Abstract
The elongation competence of the RNA polymerase II complex is critically dependent on the positive transcription elongation factor b (P-TEFb). P-TEFb exists in two forms in cells, an active form composed of cyclin T1 and CDK9 and an inactive form, in which cyclin T1/CDK9 is sequestered by Hexim1 and 7SK snRNA. Here, we report that partitioning of active and inactive P-TEFb is regulated by acetylation of cyclin T1. Cyclin T1 acetylation triggers dissociation of Hexim1 and 7SK snRNA from cyclin T1/CDK9 and activates the transcriptional activity of P-TEFb. This activation is lost in P-TEFb complexes containing cyclin T1 that can no longer be acetylated. An acetylation-deficient cyclin T1 mutant dominantly suppresses NF-κB-mediated activation of the interleukin-8 promoter but continues to synergize normally with the HIV Tat protein to transactivate the HIV long terminal repeat. These findings support the model that acetylation of cyclin T1 serves as a physiological switch that liberates P-TEFb from its endogenous inhibitors Hexim1 and 7SK snRNA, but is not required for the cooperative action with HIV Tat.
Keywords: Hexim1, HIV, p300, P-TEFb, transcription elongation
Introduction
The transcription of eukaryotic genes is tightly regulated at the elongation step (Wade and Struhl, 2008). Recent genome-wide chromatin immunoprecipitation assays revealed that the transition from initiation to elongation is a major rate-limiting step in transcription in flies and humans (Nechaev and Adelman, 2008). The hallmark of transcription elongation is the phosphorylation of the largest subunit of the RNA polymerase II complex. This subunit contains a species-specific number of heptad repeats (YSPTSPS), which undergo sequential phosphorylation at serines 5 and 2. Phosphorylation of serine 2, the final step in the phosphorylation cascade, is carried out by the positive elongation factor b (P-TEFb; Price, 2008). About 80% of cellular P-TEFb is composed of cyclin T1 and the cyclin-dependent kinase 9 (CDK9), and about 20% of cellular CDK9 is complexed to other C-type cyclins, such as cyclin T2A, T2B and cyclin K.
Approximately half of P-TEFb in HeLa cells is sequestered in a large ribonucleoprotein complex containing the Hexim1 protein and 7SK small nuclear RNA (7SK snRNA; Zhou and Yik, 2006). This complex also contains the 7SK snRNA capping enzyme and the La-related protein 7 responsible for 7SK snRNA stability (Jeronimo et al, 2007; He et al, 2008; Krueger et al, 2008). Within the 7SK ribonucleoprotein complex, Hexim1 associates with cyclin T1 and inhibits the kinase activity of CDK9. The interaction requires 7SK snRNA as a scaffold, which binds both Hexim1 and cyclin T1 (Barboric et al, 2005; Dulac et al, 2005; Li et al, 2005; Schulte et al, 2005; Dames et al, 2007). The other half of cellular P-TEFb is free of Hexim1 and 7SK snRNA and is catalytically active. The bromodomain protein Brd4 interacts with active P-TEFb and is believed to tether it to actively transcribed genes (Jang et al, 2005; Yang et al, 2005; Bisgrove et al, 2007).
The relative partitioning of P-TEFb into active and inactive complexes is a highly dynamic process that is central to the global regulation of cell growth and differentiation (Zhou and Yik, 2006). Agents that induce stress or cell differentiation and drastically modify gene expression, such as ultraviolet light or treatment with actinomycin D, 5, 6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB), or hexamethylene bisacetamide (HMBA), cause rapid changes in the equilibrium of active and inactive P-TEFb (Nguyen et al, 2001; He et al, 2006; Contreras et al, 2007). These agents cause the acute dissociation of Hexim1 and 7SK snRNA from P-TEFb and globally increase the pool of active P-TEFb. Although the physiological mechanisms that shift P-TEFb from inactive to active complexes remain unknown, recent evidence implicates calcium ion-calmodulin-protein phosphatase 2B-dependent signalling as well as the phosphatidylinositol-3-kinase (PI3K)/Akt signal transduction pathway in HMBA-mediated activation of P-TEFb (Contreras et al, 2007; Chen et al, 2008). It was also shown that 7SK snRNA not occupied by P-TEFb and Hexim1 binds RNA helicase A and heterogeneous nuclear ribonucleoproteins A1, A2/B1, R and Q. This binding is enhanced upon treatment with DRB or actinomycin D and supports dissociation of 7SK snRNA from P-TEFb (Barrandon et al, 2007; Van Herreweghe et al, 2007).
Acetylation is a reversible post-translational protein modification that takes place at the ɛ-amino group of lysine residues in core histones (Shahbazian and Grunstein, 2007). Levels of histone acetylation are determined by the competing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs) and correlate with the transcriptional activity of individual genes. In addition, a growing number of nonhistone, often transcription-related, proteins have been identified as targets of HATs and HDACs (Yang and Seto, 2008). Although histone acetylation is commonly associated with transcription initiation, a recent report showed that the HAT activity of p300 has an important function during a post-initiation step of transcription through an as yet unknown mechanism (Guermah et al, 2006).
We and others observed earlier that acetylation of the HIV Tat protein by p300 regulates the interaction of Tat with TAR RNA and cyclin T1 (Kiernan et al, 1999; Ott et al, 1999). Tat is a key regulator of HIV transcription elongation and recruits P-TEFb to the TAR element, an RNA stem–loop structure that forms spontaneously at the 5′ extremities of all HIV transcripts. Cyclin T1 is an essential cofactor of Tat and binds cooperatively with Tat to TAR RNA to induce phosphorylation of the CTD by CDK9 (Zhu et al, 1997; Wei et al, 1998). Tat acetylation by p300 leads to a disruption of the Tat/TAR/cyclin T1 complex and recruits the p300-associated factor (PCAF) to acetylated Tat through the PCAF bromodomain (Hetzer et al, 2005).
We were intrigued by the similar architectures of the Tat/TAR RNA/cyclin T1 and Hexim1/7SK snRNA/cyclin T1 ribonucleoprotein complexes and speculated that acetylation of one of the components of the 7SK snRNA complex could alter its integrity. Here, we report that p300 acetylates cyclin T1. Acetylated cyclin T1 marks the active (Hexim-free) fraction of cellular P-TEFb and interferes with the association of Hexim1 and 7SK snRNA. Our findings support a model in which reversible cyclin T1 acetylation serves as an important mechanism to maintain the physiological equilibrium between active and inactive P-TEFb in cells.
