Abstract
Syntaxin 18, a soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptor (SNARE) protein implicated in endoplasmic reticulum (ER) membrane fusion, forms a complex with other SNAREs (BNIP1, p31, and Sec22b) and several peripheral membrane components (Sly1, ZW10, and RINT-1). In the present study, we showed that a peripheral membrane protein encoded by the neuroblastoma-amplified gene (NAG) is a subunit of the syntaxin 18 complex. NAG encodes a protein of 2371 amino acids, which exhibits weak similarity to yeast Dsl3p/Sec39p, an 82-kDa component of the complex containing the yeast syntaxin 18 orthologue Ufe1p. Under conditions favoring SNARE complex disassembly, NAG was released from syntaxin 18 but remained in a p31-ZW10-RINT-1 subcomplex. Binding studies showed that the extreme N-terminal region of p31 is responsible for the interaction with NAG and that the N- and the C-terminal regions of NAG interact with p31 and ZW10-RINT-1, respectively. Knockdown of NAG resulted in a reduction in the expression of p31, confirming their intimate relationship. NAG depletion did not substantially affect Golgi morphology and protein export from the ER, but it caused redistribution of Golgi recycling proteins accompanied by a defect in protein glycosylation. These results together suggest that NAG links between p31 and ZW10-RINT-1 and is involved in Golgi-to-ER transport.
INTRODUCTION
In the eukaryotic secretory pathway, newly synthesized proteins are exported from the endoplasmic reticulum (ER) and transported to the Golgi apparatus, in which they are sorted according to their destination (Palade, 1975). Protein transport is mediated by vesicles or membrane carriers that bud from the donor compartment and then tether to and fuse with the acceptor compartment (Bonifacino and Glick, 2004). Some of the vesicular components that have been delivered to the acceptor compartment concomitant with transport are recycled back to the donor compartment through the retrograde pathway. The anterograde and retrograde membrane flow is balanced, which ensures steady-state distribution of proteins and may allow the formation of new transport vesicles from the donor membrane (Sannerud et al., 2003).
Soluble N-ethylmaleimide-sensitive factor attachment protein (SNAP) receptors (SNAREs) play a pivotal role in membrane fusion of transport vesicles with the acceptor compartment (Jahn and Scheller, 2006). In mammalian cells, there are at least 36 SNARE members that are uniquely localized in different membrane compartments (Hong, 2005). Most SNAREs contain a single SNARE motif (a coiled-coil domain of 60–70 amino acids), which is followed by a C-terminal transmembrane domain (TMD) (Weimbs et al., 1997). Zippering of four SNARE motifs, one provided by vesicles and three by the target membrane (Sutton et al., 1998), seems to overcome the energy barrier and trigger membrane fusion (Weber et al., 1998; McNew et al., 2000). Depending on their function and localization on two opposing membranes, i.e., vesicles and target membranes, SNAREs can be classified into vesicle (v)-SNARE and target membrane (t)-SNARE, respectively (Söllner et al., 1993). Alternatively, SNAREs are categorized as R- and Q-SNAREs due to the presence of Arg and Gln, respectively, in the core binding domains of the four SNARE motifs (Fasshauer et al., 1998).
Syntaxins are members of the t/Q-SNARE family of proteins. Many of them have a three helical bundle in their N-terminal region, called the Habc domain (Dietrich et al., 2003; Gerst, 2003). The Habc domain in several syntaxins can fold back to interact with the C-terminal SNARE motif, generating a closed conformation, whereas the Habc domain in others does not interact with the C-terminal region. Preceding the Habc domain, some syntaxin species have an extended N-terminal region that interacts with the Sec1/Munc18-like (SM) family of proteins, containing Sec1/Munc 18, Vps45, and Sly1. Although discrete modes of binding had been described for various SM–SNARE complexes (Toonen and Verhage, 2003), several recent findings revealed a common mode of binding for these complexes (for references, see Burkhardt et al., 2008). In addition to the regulation of conformation, SM proteins have been reported to contribute to the stability of syntaxins (Bryant and James, 2001; Toonen et al., 2005; Braun and Jentsch, 2007; Carpp et al., 2007).
