken together, these data indicated telomerase activity is affected in sua5Δ cells and limited telomerase activity is allocated to the shorter telomeres (Chang et al, 2007), and longer telomeres are less frequently elongated.
Sua5 functions in telomerase pathway
As Sua5p appears not to be involved in telomere capping, it is possible that Sua5p functions in the telomerase pathway. We then examined the telomere length in the continuously passaged est1Δ and est1Δsua5Δ cells. The telomere shortening rate of sua5Δest1Δ was similar to that of est1Δ cells (Figure 3A). The result showed that deletion of SUA5 does not accelerate the telomere shortening caused by EST1 deletion, and SUA5 regulates telomere replication through the telomerase pathway.
Figure 3.
SUA5 deletion does not affect telomerase expression and assembly. (A) SUA5 deletion does not increase the telomere shortening rate in the telomerase-deficient cells. The isogenic strain of est1Δ or est1Δ sua5Δ was continuously passaged and the genomic DNAs were digested with XhoI and subjected to telomere blot. (B) SUA5 is required for the elevated gross chromosomal rearrangements (GCRs) rates observed in pif1Δ cells. GCR rates in pif1Δ, sua5Δ and pif1Δsua5Δ strains were analysed as reported (Myung et al, 2001). (C) The protein levels of telomerase components and Cdc13p are not affected by SUA5. The Myc-tagged proteins were examined by western blot with an anti-Myc antibody. The triangle (Δ) indicates unknown proteins cross-reacted with the antibody, which also serve as loading controls. The asterisked (*) bands might be the degradation products of Cdc13p. (D) The expression of TLC1 is not affected by SUA5. Northern blot analysis of total RNAs from the indicated strains with TLC1 and snoRNA probes. (E) Sua5p is not associated with TLC1. The myc-tagged proteins were immunoprecipitated with the anti-myc antibody, and TLC1 was detected with semiquantitative PCR. (F) SUA5 is not involved in TLC1 biogenesis. TLC1 RNA was over-expressed under the control of GPD promoter. The mtr10Δ mutant, which has a defect in TLC1 trafficking, serves as a positive control. (G) SUA5 does not affect telomerase assembling. The myc-tagged Est2p was immunoprecipitated, and co-immunoprecipitated TLC1 was detected with semiquantitative PCR.
To further test whether Sua5p is required for the in vivo telomerase activity, we used the gross chromosomal rearrangement (GCR) assay (Myung et al, 2001) to measure the rate of telomere healing when telomerase acts on DNA double-strand breaks. In pif1Δ cells, the GCR rate is dramatically increased through efficient telomere healing (Myung et al, 2001). SUA5 deletion significantly suppressed the GCR rate in pif1Δ cells (Figure 3B), suggesting that SUA5 affects telomerase activity in vivo.
Sua5 does not affect telomerase expression and assembling
Earlier studies have suggested that SUA genes might be involved in gene transcription and RNA metabolism (Pinto et al, 1992, 1994; Cui et al, 1995), we wondered whether the shorter telomeres in sua5Δ cells are caused by low level of telomerase or defect of telomerase assembling. With this in mind, we first examined the protein levels of telomerase components (Est2p, Est1p and Est3p) and Cdc13p in SUA5 and sua5Δ cells. Neither telomerase components nor Cdc13p showed any significant changes in sua5Δ cells (Figure 3C). Then, we checked the TLC1 RNA. Northern blot analysis showed that TLC1 RNA level in sua5Δ cells was comparable to that in wild-type cells (Figure 3D). In addition, Sua5p appeared not to associate with TLC1 in vivo as revealed by the co-immunoprecipitation-PCR experiment (Figure 3E). Moreover, over-expression of TLC1 could not elongate the short telomeres in sua5Δ cells (Figure 3F; Supplementary Figure 5). Furthermore, the interaction of Est2p and TLC1 is not affected in sua5Δ cells (Figure 3G). These data suggested that Sua5p does not affect expression of telomerase subunits and TLC1, nor influences telomerase assembling.