Results
Cyclin T proteins are acetylated by p300
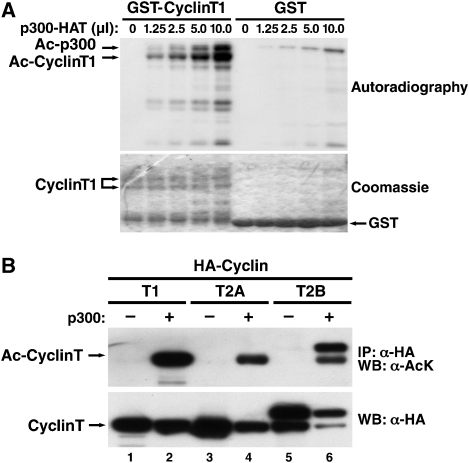
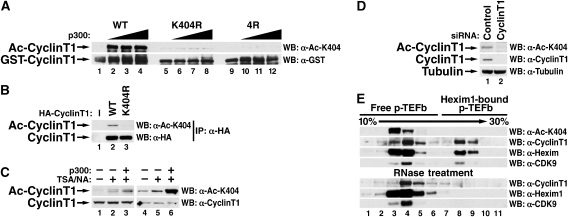
To determine whether cyclin T1 is a substrate of p300, we overexpressed full-length recombinant GST-cyclin T1 and the HAT domain of p300 in Escherichia coli and performed in vitro HAT assays in the presence of radioactively labelled acetyl-coenzyme A. Reaction products were resolved by SDS–PAGE and examined by autoradiography. GST-cyclin T1, but not GST alone, was acetylated by p300 in a dose-dependent manner (Figure 1A). In both reactions, we also detected a higher molecular mass band corresponding to the autoacetylated p300 HAT protein as described earlier (Thompson et al, 2004). In acetylation reactions of GST-cyclin T1, we detected additional lower molecular mass bands that are likely degradation products of cyclin T1. Equal loading and partial degradation of GST-cyclin T1 were confirmed by coomassie staining of the gel (Figure 1A).
Figure 1.
Cyclin T acetylation by p300. (A) In vitro acetylation assay of purified GST-cyclin T1 (amino acids 1–726) or GST by p300 HAT in the presence of [acetyl-14C] coenzyme A visualized by autoradiography or coomassie-staining. (B) Immunoprecipitation of HA-tagged cyclin T1, T2A or T2B from 293 cells overexpressing p300. Immunoprecipitated material was examined by western blotting with antibodies against acetyl-lysine (AcK) or HA. All reactions were treated with trichostatin A (TSA; 400 nM) and nicotinamide (NA; 5 mM) to block endogenous deacetylase activity.
Next, we isolated cyclin T1 from 293 cells expressing an epitope-tagged version of cyclin T1 (HA–cyclin T1). After immunoprecipitation with HA antibodies, acetylated cyclin T1 was detected by western blotting with a pan acetyl-lysine antibody when p300 was coexpressed (Figure 1B). Thus, cyclin T1 can be acetylated by p300 in vitro and in cells. We performed similar experiments with another HAT enzyme, PCAF, and did not observe any acetylation of cyclin T1 in vitro or in cells (data not shown). We also tested acetylation of other C-type cyclins in 293 cells. Cyclin T2A is a shorter splice variant of cyclin T2B and is also produced after transfection of the cyclin T2B-expressing construct. Both cyclin T2 proteins were acetylated by p300 raising the possibility that cyclin acetylation might be a universal mechanism to regulate the activity of P-TEFb (Figure 1B). Western blotting with antibodies against the HA epitope confirmed similar levels of immunoprecipitated cyclin T proteins in all reactions.
In vitro acetylation of cyclin T1 deletion mutants
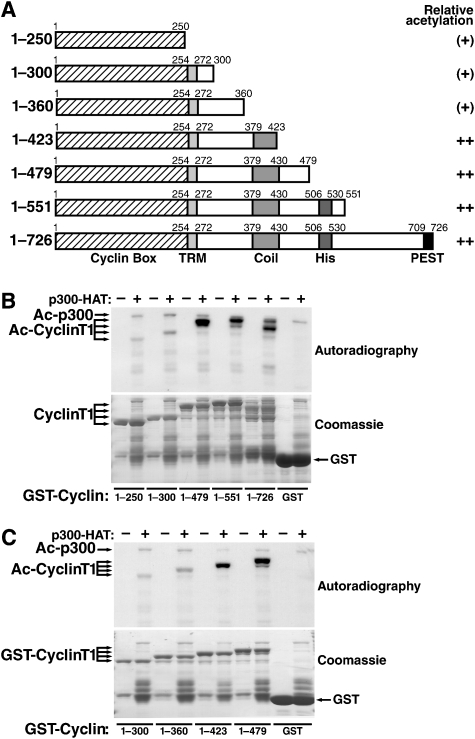
Cyclin T1 contains an N-terminal cyclin box that interacts with CDK9 and is highly conserved among C-type cyclins (Figure 2A). Downstream of the cyclin box is an 18-amino acid Tat recognition motif (TRM) that binds Tat and is unique to cyclin T1 (Bieniasz et al, 1998; Garber et al, 1998). The C terminus of cyclin T1 harbours a predicted coiled-coil region with no assigned function and a so-called histidine-rich domain that binds the RNA polymerase II CTD and is linked to the interaction with Brd4 (Taube et al, 2002; Jang et al, 2005).
Figure 2.
Acetylation of cyclin T1 by p300 in vitro. (A) Schematic representation of GST-cyclin T1 deletion proteins used in the study and relative acetylation by p300 HAT. (B, C) In vitro acetylation assays of GST-cyclin T1 deletion mutants or GST by p300 HAT. Autoradiography and coomassie staining of SDS–PAGE gels are shown.
To identify the region in cyclin T1 that is acetylated by p300, we performed in vitro HAT assays with C-terminal deletion mutants of GST-cyclin T1. Acetylation was low when only the N-terminal 300 amino acids (corresponding to the cyclin box and the TRM) were incubated with p300 HAT (Figure 2B). The acetylation signal was strongly enhanced when the next 179 amino acids (harbouring the coiled-coil region) were included. No further enhancement was observed with longer proteins containing the histidine-rich region and the C-terminal PEST domain. Analysis of two additional deletion mutants (amino acids 1–360 and 1–423) showed a strong acetylation signal only with a protein corresponding to amino acids 1–423 in cyclin T1 (Figure 2C). On the basis of these findings, we conclude that amino acids 361–423 in cyclin T1 harbour the main recognition motif for p300.