We reported previously that syntaxin 18 is located in the ER and forms a large complex containing three other SNAREs (p31, BNIP1, and Sec22b) and three peripheral membrane proteins (Sly1, ZW10, and RINT-1) (Hatsuzawa et al., 2000; Hirose et al., 2004; Nakajima et al., 2004). Syntaxin 18 is the mammalian orthologue of yeast Ufe1p implicated in retrograde transport from the Golgi to the ER (Lewis and Pelham, 1996) and homotypic ER membrane fusion (Patel et al., 1998). The ER membrane fusion machinery seems to be conserved during evolution (Kraynack et al., 2005) because almost all equivalent components are present in the syntaxin 18 and Ufe1p complexes: p31, BNIP1, Sec22b, Sly1, ZW10, and RINT-1 in mammals correspond to Use1p/Slt1p, Sec20p, Sec22p, Sly1p, Dsl1p, and Tip20p in yeast, respectively (Sweet and Pelham, 1992, 1993; Lewis et al., 1997; Andag et al., 2001; Reilly et al., 2001; VanRheenen et al., 2001; Belgareh-Touze et al., 2003; Burri et al., 2003; Dilcher et al., 2003; Hirose et al., 2004; Nakajima et al., 2004). Our functional analyses suggested that ZW10 and RINT-1 are involved in membrane traffic between the ER and Golgi apparatus (Hirose et al., 2004; Arasaki et al., 2006, 2007; Inoue et al., 2008) and that BNIP1 participates in the formation of the three-way junctions of the ER network (Nakajima et al., 2004). Sun et al. (2007) demonstrated that ZW10 and RINT-1 play a role in a Rab6-dependent recycling pathway from the Golgi apparatus to the ER. Another group reported that syntaxin 18 and p31 participate in phagocytosis and post-Golgi transport, respectively (Hatsuzawa et al., 2006; Okumura et al., 2006). The versatile ability of the syntaxin 18 complex may be related to a unique mechanism of SNARE core complex assembly (Aoki et al., 2008).
One missing component of the mammalian ER fusion machinery is the equivalent of yeast Dsl3p/Sec39p, which has been reported to be required for the stability of t/Q-SNARE complex at the ER (Kraynack et al., 2005). In the present study, we demonstrated that a peripheral membrane protein encoded by the neuroblastoma-amplified gene (NAG) is a component of the syntaxin 18 complex. Despite a remarkable difference in molecular size, NAG (270 kDa) exhibits low sequence identity to Dsl3p/Sec39p (82 kDa). Our results showed that NAG serves as a link between p31 and ZW10-RINT-1 by interacting with the extreme N-terminal region of p31.
MATERIALS AND METHODS
Antibodies and Proteins
A polyclonal antibody against human NAG was raised against a bacterially expressed His-tagged NAG fragment (amino acids 2125-2371) and affinity purified using antigen-coupled beads. Rabbit polyclonal antibodies against vesicular stomatitis virus-encoded glycoprotein (VSVG) (amino acids 501-511), human Sly1 (amino acids 1-146 and 147-396), and human Rer1 (amino acids 1-14 and 183-196) were prepared and affinity purified. A monoclonal antibody (mAb) against VSVG (8G5) was prepared from ascites. mAbs against calnexin, GM130, GS15, GS27, GOS28, and Bet1 were obtained from BD Biosciences Transduction Laboratories (Lexington, KY). mAbs against α-tubulin and CD44 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). mAbs against protein disulfide isomerase, lysosome-associated membrane protein (LAMP)-2, mannosidase IB, and FLAG M2 were purchased from Daiichi Fine Chemical (Tokyo, Japan), Abcam (Cambridge, United Kingdom), Abnova (Taipei City, Taiwan), and Sigma-Aldrich (St. Louis, MO), respectively. A mAb against ER-Golgi intermediate compartment (ERGIC)-53 and a rabbit polyclonal antibody against KDEL receptor (KDEL-R) were generous gifts from Dr. Hans-Peter Hauri (University of Basel, Basel, Switzerland) and Dr. Hans-Dieter Söling (Max-Planck-Institute, Göttingen, Germany), respectively. Rabbit polyclonal antibodies against ERGIC-53 and FLAG were purchased from Sigma-Aldrich. Rabbit polyclonal antibodies against GPP130, Sec61β, and mannosidase II (Man II) were purchased from Covance Research Products (Berkeley, CA), Millipore (Billerica, MA), and Millipore Bioscience Research Reagents (Temecula, CA), respectively. Sheep antisera against TGN46 were purchased from AbD Serotec (Oxford, United Kingdom). Rabbit immunoglobulin G was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Other antibodies were described previously (Hirose et al., 2004; Nakajima et al., 2004; Shimoi et al., 2005; Wakana et al., 2008).