Sua5 functions downstream telomerase recruitment
Telomerase-dependent telomere elongation is mainly achieved through telomerase recruitment and activation (Smogorzewska and de Lange, 2004). The G1-phase telomere association of telomerase is abolished in Ku–Tlc1 interaction-deficient mutants (yku80-135i or tlc1-Δ48) (Peterson et al, 2001). We examined the telomere lengths of sua5Δ yku80-135i and sua5Δ tlc1-Δ48 double mutants, and found that SUA5 deletion further shortened the telomeres of the Ku–Tlc1 interaction-deficient mutants (Figure 4A), indicating that SUA5 functions independent of G1-phase telomerase recruitment. The S-phase telomerase recruitment can be bypassed by the introduction of Cdc13–Est2p or Cdc13–Est1p fusion protein (Evans and Lundblad, 1999; Schramke et al, 2004). If Sua5 participates in this or upstream step of telomerase recruitment, it would be bypassed by the presence of Cdc13–Est2p fusion protein. Accordingly, we observed the same dramatic telomere length increase in the wild-type cells, which harbour the fusion Cdc13–Est2p or Cdc13–Est1p proteins (Figure 4B). Surprisingly, telomere over-elongations by fusion proteins were compromised in sua5Δ cells (Figure 4B). In addition, the chromatin immunoprecipitation (ChIP) results showed that bindings of telomerase components Est2p, Est1p and Est3p to telomere were not changed in sua5Δ cells (Figure 4C). We, therefore, concluded that Sua5p functions downstream telomerase recruitment.
Figure 4.
Sua5p does not affect telomerase recruitment. (A) Deletion of SUA5 shortens the telomere lengths of the Ku–Tlc1 interaction-deficient mutants (tlc1Δ-48 or yku80-135i). The genomic DNAs were digested with XhoI and subjected to telomere blot. (B) Cdc13–Est2p or Cdc13–Est1p fusion protein cannot over-elongate the shortened telomeres in sua5Δ cells. (C) The association of telomerase to telomeres is not affected. The association of telomerase to telomeres was detected with ChIP-qPCR in sua5Δ and wild-type cells. (D) Sua5 functions in the same pathway as Pif1, but not Rif1. The isogenic strains are labelled on the top of the panel, and the numbers indicate the restreaked times on YPD plate. (E) Sua5p does not affect telomerase activity in vitro. Purified telomerase (Liao et al, 2005) was incubated with 1 μM Tel15 (5′-TGTGGTGTGTGTGGG-3′) and indicated amount of recombinant Sua5p. LC, loading control.
Sua5 functions epistatic to Pif1 in the telomerase pathway
Both Rif1 and Pif1 negatively regulate telomere elongation, and their deletion resulted in longer telomeres (Wotton and Shore, 1997; Zhou et al, 2000). Rif1 is an essential component of telomere length counting machinery (Marcand et al, 1997), whereas Pif1p inhibits telomerase activity through disassociating telomerase from telomeric DNA (Zhou et al, 2000; Boule et al, 2005). To further validate that Sua5p affects telomerase pathways, we examined the telomere lengths of sua5Δpif1Δ and sua5Δrif1Δ double-mutant cells when they were passaged about 350 and 275 generations (i.e. 14 and 11 times restreaking on plate), respectively. The long telomeres caused by PIF1 or RIF1 deletion were gradually shortened when Sua5p is absent (Figure 4D). Interestingly, Compared with telomeres of the sua5Δrif1Δ cells, which finally kept at wild-type length, telomeres of sua5Δ pif1Δ cells had similar telomere length to sua5Δ cells (Figure 4D). This result indicated that SUA5 functions epistatic to PIF1 in the telomerase pathway. To ask whether Sua5p has any direct influence on telomerase activity in vitro, we purified recombinant Sua5p (Figure 5C), and added it into the reaction mixture of telomerase activity assay (Liao et al, 2005). Neither the nucleotide addition activity nor the processivity of telomerase core enzyme was affected (Figure 4E).
Figure 5.