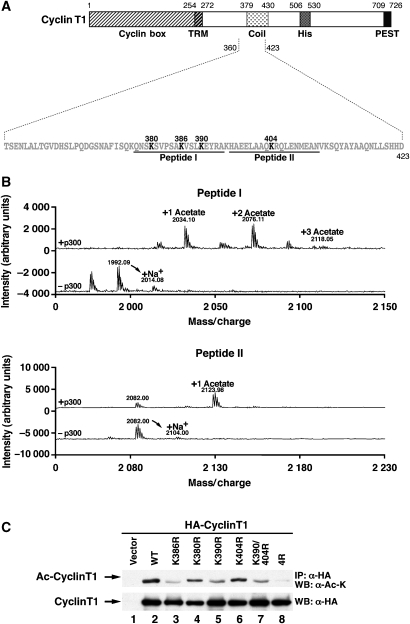
Identification of four acetylation sites in cyclin T1
Amino acids 361–423 overlap with the predicted coiled-coil region in cyclin T1 (Figure 3A). We focused on four lysines in this region (K380, 386, 390 and 404). To determine whether these lysines are acetylated by p300, we generated two synthetic peptides spanning K380, K386 and K390 (peptide I) and K404 (peptide II). Both peptides were subjected to in vitro HAT assays with p300 and nonradioactive acetyl-coenzyme A and were analysed by MALDI-TOF mass spectrometry. After acetylation of peptide I, three peaks were identified that correspond to masses of mono-, di- and triacetylated peptides (Figure 3B, peptide I, +p300). Nonacetylated peptide (1992 Da) was not detected, indicating that it is an avid substrate of p300 and was entirely consumed in the reaction.
Figure 3.
Mapping of the acetylation sites in cyclin T1. (A) Sequence of the p300 acetylation domain in cyclin T1. Candidate acetyl-acceptor sites are in bold. Synthetic peptides I and II used in mass spectrometry are shown. (B) MALDI TOF mass spectrometry of peptides I and II after incubation with recombinant p300 HAT and nonradioactive acetyl-coenzyme A. Control reactions were performed in the presence of acetyl-coenzyme A without p300 HAT. (C) Immunoprecipitation of wild-type or lysine-to-arginine mutant HA-cyclin T1 in 293 cells cotransfected with p300 and treated with TSA (400 nM) and NA (5 mM). Immunoprecipitated material was examined by western blotting with pan acetyl-lysine (AcK) or HA antibodies.
Acetylation of peptide II produced two peaks corresponding to the predicted mass of the nonacetylated (2082 Da) and the monoacetylated (2124 Da) peptides (Figure 3B, peptide II, +p300). Control reactions with each peptide and acetyl-coenzyme A in the absence of p300 produced the expected nonacetylated peaks and respective sodium adducts (Figure 3B, −p300). These data show that K380, K386, K390 and K404 in cyclin T1 are targets of p300 in vitro.
To assess whether these lysines were also acetylated in cells, we replaced each lysine within full-length HA-cyclin T1 with an arginine. This mutation preserves the basic charge of a lysine and mimics the nonmodified residue. Wild-type and mutated proteins were coexpressed with p300 in 293 cells, immunoprecipitated and analysed with the pan acetyl-lysine antibody (Figure 3C). Acetylation was abrogated by the mutation of all four lysines (4R). Individual mutations of all candidate lysines slightly decreased acetylation of cyclin T1 with the most pronounced decrease observed with mutations of K386 and K390 and the combined mutation of K390 and K404 (Figure 3C). All HA-cyclin T1 proteins accumulated to equivalent levels as assessed by western blotting with HA antibodies. These results confirm that four lysines present in the coiled-coil region of cyclin T1 can be acetylated by p300 in cells.
A positive role of cyclin T1 acetylation in P-TEFb-dependent transcription
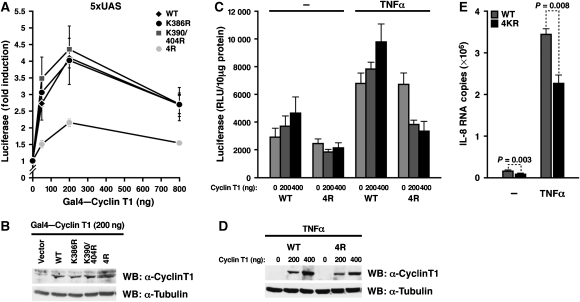
To test the influence of cyclin T1 acetylation on P-TEFb-dependent transcription, we artificially tethered acetylation-deficient cyclin T1 to a minimal promoter through the Gal4 DNA-binding system (Taube et al, 2002). The wild-type Gal4 fusion protein induced a dose-dependent activation of a promoter luciferase construct containing five Gal4 DNA-binding sites (Figure 4A). A similar activation was observed with K386R and K390/404R mutant cyclin T1. However, activation was reduced when the 4R mutant was tested, supporting the model that acetylation of the four lysines in cyclin T1 is necessary for full transcriptional activity of P-TEFb. Wild-type and mutant cyclin T1 proteins were expressed at equivalent levels as confirmed by western blotting (Figure 4B).
Figure 4.
Acetylation of cyclin T1 is required for P-TEFb transcriptional activity. (A) Transcriptional activity of increasing amounts of wild-type or acetylation-deficient Gal4–cyclin T1 fusion proteins in HeLa cells cotransfected with the 5xUAS luciferase construct containing five Gal4-binding sites upstream of the thymidine kinase promoter (200 ng). Results (mean±s.e.m.) are expressed as fold induction above the basal activity of the 5xUAS promoter and represent three independent experiments. (B) Western blotting of HeLa cell lysates transfected with constructs expressing wild-type or mutant Gal4-cyclin T1 proteins (200 ng) with antibodies specific for cyclin T1 or tubulin. (C) Transcriptional activity of the IL-8 promoter luciferase construct (200 ng) in HeLa cells cotransfected with 200 or 400 ng of wild-type or 4R mutant cyclin T1. Cells were either treated with TNFα (2 ng/ml) for 5 h or left untreated. The results expressed as relative luciferase activity were normalized by protein content and represent the average (mean±s.e.m.) of three independent experiments. (D) Western blotting of HeLa cell lysates transfected with expression constructs for wild-type or 4R mutant HA-cyclin T1 proteins and treated with TNFα. (E) Endogenous IL-8 mRNA levels of HeLa cells transfected with wild-type or 4R mutant cyclin T1 and treated with or without TNFα. RNA levels were measured by real-time RT–PCR as described in Materials and methods.
Next, we studied the role of cyclin T1 acetylation in the activation of a cellular promoter. In response to treatment with TNFα, P-TEFb is recruited to the interleukin-8 (IL-8) promoter through an interaction with the p65 subunit of NF-κB (Barboric et al, 2001; Luecke and Yamamoto, 2005). To determine whether cyclin T1 acetylation is required, we coexpressed wild-type and 4R mutant cyclin T1 and a luciferase construct corresponding to the IL-8 promoter in HeLa cells. Half of the cultures were treated with a low dose of TNFα to induce NF-κB (Luecke and Yamamoto, 2005). Wild-type cyclin T1 activated the IL-8 promoter in a dose-dependent manner in the absence or presence of TNFα (Figure 4C). In contrast, 4R mutant cyclin T1 dominantly suppressed IL-8 transcription most prominently after TNFα treatment, indicating that cyclin T1 acetylation is important for the physiological activation of the IL-8 promoter. Wild-type and 4R mutant cyclin T1 proteins were expressed at similar levels as confirmed by western blot analysis (Figure 4D).