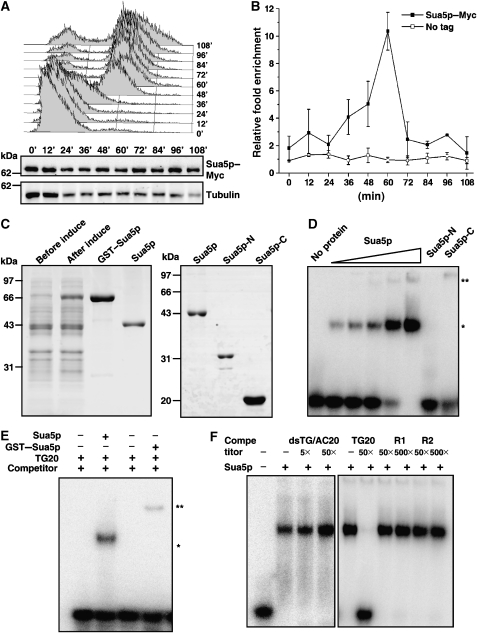
Sua5p interacts with telomeric DNA. (A) The expression of Sua5p remains unchanged during the cell cycle. The myc-tagged SUA5 cells were arrested at G1 phase with α-factor, and allowed to progress into S and G2/M phases (upper panel). Sua5p was examined by western blot (middle panel). Tubulin serves as a loading control (lower panel). (B) Sua5p preferentially binds telomeres at late S phase of cell cycle. ChIP-qPCR was performed to analyse the association of Sua5p to telomeric DNA. (C) Purification of Sua5p proteins in E. coli. The GST–fusion proteins were over-expressed, and purified with the glutathione-affinity column. The GST-tag was cleaved by PreScission protease. The purified proteins were analysed by Coomassie blue stained SDS–PAGE. (D) Full-length Sua5p, but not Sua5p-N or Sua5p-C, binds ssTG DNA. Radioactively labelled oligo TG20 (2.4 nM) was incubated with increasing amounts of purified Sua5p (0.1, 0.27, 0.54, 1 and 2.1 μM, respectively), Sua5p-N (2 μM) or Sua5p-C (2 μM). The shifted Sua5p–DNA bands are indicated with a single asterisk, and super-shifted bands, which may represent higher-ordered protein–DNA complex, are indicated with a double asterisk. (E) GST–Sua5p–DNA complex (**) migrates slower than Sua5p–DNA complex (*). About 0.5 μM of GST–Sua5p and 1 μM of Sua5p were added in the reactions. (F) Sua5p specifically binds ssTG DNA. Competition assays were performed in the presence of the 5- or 50-fold excess of double-stranded telomeric DNA, 50-fold excess of unlabelled TG20 and 50- or 500-fold excess random-sequence oligos R1 and R2. About 2 μM Sua5p was used in each of the reactions.
Sua5p specifically interacts with telomeric DNA in vivo and in vitro
To find out whether Sua5p could specifically interact with telomere in vivo, a ChIP experiment was carried out as described earlier (Taggart et al, 2002). The chromosomal SUA5 gene was tagged with a Myc epitope, which did not affect telomere length (data not shown). The amount of Sua5p in cells remains unchanged along the cell cycle (Figure 5A), whereas the ChIP-qPCR result showed that Sua5p is associated with telomeric DNA, and the interaction peaked at late S phase (Figure 5B). These results indicated that the Sua5p interacts with telomeres in vivo, and its effect on telomere replication is direct.
The primary sequence analysis revealed that Sua5p is an evolutionarily conserved protein (Supplementary Figure 6A). Over-expression of either Candida albicans or Schizosaccharomyces pombe homolog of Sua5p in sua5Δ S. cerevisiae cells could rescue the telomere shortening phenotype (Supplementary Figure 6B), suggesting that the Sua5p's function in telomere maintenance might be conserved. Sua5p consists of N terminal yrdC and the C terminal SUA5 domains (Bateman et al, 2004). Both Escherichia coli yrdC and thermoacidophilic archaeon Sulfolobus tokodaii Sua5 have been proposed to bind nucleic acids (Teplova et al, 2000; Agari et al, 2008). However, their biochemical activity has not been formally shown. We over-expressed and purified the Sua5p, as well as the N terminal fragment (aa 1–250, named Sua5p-N), which contains yrdC domain, and the C terminal fragment (aa 251–426, named Sua5p-C), which contains SUA5 domain (Figure 5C). Interestingly, we found that the recombinant full length Sua5p, but not Sua5p-N or Sua5p-C, could bind single-stranded yeast telomeric DNA (Figure 5D), suggesting that the DNA binding of Sua5p requires both the yrdC and Sua5 domains. The amount of shifted DNA increased with higher concentrations of Sua5p (Figure 5D, asterisked) and a higher-shift band was detected when the GST–Sua5p was added in the reaction (Figure 5E, double asterisked), indicating that the shifted band is Sua5p–DNA complex. The ssTG DNA binding was completely competed out by the excess amount of unlabelled ssTG DNA, but not the dsTG/AC DNA or nontelomeric single-stranded DNA oligos (Figure 5F). Sua5p could not bind either ssCA or dsTG/AC DNA (Supplementary Figure 7). These results showed that Sua5p specifically binds ssTG DNA in vitro. To determine the consensus sequence for Sua5p binding, we made a series mutant oligos based on TG20 sequence (Table I), and carried out a systematical-affinity analysis. The binding affinity was significantly decreased when the core telomere sequence was mutated (Table I), which indicated that Sua5p binds to the core telomere sequence TGGGTGT.
Table 1.
Affinity changes for binding of the Sua5p to mutated TG20 oligos
| Primer | (TG)0–6TGGGTGT(G)0–1 | Relative Kd | ΔΔG (kcal/mol) ensemble |
|---|---|---|---|
| TG20 | TGGTGTGTGTGGGTGTGGTG | 1 | |
| TG201 | TaaTGTGTGTGGGTGTGGTG | 3.7 | 0.79 |
| TG202 | TGGTaaGTGTGGGTGTGGTG | 7.4 | 1.20 |
| TG203 | TGGTGTaTGTGGGTGTGGTG | 1.2 | 0.14 |
| TG204 | TGGTGTGTaTGGGTGTGGTG | 3.6 | 0.78 |
| TG205 | TGGTGTGTGaGGGTGTGGTG | 1.1 | 0.034 |
| TG214 | TGGTGTGTGTaGGTGTGGTG | 38 | 2.19 |
| TG206 | TGGTGTGTGTGaaTGTGGTG | 175 | 3.11 |
| TG215 | TGGTGTGTGTGaGTGTGGTG | 439 | 3.66 |
| TG216 | TGGTGTGTGTGGaTGTGGTG | 36.8 | 2.16 |
| TG207 | TGGTGTGTGTGGGTaTGGTG | 1.1 | 0.065 |
| TG208 | TGGTGTGTGTGGGTGTaaTG | 0.78 | −0.15 |
The DNA-binding ability of Sua5p is crucial for its functions
The 3D structures of E. coli yrdC protein, S. tokodaii Sua5 protein (StSua5p) and the SUA5 domain of Pyrococcus horikoshii Sua5 protein (PhSua5p) have been solved (Teplova et al, 2000; Agari et al, 2008; Agari Y, unpublished data). The sequence alignment of S. cerevisiae Sua5p (ScSua5p) and StSua5p revealed 37% identity and 50% similarity (Supplementary Figure 8A). According to structure of StSua5p (PDB: 2EQA), we modelled the structure of ScSua5p (Figure 6A). The surface potential prediction had revealed a positive-charged region at the interface of these two domains (Figure 6B). As neither yrdC nor SUA5 domain alone could bind telomeric DNA (Figure 5D), we speculated that the DNA-binding region of Sua5p may be within the positive-charged region. To further examine the DNA-binding ability and validate the modelled Sua5p structure, we constructed 48-point mutants of SUA5. Most of the mutated residues are conserved among species or on the surface of Sua5p. In all, 19 of 48 alleles failed to restore telomere length (Supplementary Table III), and most of these lost-of-function mutations (15/19) located at the positive-charged region (Figure 6C; Supplementary Figure 8B).
Figure 6.
A positive-charged region of Sua5p is crucial for its function. (A) Modelled 3D structure of S. cerevisiae Sua5p. The modelled structure of S. cerevisiae Sua5p (green) is aligned with the template structure of S. tokodaii Sua5p (grey). (B) The relative protein surface potential is predicted and displayed with PyMOL software. A positive-charged region at the interface of yrdC and SUA5 domains is shown. (C) The point mutation in the positive-charged surface region of Sua5p affects its telomere maintenance function. Representative mutants, including K93A, R95A, D98K, R248A and R414D in the positive-charged region and S107F outside the positive-charged region, are pointed out in the upper panel. The mutant telomere lengths were examined (lower panel). (D) The point mutation in the positive-charged region of Sua5p abolishes its DNA-binding ability. 1 μM of wild-type or mutant Sua5 protein was used, and 50-fold excess of the random-sequence oligos were added into each reaction as competitor (upper panel). The purified wild-type and mutant Sua5 proteins are shown by Coomassie blue stained gel (lower panel).
To examine the in vitro activity of these mutants, six recombinant mutant proteins were purified as wild-type Sua5p (Figure 6D, lower panel). The mutants at the positive-charged region (K93, R95, D98, R248 and R414) (Figure 6C, upper panel) exhibited little DNA-binding activity (Figure 6D, upper panel), whereas the sua5-1 mutant protein (S107F, outside the positive-charged region), which does not affect telomere length (Figure 6C), exhibited wild-type level of telomeric DNA-binding ability (Figure 6D). These results confirmed that the positive-charged region in Sua5p is responsible for its DNA binding, and the DNA-binding activity is essential for its positive regulatory function in telomere replication.
Discussion
In this study, we have found that sua5Δ cells display progressively shortened telomeres during early passages in a large-scale screening. SUA5 functions downstream the telomerase recruitment and may regulate the telomerase activity. Further analyses have revealed that Sua5p specifically interacts with ssTG DNA in vitro through a distinct ssTG-binding region. Sua5p directly binds telomeres in vivo. The DNA-binding ability of Sua5p is essential for its telomere function.