We also measured endogenous IL-8 mRNA levels with real-time RT–PCR after transfection of wild-type or 4R mutant cyclin T1 in HeLa cells. Consistent with our model that acetylation of cyclin T1 is required for IL-8 transcription, IL-8 mRNA levels were significantly lower in cells expressing the 4R mutant than in cells expressing wild-type cyclin T1 (Figure 4E). Transfected cells were not sorted in this experiment; therefore, values also include IL-8 mRNA levels in nontransfected cells (∼40–50%). These results collectively show that acetylation of cyclin T1 supports full transcriptional activity of P-TEFb.
Detection of endogenous acetylated cyclin T1
Next, we immunized rabbits with a synthetic peptide carrying acetylated lysine 404 in cyclin T1 to generate antibodies specific for acetylated cyclin T1 (AcK404). We speculated that lysine 404 is a distinct site that lies apart from the cluster of lysines 380/386/390 and is less likely corrupted by epitope exclusion caused by differential acetylation of these lysines. The resulting antiserum did not recognize recombinant GST-cyclin T1 by western blotting unless it was acetylated by p300 HAT confirming that it was specific for the acetylated form of cyclin T1 (Figure 5A). Similarly, recognition by the antiserum was abrogated when mutant cyclin T1 proteins (K404R or 4R) were analysed (Figure 5A). The antibody also recognized wild-type, but not K404R mutant, HA-cyclin T1 immunoprecipitated from 293 cells in the presence of p300 (Figure 5B).
Figure 5.
Endogenous cyclin T1 is acetylated. (A) Western blot analysis of wild-type recombinant GST-cyclin T1 (amino acid 1–479) or K404R or 4R mutants incubated with increasing amounts of recombinant p300 HAT and nonradioactive acetyl-coenzyme A. The antibodies specific for acetylated K404 (AcK404) only recognize wild-type cyclin T1 when acetylated by p300, while no difference is observed with antibodies specific for GST. (B) Western blot analysis of HA-cyclin T1 proteins (wild-type and 4R mutant) with AcK404-specific and HA-specific antibodies. Expression constructs were cotransfected with a p300-expressing construct into 293 cells, treated with TSA/NA, and western blotting was performed after immunoprecipitation with H agarose. (C) Detection of endogenous cyclin T1. Cell lysates of 293 cells, either untreated, treated with TSA/NA or treated with TSA/NA after transfection of a p300 expression construct, were analysed by western blotting with AcK404-specific antiserum (lanes 1–3) or antigen-purified antibodies (lanes 4–6). (D) Western blot analysis of 293 cells treated with siRNAs directed against cyclin T1 or control siRNAs for 3 days. (E) Glycerol gradient sedimentation of Hexim1-free (active) and Hexim1-bound (inactive) P-TEFb complexes in HeLa cells followed by western blotting with indicated antibodies. To disrupt 7SK ribonucleoprotein complexes containing Hexim1, cell lysates were treated with RNase A as described earlier (Blazek et al, 2005). The Hexim1 fraction co-sedimenting with active P-TEFb represents free Hexim1, which is not associated with cyclin T1/CDK9 (Zhou and Yik, 2006).
To examine whether the AcK404 antiserum recognizes endogenous acetylated cyclin T1 in cells, we performed direct western blotting of cell extracts derived from 293 cells. We detected a band at the expected size of cyclin T1 when cells were treated with HDAC inhibitors trichostatin A (TSA) and nicotinamide (NA) or were transfected with the p300 expression construct and treated with TSA/NA (Figure 5C, lanes 1–3). This signal was enhanced after antigen purification of the AcK404 antiserum, and acetylated cyclin T1 was now also clearly detected in cells in the absence of HDAC inhibitor treatment or p300 overexpression (Figure 5C, lane 4). This signal increased after HDAC inhibitor treatment or p300 overexpression (Figure 5C, lanes 5 and 6) and disappeared when cells were treated with siRNAs directed against endogenous cyclin T1 excluding cross-reactivity of AcK404 antibodies with another cellular acetylated protein (Figure 5D). These data indicate that a substantial amount of cyclin T1 is acetylated in cells during regular cell growth.
We performed glycerol gradient sedimentation analysis to test whether acetylated cyclin T1 is present in active or inactive P-TEFb complexes. Interestingly, acetylated cyclin T1 was exclusively detected in the small, Hexim1-free fraction of P-TEFb, indicating that cyclin T1 acetylation marks the active form of P-TEFb (Figure 5E). This form of P-TEFb is resistant to treatment with RNase A, while the large, 7SK ribonucleoprotein complex disintegrates after RNase treatment as expected (Figure 5E).
Cyclin T1 acetylation dissociates Hexim1 from P-TEFb
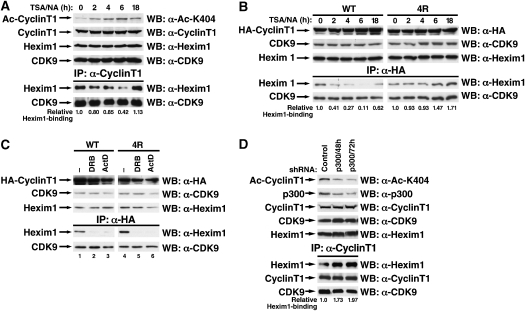
Because Hexim1 is a critical component of the 7SK ribonucleoprotein complex and directly interacts with cyclin T1, we examined whether cyclin T1 acetylation regulates the dynamics of this association. We treated 293 cells with TSA/NA to block endogenous cyclin T1 deacetylation and analysed the fraction of endogenous Hexim1 that coimmunoprecipitated with cyclin T1. Maximal hyperacetylation of cyclin T1 was observed at 4–6 h after treatment with TSA/NA and decreased after 18 h of treatment (Figure 6A). At the time of maximal cyclin T1 hyperacetylation association of endogenous Hexim1 to cyclin T1 was progressively disrupted, indicating that acetylation of cyclin T1 interferes with binding of Hexim1 to P-TEFb (Figure 6A). Accordingly, cellular P-TEFb complexes shifted into the small, Hexim1-free form as determined by glycerol gradient sedimentation analysis (Supplementary Figure 1). Re-association of the Hexim1/cyclin T1 complex was observed when cyclin T1 acetylation decreased at the 18 h time point (Figure 6A). Similar results were obtained when the immunoprecipitated material was analysed by real-time RT–PCR for the presence of 7SK snRNA; 7SK snRNA dissociated from cyclin T1 at 4 and 6 h of treatment while re-association was observed after 18 h (Supplementary Figure 2). Binding of CDK9 to cyclin T1 was not affected supporting the model that the observed dissociation of Hexim1/7SK snRNA from P-TEFb results in activation of the cyclin T1/CDK9 heterodimer.
Figure 6.
Modulation of Hexim1 binding through cyclin T1 acetylation. (A) Coimmunoprecipitation experiments of endogenous Hexim1 and CDK9 proteins with endogenous cyclin T1 in 293 cells treated with TSA/NA for indicated times. (B) Coimmunoprecipitation experiments of endogenous Hexim1 and CDK9 proteins with wild-type or 4R mutant HA-cyclin T1 in 293 cells treated with TSA/NA for indicated times. (C) Coimmunoprecipitation of endogenous Hexim1 or CDK9 with wild-type or 4R mutant HA-cyclin T1 in 293 cells after treatment with DRB (100 mM) or actinomycin D (1 mg/ml) for 2 h. (D) Coimmunoprecipitation of endogenous Hexim1 or CDK9 with endogenous cyclin T1 in 293 cells after transfection of shRNAs directed against p300 or control transfection at different time points.
To determine whether treatment with HDAC inhibitors induces dissociation of Hexim1 through hyperacetylation of cyclin T1, we performed the above experiment with cells expressing wild-type or 4R mutant cyclin T1. Treatment with TSA/NA caused a time-dependent dissociation of Hexim1 from wild-type HA-cyclin T1 (Figure 6B). In parallel, it activated the CDK9 kinase activity as determined in ex vivo kinase assays with recombinant GST-CTD (Supplementary Figure 3). 4R mutant cyclin T1 interacted efficiently with Hexim1 and CDK9. However, treatment with TSA/NA did not induce any changes in the interaction with both binding partners (Figure 6B) and did not activate the CDK9 kinase activity (Supplementary Figure 3). In fact, the fraction of Hexim1 bound to 4R mutant cyclin T1 increased modestly during the 18-h observation period in agreement with the concept that defective cyclin T1 acetylation ‘traps' P-TEFb in the Hexim1-associated inactive form (Figure 6B).
In contrast, Hexim1 dissociated from wild-type and 4R mutant cyclin T1 efficiently in cells treated with DRB, actinomycin D (Figure 6C) or HMBA (Supplementary Figure 4). These data indicate that cyclin T1 acetylation is not involved in Hexim1 dissociation from P-TEFb in response to these agents. However, the endogenous equilibrium between Hexim1-bound and Hexim1-free P-TEFb was perturbed when cyclin T1 acetylation was suppressed with small hairpin RNAs directed against p300 (Figure 6D). Levels of endogenous P-TEFb associated with Hexim1 almost doubled when cellular cyclin T1 acetylation was downregulated; no change in CDK9 association was observed (Figure 6D). The same result was obtained when p300 expression was knocked down with siRNA oligonucleotides targeting a different region in the p300 mRNA independently confirming this result (data not shown). Thus, acetylation of cyclin T1 by p300 is required to maintain the distribution of Hexim1-bound and Hexim1-free P-TEFb complexes in cells.
Tat activates HIV transcription independently from cyclin T1 acetylation
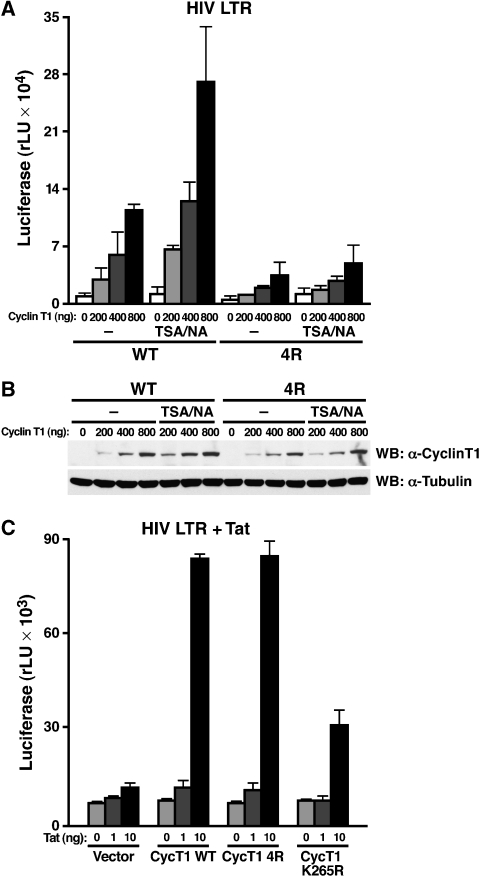
The transcriptional activity of the HIV long terminal repeat (LTR) is uniquely dependent on P-TEFb, which is usually recruited to TAR RNA by Tat. In the early stages of HIV infection when Tat is absent, P-TEFb also associates with the LTR through alternative activators such as NF-κB/p65, Brd4 or Sp1 (Barboric et al, 2001; Jang et al, 2005; Choudhary et al, 2008). To test the effect of cyclin T1 acetylation on the basal activity of the HIV LTR in the absence of Tat, wild-type or 4R mutant cyclin T1 was coexpressed with an HIV LTR luciferase construct in HeLa cells. Wild-type cyclin T1 induced LTR activity in a dose-dependent manner, but activation by 4R mutant cyclin T1 was markedly impaired (Figure 7A). Treatment with TSA and NA further activated wild-type, but not mutant cyclin T1 activity supporting the notion that cyclin T1 hyperacetylation induces P-TEFb activation and regulates basal HIV transcription. Wild-type and 4R mutant cyclin T1 proteins were equivalently expressed in cells treated with TSA/NA as determined by western blotting (Figure 7B).
Figure 7.
Differential role of cyclin T1 acetylation in HIV transcription. (A) Cotransfection of wild-type or mutant cyclin T1 at indicated amounts with the HIV LTR luciferase construct (200 ng) into HeLa cells treated with TSA/NA or with DMSO for 8 h. Average (mean±s.e.m.) of three independent experiments is shown. (B) Western blot analysis of wild-type or 4R mutant HA-cyclin T1 in HeLa cells with HA- or tubulin-specific antibodies. (C) Transcriptional synergy between HIV Tat (1 and 10 ng) and cyclin T1 (50 ng) in NIH3T3 cells transiently transfected with the HIV LTR-luciferase reporter plasmid (200 ng). The K265R mutant contains a mutation in the Tat-responsive motif of cyclin T1. Results represent average (mean±s.e.m.) of three independent experiments.