A large-scale screening of new telomere maintenance genes
In a genome-wide characterization of essential genes (total 1094 genes) in S. cerevisiae, we unexpectedly found out that 64 genes were ‘mis-classified' as essential genes. The earlier genome-wide screenings of telomere maintenance genes in the nonessential gene deletion set have identified about 280 genes (Askree et al, 2004; Gatbonton et al, 2006), and our current study has added 29 more genes in this group. Some of the genes, for example, SWC4, HRR25 and PDS5, function in the process of chromatin remodelling or DNA metabolism (Supplementary Table II), and it is unsurprising that they play roles in telomere length regulation. Our screening has recovered the MTR10 and KAE1, which have been reported to regulate telomere length (Figure 1A). The INO80, which regulates telomere structure (Yu et al, 2007), has also been found to regulate telomere length (Supplementary Table II). Therefore, the new genes uncovered in our screening would provide new clues to understand telomere replication.
In the earlier screenings, nearly 5% of the yeast mutants were found to have abnormal telomeres (Askree et al, 2004; Gatbonton et al, 2006). Interestingly, in our current screening, 29 out of 64 nonessential genes (45%) appear to affect telomeres. A possible explanation is that most of the recovered mutants have severe growth defect, and they may affect telomeres indirectly. To find out the genes that are directly relevant to telomere replication, we examined the telomere changes during the early passages after sporulation. Sua5 particularly caught our attention because like mtr10Δ or kae1Δ cells, sua5Δ cells showed progressively shortened telomeres in their early passages (Figure 1), which is reminiscent of the EST phenotype seen in telomerase-deficient cells. MTR10 and KAE1 have been shown to regulate telomerase activity (Ferrezuelo et al, 2002; Downey et al, 2006), and the progressively shortening telomere phenotype of sua5Δ cells led us to suspect that SUA5 may regulate telomerase activity.
Sua5 may positively regulate telomerase activity
The imbalance between telomere lengthening and shortening in sua5Δ cells suggested SUA5 may function in telomere protection or telomerase activity regulation. Several lines of evidence argue against the telomere protection hypothesis: (1) sua5Δ mutant cell did not display temperature sensitivity, which is frequently observed in the telomere uncapping mutants (Supplementary Figure 4A); (2) the amount of single-stranded TG DNA in sua5Δ cells has not increased (Supplementary Figure 4B); (3) sua5Δ exo1Δ cells does not exhibit any growth advantage to sua5Δ cells, or telomere length restoration (Supplementary Figure 4C); (4) the telomere shortening rate of est1Δsua5Δ cells is comparable to that of est1Δ cells (Figure 3A) and (5) the APD of the first-restreaked sua5Δ cell is comparable to that of the wild-type cells (Figure 2B). These observations encouraged us to postulate that Sua5 may participate in the regulation of telomerase activity.
Telomerase recruitment and activation are two important steps for telomerase activity. SUA5 and Ku–Tlc1 interaction-deficient mutant function in different epistasis groups (Figure 4A). CDC13–EST2 and CDC13–EST1 fusion proteins cannot bypass the requirement of Sua5p for full telomerase activity (Figure 4B). In addition, the association of telomerase to telomeres is not changed in sua5Δ cells (Figure 4C). Therefore, Sua5p appears not to affect telomerase recruitment through Ku–Tlc1 at G1 phase, or through Cdc13–Est1 and RPA–Est1 at late S phase (Evans and Lundblad, 1999; Peterson et al, 2001; Taggart et al, 2002; Stellwagen et al, 2003; Fisher et al, 2004; Schramke et al, 2004).
In the absence of Sua5p, the elevated de novo telomere addition events in pif1Δ cells are dramatically decreased (Figure 3B), and the longer telomeres are less frequently elongated likely due to the limited telomerase activity (Figure 2). Apparently, the telomerase protein/RNA levels and assembling are not affected in sua5Δ cells (Figure 3). Sua5 functions epistatic to Pif1 in the telomerase pathway (Figure 4D), and Sua5p preferentially binds telomeres at late S phase (Figure 5B) as the telomerase does (Taggart et al, 2002). On the basis of these findings, we hypothesize that Sua5p may synergize the function of telomere-bound telomerase. In vitro, Sua5p seems not to affect telomerase activity (Figure 4E), raising the possibility that Sua5p may regulate telomerase activity through interaction with other telomere-binding proteins or telomeric DNA in vivo (Figures 5 and 6). It will be of great interest to investigate the genetic and physical interaction between Sua5 and the other telomere maintenance genes. The identification of Sua5p interacting proteins will help to explain its molecular mechanism.
Sua5p possesses a distinct ssTG DNA-binding structure
The OB-fold domain appears to be the fingerprint to many single-stranded nucleotide-binding proteins, including several ssTG DNA-binding proteins, that is, TEBPβ, Cdc13p, Stn1p, Pot1 and TPP1 (Horvath et al, 1998; Baumann and Cech, 2001; Lei et al, 2003; Mitton-Fry et al, 2004; Gao et al, 2007; Wang et al, 2007). Like these proteins, Sua5p could specifically bind ssTG DNA (Figure 5A). However, no OB-fold domain was founded in the Sua5p 3D structure (Figure 6A). Instead, both the yrdC and the SUA5 domains of Sua5p contributed to its interaction with single-stranded TG DNA (Figure 5D). Mutations in DNA-binding region at the interface abolished the interaction between Sua5p and ssTG DNA (Figure 6D), and caused the defects in telomere elongation (Figure 6C). The binding affinity of Sua5p (Kd=615 nM) is relatively low compared with Cdc13p (Kd=28 nM) under the same reaction condition. These data strongly suggested that the specific TG-rich DNA-binding mode of Sua5p represents a novel structure for ssTG-DNA recognition and/or binding.
One molecule of both AMP and Mg2+ ion were found in the crystal structure of StSua5p (Agari et al, 2008). The amino acids responsible for AMP and Mg2+ bindings are highly conserved in S. tokodaii and S. cerevisiae Sua5 proteins. Mutations of these amino acids cause loss of SUA5 function (Supplementary Table III). It has been reported that StSua5p has ATPase activity (Agari et al, 2008), but our recombinant fungal Sua5p (Supplementary Figure 9A) did not show any ATPase (Supplementary Figure 9B) or GTPase (Supplementary Figure 9C) activity. This discrepancy makes the function of AMP and Mg2+ confusing. It remains elusive whether and how AMP and Mg2+ contribute to the Sua5p function.
Evolutionary conservation of SUA proteins
The conserved yrdC domain by itself as well as conjugated with other motifs has been found in a broad-spectrum species (Teplova et al, 2000; Chen et al, 2003), and SUA5 domain exists in its self or at the C terminus of yrdC domain (Bateman et al, 2004). The homologs of Sua5p have been found in archaea, bacteria and eukaryotes. Bacterial yrdC protein is suggested to be involved in rRNA maturation (Kaczanowska and Ryden-Aulin, 2005). However, the rRNA maturation was not affected in S. cerevisiae when Sua5p was absent (Supplementary Figure 2D), and the shorter telomeres in sua5Δ mutant are not caused by an indirect translation defect.
In eukaryotes, most Sua5 homologs exist in Fungus Kingdom (Bateman et al, 2004). Therefore, it will be of great interest to investigate whether fungal Sua5p binds telomeric DNA and regulates telomeres in other yeast such as C. albicans and S. pombe. Unfortunately, Sua5 is essential for the growth of S. pombe and C. albicans, and we could not get the deletion mutant (data not shown). The over-expression of either S. pombe or C. albicans Sua5 protein in budding yeast can rescue the telomere shortening caused by the S. cerevisiae SUA5 deletion (Supplementary Figure 6B). In addition, the purified recombinant SpSua5p and CaSua5p are able to specifically bind ssTG DNAs (data not shown), suggesting Sua5p's function in telomere maintenance may be conserved among fungi.
It will be intriguing to test whether the human yrdC protein (Chen et al, 2003; Jiang et al, 2005) also affect telomere length. However, the function of Sua5p at telomere seems to be difficult to spread to its archaeobacterial or bacterial homologs as there is not DNA end replication problem in these organisms. Therefore, it is hard to interpret why fungal Sua5p has adapted a specific single-strand telomeric DNA-binding activity. One possibility is that Sua5p and its homologs may have a higher affinity to T(U)G-rich nucleotides. Another possibility is that the fungal Sua5p have gained a telomere regulation function during the evolution.
In conclusion, the results presented here have identified Sua5 as a novel ssTG DNA-binding protein, which is important for telomere length homeostasis. Elucidating the molecular mechanism of Sua5p function will shed light on the understanding of telomere replication.
Materials and methods
Yeast strains and plasmids
Strains used in this work are derivatives of BY4743 or YPH501 as indicated in Supplementary Table I. Yeast strains were constructed using standard genetic procedures either by genetic cross or by homologous recombination. Systematic deletion strains are from EUROSCARF. All gene disruptions and taggings were verified by PCR or gene-specific phenotypes. SUA5 CEN plasmids were constructed by cloning a 2.3 kb (from upstream 800 bp to downstream 200 bp of SUA5 open reading frame) PCR-amplified fragment of SUA5 in the BamHI-XhoI site of pRS316. The point mutations were introduced by site-directed mutagenesis.
Telomere blot
Genomic DNA was digested with XhoI or PstI, separated on agarose gels, transferred to Hybond-N membrane (GE Healthcare), and hybridized to a telomere probe. The screening experiments for telomere-length changes were started with two asci; each spore was restreaked five times on YPD plates after sporulation. Genomic DNAs of the first- and fifth-restreaked cells were subjected to telomere blot. Telomere length was determined with Image Quant TL software. Linear log curve stimulation was used to calibrate the molecular weight and Student's t test was applied to state the significant changes.