To examine the effect of acetylated cyclin T1 on Tat transactivation of the HIV LTR, we expressed low levels of Tat together with the HIV LTR luciferase reporter in NIH3T3 cells. Tat transactivation is impaired in murine cells because of the reduced efficiency of murine cyclin T1 to interact with Tat (Bieniasz et al, 1998; Garber et al, 1998). Coexpression of human cyclin T1 increased Tat transactivation in murine cells, but a mutant cyclin T1 harbouring a lysine-to-arginine mutation (K265R) in the TRM was deficient in supporting Tat transactivation as expected (Figure 7C). Notably, 4R mutant cyclin T1 synergized with Tat as efficiently as wild-type cyclin T1, indicating that Tat function does not require cyclin T1 acetylation (Figure 7C). Importantly, these data also show that the lysine-to-arginine mutations introduced into the coiled-coil region in the 4R mutant do not interfere with the ability of cyclin T1 to support CDK9 activity.
Discussion
These findings characterize cyclin T1 as a novel substrate of the p300 acetyltransferase activity. P300 acetylates cyclin T1 at four lysines in a predicted coiled-coil domain with no previously assigned function. P300 acetylation is conserved among three members of the cyclin T protein family suggesting that it might represent a universal mechanism to regulate P-TEFb function.
Three independent observations support the model that cyclin T1 acetylation is a mark of active P-TEFb. First, we find that acetylated cyclin T1 accumulates exclusively in the small, Hexim1-free form of P-TEFb by glycerol gradient sedimentation analysis. Although other post-translational modifications, such as phosphorylation of threonine 186 and acetylation of lysine 44 in CDK9 have been linked to P-TEFb activity, these modifications do not associate exclusively with active P-TEFb (Fu et al, 2007; Chen et al, 2008). Phosphorylation of T186 in CDK9 occurs at the tip of a flexible loop that is conserved among all CDK family members and induces a conformational change that allows entry of substrate and ATP into the active site of CDK enzymes (Russo et al, 1996). In CDK9, T186 phosphorylation is required for interaction with Hexim1 and 7SK snRNA and is enriched in the Hexim1-bound P-TEFb fraction (Li et al, 2005; Chen et al, 2008).
CDK9 is also subject to protein acetylation, but the functional outcome of this acetylation remains controversial. Although acetylation of K44 by p300 activated CDK9 activity through the dissociation of HDAC3 (Fu et al, 2007), acetylation of K44 and K48 by PCAF inhibited CDK9 activity by interfering with the phosphate transfer reaction (Sabo et al, 2008). In both cases, CDK9 acetylation did not affect the association of P-TEFb with Hexim1 or 7SK snRNA. Functional interaction may exist between p300-mediated acetylation of K44 in CDK9 and four lysines in cyclin T1. Although acetylation of cyclin T1 liberates P-TEFb from its physiological inhibitors Hexim1 and 7SK snRNA, acetylation of CDK9 modulates its kinase activity directly or prevents access of deacetylases to CDK9 and possibly cyclin T1.
Second, we show that the balance between Hexim1-bound (inactive) and Hexim1-free (active) P-TEFb is disturbed in the absence of cyclin T1 acetylation. When we blocked cyclin T1 deacetylation with HDAC inhibitors, this treatment decreased the fraction of cellular P-TEFb bound to Hexim1 and 7SK snRNA and globally increased the kinase activity of CDK9. These changes were not observed with acetylation-deficient cyclin T1 excluding effects of CDK9 acetylation in this process. Interestingly, we observed that the P-TEFb kinase activity remained high after 18 h of TSA/NA treatment although cyclin T1 acetylation decreased (Supplementary Figure 3). We speculate that acetylation of K44 in CDK9 could account for this increase and could explain why CTD phosphorylation remains active despite beginning re-association of P-TEFb with Hexim1.
The opposite results were obtained when acetylation of cyclin T1 was suppressed by shRNAs directed against p300 demonstrating that the acetylation status of cyclin T1, and not an independent stress pathway induced by HDAC inhibitors, regulates association of Hexim1 with cyclin T1. This model is further supported by the observation that a large fraction of the acetylation-deficient cyclin T1 mutant accumulates in the Hexim1-bound form, indicating that this mutant cannot properly shuttle between active and inactive P-TEFb complexes.
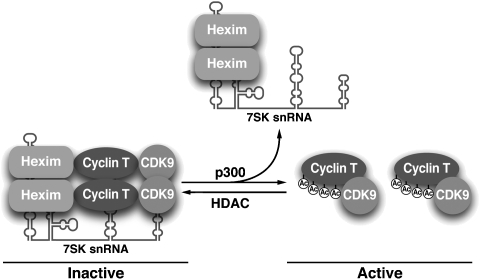
Interestingly, we show that cyclin T1 acetylation is not required for the acute mobilization of active P-TEFb after treatment with DRB, actinomycin D, or HMBA. Accordingly, no changes in acetylation levels of endogenous cyclin T1 were detected in response to treatment with these agents (data not shown). Treatment with UV light or HMBA was previously linked to the dephosphorylation of T186 in CDK9 by protein phosphatases 2B and 1α, a process causing the release of P-TEFb from the 7SK ribonucleoprotein complex (Chen et al, 2008). HMBA treatment was also shown to activate the PI3K/Akt pathway, which led to the phosphorylation of Hexim1 and also dissociated the 7SK ribonucleoprotein complex (Contreras et al, 2007). On the basis of our results, we propose that cyclin T1 acetylation fulfills an important regulatory function in maintaining the physiological partitioning of active and inactive P-TEFb but is not involved in pathways induced by acute transcriptional arrest or cell differentiation. To maintain this partitioning p300 acetylates a fraction of cellular cyclin T1 thereby inducing the dissociation of Hexim1 and 7SK snRNA and activating P-TEFb. Cyclin T1 acetylation is reversed by the action of an as yet unknown HDAC, which renews Hexim1 binding and silences activity of a fraction of P-TEFb (Figure 8).
Figure 8.
Reversible cyclin T1 acetylation regulates the physiological partitioning of P-TEFb in inactive (Hexim1 bound) and active (Hexim1 free) complexes in cells. See text for details.
In a third approach, we show that acetylation-deficient cyclin T1 dominantly suppresses activation of IL-8 transcription. The P-TEFb activity is normally recruited to the IL-8 promoter through the p65 subunit of NF-κB; the fact that acetylation-deficient cyclin T1 dominantly suppressed this activity supports the model that physiological P-TEFb activation depends on active cyclin T1 acetylation. In addition, we show that the HIV LTR in the absence of Tat is strongly activated by cyclin T1 acetylation. The C terminal half of cyclin T1, including the coiled-coil region that carries the four acetylation sites, was previously found essential for basal HIV transcription by mediating interaction of P-TEFb with Tat-SF1, the RNA polymerase II and other components of the elongation apparatus (Fong and Zhou, 2000). We speculate that this process could be affected by cyclin T1 acetylation. It is important to note that HDAC inhibitors generally activate cellular transcription and the activity of the HIV LTR by inducing hyperacetylation of histones, NF-κB or Tat (Hetzer et al, 2005). This is in contrast to agents such as actinomycin D, UV and DRB that induce transcriptional arrest and are thought to mobilize P-TEFb as a reaction to this change (Zhou and Yik, 2006). Our finding that HDAC inhibitors synergize with wild-type, but not acetylation-deficient, cyclin T1 to activate the HIV LTR suggests that cyclin T1 may be a novel effector of HDAC inhibitor-mediated activation of the HIV promoter.
In contrast, the finding that acetylation-deficient cyclin T1 synergizes normally with the HIV Tat protein to transactivate the HIV LTR implicates that acetylated cyclin T1 is not necessary for the Tat-mediated stage of HIV transcription. Human cyclin T1 was isolated as a Tat cofactor that bound TAR RNA cooperatively with Tat (Wei et al, 1998) and that was missing in the correct form in murine cells (Bieniasz et al, 1998; Garber et al, 1998). Tat competes directly with Hexim1 for cyclin T1 binding suggesting that Tat can recruit cyclin T1/CDK9 from the Hexim1-bound P-TEFb complex to the HIV LTR (Schulte et al, 2005; Barboric et al, 2007; Sedore et al, 2007). Our data support the model that Tat directly taps into the pool of nonacetylated (Hexim-bound) cyclin T1/CDK9. In contrast, the HIV promoter in the absence of Tat competes with cellular promoters for the tightly controlled supply of acetylated (Hexim-free) cyclin T1/CDK9 in cells.
Notably, p300 associates with the IL-8 and the HIV promoters, the two genes that we find are regulated by cyclin T1 acetylation (Perkins et al, 1997; Huang and McCance, 2002). Association of p300 mainly occurs at the initiation stage of transcription when it causes histone or transcription factor acetylation. After recruitment to promoters, p300 could also acetylate cyclin T1, thereby activating P-TEFb function and triggering transcription elongation (Guermah et al, 2006). At this point it is not known whether cyclin T1 acetylation creates a pool of active P-TEFb that is recruited to promoters or whether cyclin T1 acetylation occurs at the promoter level to activate P-TEFb activity. Both models are possible; the latter implies that the Hexim1-bound (inactive) P-TEFb complex associates with the initiating RNA polymerase II complex, where p300 gains access to cyclin T1. The initiating RNA polymerase II complex also interacts with Brd4, a component of the Mediator complex and a protein that associates with active P-TEFb (Jang et al, 2005; Yang et al, 2005). Although required for P-TEFb activation of the HIV LTR, Brd4 association with P-TEFb is unfavorable for Tat transactivation supporting the concept that different P-TEFb complexes are involved in the activation of the HIV LTR depending on the presence or absence of Tat (Yang et al, 2005; Bisgrove et al, 2007).
The exact mechanism how cyclin T1 acetylation interferes with the binding to Hexim1 is not known at this time. Acetylation occurs within the predicted coiled-coil domain of cyclin T1 and could thereby induce changes in protein interactions with cyclin T1. Several studies demonstrated that the inactive P-TEFb complex contains one copy of 7SK snRNA, but two molecules of Hexim1, cyclin T1 and CDK9 each (Blazek et al, 2005; Li et al, 2005). The possibility exists that homodimerization between cyclin T1 molecules stabilizes the 7SK ribonucleoprotein complex, a process disturbed by acetylation of the coiled-coil domains. Hexim1 also contains a C-terminal coiled-coil domain that is critical for oligomerization of Hexim1 (Blazek et al, 2005).
However, the interaction domain with Hexim1 does not map to the coiled-coil domain in cyclin T1; it was previously mapped to the first 250 amino acids in cyclin T1 overlapping with the cyclin box and the binding site for 7SK snRNA (Zhou and Yik, 2006). As the highly positive nature of lysine residues is necessary for nucleic acid binding in ribonucleoprotein complexes, neutralization of positive charges by acetylation might induce structural changes in cyclin T1 that trigger dissociation from 7SK snRNA and consequently from Hexim1. Future studies will determine how acetylation of cyclin T1 interferes with the formation of the inactive P-TEFb complex and whether these mechanisms apply to other ribonucleoprotein complexes in cells.
Materials and methods
Materials
Details for materials, vector constructs, and K404-acetylated peptide specific antibody generation are described in Supplementary data.
Transient transfection and luciferase assays
HeLa or NIH3T3 cells were transfected using lipofectamine reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Gal4-cyclin T1 assays were performed in HeLa cells with 200 ng of 5xUAS reporter plasmid cotransfected with increasing amounts of constructs expressing wild-type or mutant-cyclin T1 (0, 50, 200 and 800 ng). In transactivation assays with the IL-8 and HIV LTR promoter reporters, HeLa cells were cotransfected with 200 ng of reporter construct and increasing amounts of wild-type or 4R mutant cyclin T1 (0, 200 and 400 ng). After 18 h, the cells were treated for 5 h with 2 ng/ml TNFα as reported (Luecke and Yamamoto, 2005) or with TSA/NA as indicated. Tat transactivation assays were performed in NIH3T3 cells, and 200 ng of HIV LTR luciferase reporter plasmid were cotransfected with 1 or 10 ng of Tat and 50 ng of wild-type cyclin T1, 4R mutant cyclin T1, K265R mutant cyclin T1, or the empty vector control. Luciferase activity was measured with the Luciferase assay system kit from Promega (Madison, WI).
Real time RT–PCR
Total RNA was isolated using the RNA STAT-60 kit (TEL-TEST INC, Friendswood, TX) from HeLa cells transfected by wild-type or 4R mutant cyclin T1 with or without treatment of TNFα (2 ng/ml) for 5 h and amplification by real-time PCR are described in Supplementary data.
Immunoprecipitation assays and western blotting
In all, 293 cells transfected with wild-type or mutant HA-cyclin T1 expression vectors (1 μg) with lipofectamine reagent (Invitrogen) were lysed in a buffer containing 250 mM NaCl, 0.1% NP40, 20 mM NaH2PO4, pH 7.5, 5 mM EDTA, 30 mM sodium pyrophosphate, 10 mM NaF, and a protease inhibitor cocktail (Roche, Indianapolis, IN) for 1 h at 4°C. Immunoprecipitation assays were performed with rat anti-HA antibody-conjugated agarose beads for 12 h at 4°C. For assays testing endogenous cyclin T1, polyclonal cyclin T1 antibodies together with protein G sepharose were used. Levels of the coimmunoprecipitated Hexim1 protein were quantified using the ImageJ software available at http://rsb.info.nih.gov/ij.
Glycerol gradient sedimentation analysis
HeLa cells, either untreated or treated with TSA/NA as indicated, were lysed in 1 ml of lysis buffer (20 mM HEPES pH7.9, 300 mM KCl, 0.2 mM EDTA, and 0.1% NP-40) containing RNase inhibitor or 100 μg/ml of RNase A (Qiagen, Valencia, CA) for 1 h at 4°C. The lysates were processed as previously described (Blazek et al, 2005).