Telomere PCR
Telomere PCR was performed as described (Forstemann et al, 2000; Lee et al, 2007). The telomere ends of chromosome I-L were amplified using ‘o286S MluI' and ‘G18 BamHI' primers (Lee et al, 2007). PCR products were separated on 2% agarose gel, and cloned into the pMD18-T simple vector (Takara). Individual clones were sequenced by Majorbio.
Primer extension assay for telomerase activity
Telomerase purification and primer extension were performed as described earlier (Liao et al, 2005). Purified Sua5p was pre-incubated with telomerase at 30°C for 20 mins before initiating the reaction by adding dNTPs. Reaction products were analysed by 18% polyacrylamide-urea gel electrophoresis.
Recombinant protein purification and gel shift assay
SUA5 ORF and truncated fragments were cloned into pGEX-6p-1 vector. The GST fusion proteins were over-expressed and purified according to the manufacturer's instructions (GE Healthcare). GST tag was cut by PreScission protease, and Sua5p was further purified with Q-sepharose and Superdex-200 columns (GE Healthcare). About 2.4 nM 32P-labelled TG20 DNA (5′-TGGTGTGTGTGGGTGTGGTG-3′) was mixed with indicated amount of full length or fragments of Sua5p in 50 mM K-HEPES buffer (pH 7.5) containing 100 mM KCl, 5 mM MgCl2. The reaction mixture was incubated at 30°C for 40 min, and analysed with a 6% nondenaturing polyacrylamide gel. Oligonucleotide CA20 (5′-CACCACACCCACACACACCA-3′) was annealed with TG20 to form double-strand TG20/CA20. Two random-sequence oligonucleotides R1 (5′-TCAATGTTATACTTGACTACTTGC-3′) and R2 (5′-AGTACTAGGATATATCAGACTATG-3′) were used as nonspecific binding competitors.
ChIP analysis
ChIP was performed according to Taggart et al (2002). Sua5p was tagged with Myc epitopes, and Sua5p–Myc strain exhibits wild-type telomere length (data not shown). No-tag or Sua5p–Myc strain, which harbour bar1Δ was arrested at G1 phase by α-factor, and then released to allow cell-cycle progression. Cells were harvested at indicated time points and subjected to FACS and ChIP analysis. Myc-tagged proteins were immunoprecipitated with monoclonal α-Myc antibody (Sigma). Quantitative PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems).
Structure modelling of Sua5p
Initial models of ScSua5p were obtained with the MODELLER program (Sanchez and Sali, 2000) by using the structure of StSua5p (PDB: 2EQA) as template. Structures of E. coli yrdC (PDB: 1HRU), PhSua5p (PDB: 2YV4) were also used as references. Buried side chains were manually adjusted to avoid steric conflict or have favourable interactions with neighbouring residues using the graphics program Coot (Emsley and Cowtan, 2004), consulting reference databases of known main chain and side chain conformations and preferred side chain rotamers. The regions of ScSua5 residues aa 1–17, 27–44, 120–131, 267–291, 304–313 and 338–355 have no equivalent in the known structures and were excluded from the model. The quality and stereochemistry of the model were evaluated using the program PROCHECK (Laskowski et al, 1993). The main chain conformations for 99.07% of amino-acid residues were within the favoured or allowed regions of the Ramachandran plot. Models of AMP molecule and Mg2+ were based on the template structure of StSua5p.
Supplementary Material
Supplementary Figures 1–9
Supplementary Figure Legends
Supplementary Table I
Supplementary Table II
Supplementary Table III
Acknowledgments
We thank Daniel E Gottschling, Yasumasa Tsukamoto and Daochun Kong for strains and plasmids, and members of Zhou lab for discussion and critical reading of the paper. This work was supported by National Science Foundation of China (30630018) and Ministry of Science and Technology (2005CB522402, 2007CB914502) grants to JQZ, and Ministry of Science and Technology (2007CB914302) grant to JW.