SiRNA experiments
Double-stranded siRNA oligonucleotides directed against GL3 luciferase or cyclin T1 (Dharmacon Research, Lafayette, CO) were transfected into HeLa cells using oligofectamine reagent (Invitrogen). To knock down p300 expression, a p300-specific shRNA was expressed in 293 cells from the pKd-p300-v1 vector (Millipore, Billerica, MA) containing a short hairpin sequence. Two consecutive transfections of 5 μg of pKd-p300-v1 vector each were performed in 24 h intervals using Fugene6 transfection reagents (Roche). At indicated time points, cells were harvested and processed for immunoprecipitation or western blot experiments.
In vitro acetylation assays
Plasmids expressing GST-cyclin T1 proteins were transformed into competent E. coli (strain BL21), and expression was induced by treatment with 0.1 mM IPTG. Solubilized proteins were purified with glutathione-coupled beads (GE Healthcare, Chalfont St Gilles, UK) and incubated with [acetyl-14C] Coenzyme A (GE Healthcare) and purified HAT domain of p300 as described (Ott et al, 1999).
Mass spectrometry
Details for MALDI-TOF MS nonacetylated and in vitro acetylated peptides are described in Supplementary data.
Supplementary Material
Supplementary Data
Acknowledgments
We thank Q Zhou and O Bensaude for sharing Hexim1 antibodies, K Jones, D Price, A Rice, B Spiegelman, M Stallcup and BM Peterlin for providing DNA constructs, D Bisgrove, E Verdin, and W Greene for helpful discussions, A Wilson and J Carroll for graphics, S Ordway and G Howard for editorial assistance, Veronica Fonseca for administrative assistance and Lily Mak for technical support. This work was supported by funds from the Gladstone Institutes and the NIH (R56 AI067118-01A2; MO). We gratefully acknowledge support through fellowships from the California HIV/AIDS Research Program (SC and HSK), from the Boehringer Ingelheim Fonds (SS), and from the Human Frontiers Science Program (EH).
References
- Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM (2005) Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J 24: 4291–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM (2001) NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 8: 327–337 [DOI] [PubMed] [Google Scholar]
- Barboric M, Yik JH, Czudnochowski N, Yang Z, Chen R, Contreras X, Geyer M, Matija Peterlin B, Zhou Q (2007) Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res 35: 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O (2007) The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol Cell Biol 27: 6996–7006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR (1998) Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J 17: 7056–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, Verdin E (2007) Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci USA 104: 13690–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D, Barboric M, Kohoutek J, Oven I, Peterlin BM (2005) Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res 33: 7000–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, Ai N, Luo H, Zhu Y, Zhou N, Zhou Q (2008) PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev 22: 1356–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary SK, Archin NM, Margolis DM (2008) Hexamethylbisacetamide and disruption of human immunodeficiency virus type 1 latency in CD4(+) T cells. J Infect Dis 197: 1162–1170 [DOI] [PubMed] [Google Scholar]
- Contreras X, Barboric M, Lenasi T, Peterlin BM (2007) HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog 3: 1459–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dames SA, Schonichen A, Schulte A, Barboric M, Peterlin BM, Grzesiek S, Geyer M (2007) Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc Natl Acad Sci USA 104: 14312–14317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, Michels AA, Fraldi A, Bonnet F, Nguyen VT, Napolitano G, Lania L, Bensaude O (2005) Transcription-dependent association of multiple positive transcription elongation factor units to a HEXIM multimer. J Biol Chem 280: 30619–30629 [DOI] [PubMed] [Google Scholar]
- Fong YW, Zhou Q (2000) Relief of two built-In autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol Cell Biol 20: 5897–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Yoon HG, Qin J, Wong J (2007) Regulation of P-TEFb elongation complex activity by CDK9 acetylation. Mol Cell Biol 27: 4641–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, Jones KA (1998) The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev 12: 3512–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG (2006) Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell 125: 275–286 [DOI] [PubMed] [Google Scholar]
- He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q (2008) A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell 29: 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Pezda AC, Zhou Q (2006) Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation. Mol Cell Biol 26: 7068–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer C, Dormeyer W, Schnolzer M, Ott M (2005) Decoding Tat: the biology of HIV Tat posttranslational modifications. Microbes Infect 7: 1364–1369 [DOI] [PubMed] [Google Scholar]
- Huang SM, McCance DJ (2002) Down regulation of the interleukin-8 promoter by human papillomavirus type 16 E6 and E7 through effects on CREB binding protein/p300 and P/CAF. J Virol 76: 8710–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K (2005) The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19: 523–534 [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, Coulombe B (2007) Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 27: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C (1999) HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J 18: 6106–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, Coulombe B, Price DH (2008) LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res 36: 2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH (2005) Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem 280: 28819–28826 [DOI] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR (2005) The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev 19: 1116–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K (2008) Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle 7: 1539–1544 [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O (2001) 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414: 322–325 [DOI] [PubMed] [Google Scholar]
- Ott M, Schnolzer M, Garnica J, Fischle W, Emiliani S, Rackwitz HR, Verdin E (1999) Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr Biol 9: 1489–1492 [DOI] [PubMed] [Google Scholar]
- Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, Nabel GJ (1997) Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science 275: 523–527 [DOI] [PubMed] [Google Scholar]
- Price DH (2008) Poised polymerases: on your mark…get set…go!. Mol Cell 30: 7–10 [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Pavletich NP (1996) Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol 3: 696–700 [DOI] [PubMed] [Google Scholar]
- Sabo A, Lusic M, Cereseto A, Giacca M (2008) Acetylation of conserved lysines in the catalytic core of cyclin-dependent kinase 9 inhibits kinase activity and regulates transcription. Mol Cell Biol 28: 2201–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte A, Czudnochowski N, Barboric M, Schonichen A, Blazek D, Peterlin BM, Geyer M (2005) Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J Biol Chem 280: 24968–24977 [DOI] [PubMed] [Google Scholar]
- Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH (2007) Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res 35: 4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M (2007) Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 76: 75–100 [DOI] [PubMed] [Google Scholar]
- Taube R, Lin X, Irwin D, Fujinaga K, Peterlin BM (2002) Interaction between P-TEFb and the C-terminal domain of RNA polymerase II activates transcriptional elongation from sites upstream or downstream of target genes. Mol Cell Biol 22: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, Levrero M, Sartorelli V, Cotter RJ, Cole PA (2004) Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol 11: 308–315 [DOI] [PubMed] [Google Scholar]
- Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T (2007) Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J 26: 3570–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Struhl K (2008) The transition from transcriptional initiation to elongation. Curr Opin Genet Dev 18: 130–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA (1998) A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92: 451–462 [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E (2008) Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 31: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q (2005) Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19: 535–545 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Yik JH (2006) The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev 70: 646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH (1997) Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev 11: 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data