References
- Agari Y, Sato S, Wakamatsu T, Bessho Y, Ebihara A, Yokoyama S, Kuramitsu S, Shinkai A (2008) X-ray crystal structure of a hypothetical Sua5 protein from Sulfolobus tokodaii strain 7. Proteins 70: 1108–1111 [DOI] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ (2004) A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA 101: 8658–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR (2004) The Pfam protein families database. Nucleic Acids Res 32: D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D (2007) Increased association of telomerase with short telomeres in yeast. Genes Dev 21: 1726–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH (1992) Telomerases. Annu Rev Biochem 61: 113–129 [DOI] [PubMed] [Google Scholar]
- Boule JB, Vega LR, Zakian VA (2005) The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438: 57–61 [DOI] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J (2007) Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev 21: 2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ji C, Gu S, Zhao E, Dai J, Huang L, Qian J, Ying K, Xie Y, Mao Y (2003) Isolation and identification of a novel cDNA that encodes human yrdC protein. J Hum Genet 48: 164–169 [DOI] [PubMed] [Google Scholar]
- Counter CM, Meyerson M, Eaton EN, Weinberg RA (1997) The catalytic subunit of yeast telomerase. Proc Natl Acad Sci USA 94: 9202–9207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Hagan KW, Zhang S, Peltz SW (1995) Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev 9: 423–436 [DOI] [PubMed] [Google Scholar]
- Dahlseid JN, Lew-Smith J, Lelivelt MJ, Enomoto S, Ford A, Desruisseaux M, McClellan M, Lue N, Culbertson MR, Berman J (2003) mRNAs encoding telomerase components and regulators are controlled by UPF genes in Saccharomyces cerevisiae. Eukaryot Cell 2: 134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, Emili A, Greenblatt JF, Harrington L, Lydall D, Durocher D (2006) A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124: 1155–1168 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (1999) Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Ferrezuelo F, Steiner B, Aldea M, Futcher B (2002) Biogenesis of yeast telomerase depends on the importin mtr10. Mol Cell Biol 22: 6046–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Taggart AK, Zakian VA (2004) Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol 11: 1198–1205 [DOI] [PubMed] [Google Scholar]
- Forstemann K, Hoss M, Lingner J (2000) Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res 28: 2690–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V (2007) RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A (2006) Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet 2: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW (2007) Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell 27: 851–858 [DOI] [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC (1998) Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95: 963–974 [DOI] [PubMed] [Google Scholar]
- Jiang W, Prokopenko O, Wong L, Inouye M, Mirochnitchenko O (2005) IRIP, a new ischemia/reperfusion-inducible protein that participates in the regulation of transporter activity. Mol Cell Biol 25: 6496–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowska M, Ryden-Aulin M (2005) The YrdC protein—a putative ribosome maturation factor. Biochim Biophys Acta 1727: 87–96 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V (2008) The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol 15: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kozak M, Martin JD, Pennock E, Johnson FB (2007) Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol 5: e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Podell ER, Baumann P, Cech TR (2003) DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature 426: 198–203 [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V (1996) Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao XH, Zhang ML, Yang CP, Xu LX, Zhou JQ (2005) Characterization of recombinant Saccharomyces cerevisiae telomerase core enzyme purified from yeast. Biochem J 390: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567 [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Krauskopf A, Blackburn EH (2000) Telomeres and their control. Annu Rev Genet 34: 331–358 [DOI] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS (2004) Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J Mol Biol 338: 241–255 [DOI] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD (2001) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076 [DOI] [PubMed] [Google Scholar]
- Na JG, Pinto I, Hampsey M (1992) Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics 131: 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE (2001) The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet 27: 64–67 [DOI] [PubMed] [Google Scholar]
- Pinto I, Ware DE, Hampsey M (1992) The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell 68: 977–988 [DOI] [PubMed] [Google Scholar]
- Pinto I, Wu WH, Na JG, Hampsey M (1994) Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J Biol Chem 269: 30569–30573 [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Zakian VA (2007) Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell 27: 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Sali A (2000) Comparative protein structure modeling. Introduction and practical examples with modeller. Methods Mol Biol 143: 97–129 [DOI] [PubMed] [Google Scholar]
- Schramke V, Luciano P, Brevet V, Guillot S, Corda Y, Longhese MP, Gilson E, Geli V (2004) RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat Genet 36: 46–54 [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T (2004) Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73: 177–208 [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE (2003) Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA (2002) Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Teplova M, Tereshko V, Sanishvili R, Joachimiak A, Bushueva T, Anderson WF, Egli M (2000) The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding. Protein Sci 9: 2557–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS (2003) Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct 32: 115–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega LR, Mateyak MK, Zakian VA (2003) Getting to the end: telomerase access in yeast and humans. Nat Rev Mol Cell Biol 4: 948–959 [DOI] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M (2007) The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Wotton D, Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11: 748–760 [DOI] [PubMed] [Google Scholar]
- Young Yu E, Wang F, Lei M, Lue NF (2008) A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol 15: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Steinberg-Neifach O, Dandjinou AT, Kang F, Morrison AJ, Shen X, Lue NF (2007) Regulation of telomere structure and functions by subunits of the INO80 chromatin remodeling complex. Mol Cell Biol 27: 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA (2000) Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science 289: 771–774 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–9
Supplementary Figure Legends
Supplementary Table I
Supplementary Table II
Supplementary Table III