Expression and purification of NSF and α-SNAP were described previously (Tani et al., 2003). Glutathione transferase (GST)-BNIP1ΔTMD (amino acids 1-202), GST-Sec22bΔTMD (amino acids 1-195), GST-p31ΔTMD (amino acids 1-231), and GST-p31ΔN15ΔTMD (amino acids 16-231) proteins were expressed in Escherichia coli and purified by glutathione-Sepharose 4B chromatography as described previously (Aoki et al., 2008).
Plasmids
pSPORT-NAG was purchased from RZPD (Deutsches Ressourcenzentrum fuer Genomforschung, Berlin, Germany). Because the clone was found to contain a frameshift and a nonsense mutation at nucleotides 1977 and 5941, respectively, the mutations were corrected as follows. The full-length NAG cDNA with mutations was amplified by polymerase chain reaction (PCR) from pSPORT-NAG and inserted into the BamH1/Sma1 site of pFLAG-CMV6 to produce pFLAG-NAG full**. The cDNA encoding amino acids 1-1035 of NAG was inserted into the Sma1 site of pFLAG-CMV6, and the point mutation at nucleotide 1977 was corrected by inverted PCR. The BamH1/Nde1 fragment of pFLAG-NAG full** was cut out and replaced with the corrected fragment to obtain pFLAG-NAG full*. Next, the cDNA encoding amino acids 1036-2371 of NAG was inserted into the EcoRV site of pFLAG-CMV6, and the point mutation at nucleotide 5941 was corrected. The Nhe1/Nhe1 fragment of pFLAG-NAG full* was replaced with the corresponding fragment of correct sequence to obtain pFLAG-NAG full. To express an NAG fragment in E. coli, the cDNA encoding amino acids 2125-2371 of NAG was amplified by PCR and inserted into the BamH1/Sma1 site of pET11d-His. The cDNAs encoding amino acids 6-259, 11-259, 16-259, and 20-259 of p31 were amplified from the cDNA of full-length p31 and inserted into the BglII/EcoRV site of pFLAG-CMV6. Other constructs were reported previously (Aoki et al., 2008). The plasmids encoding N-acetylglucosaminyltransferase I-green fluorescent protein (GFP) and β-1,4-galactosyltransferase 1-GFP were kindly supplied by Dr. Nobuhiro Nakamura (Kanazawa University, Kanazawa, Japan) and Dr. Jennifer Lippincott-Schwartz (National Institutes of Health, Bethesda, MD), respectively.
Cell Culture
293T cells were cultured in DMEM supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin, and 10% fetal calf serum. HeLa cells were cultured in Eagle's minimum essential medium supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin, 100 mg/ml l-glutamine, and 10% fetal calf serum.
RNA Interference
The following short interfering RNAs (siRNAs) were used: Lamin A/C, ctggacttccagaagaaca; NAG (4160), ctagtaaagcagtacaaga; NAG (4382), ctacagccaatgaagatct; ZW10, aagggtgaggtgtgcaatatg; RINT-1, ttagccactgatattccttgt; p31, accggcctctgaggtgatcaa; and GS15, aagcatgaccagcctgctta. siRNAs were purchased from Japan BioService (Asaka, Japan). HeLa cells were grown on 35-mm or 10-cm dishes, and siRNAs were transfected at a final concentration of 200 nM using Oligofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol.
Preparation of Cell Lysates and Binding Assay
293T cells at ∼70% confluence were transfected with 2 μg of pFLAG-NAG full or 1 μg for other constructs per 35-mm dish by using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's protocol. After 24 h of incubation, the cells were lysed with lysis buffer (20 mM HEPES-KOH, 150 mM KCl, 1% Triton X-100, 1 mM dithiothreitol, 2 mM EDTA, 10 μg/ml leupeptin, 2 μM pepstatin A, 2 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride, pH 7.2).
GST-tagged proteins (5 μg) were mixed with glutathione-Sepharose 4B in binding buffer (50 mM Na2HPO4, 100 mM NaCl, 0.1% Triton X-100, 1 mM EDTA, and 2 mM 2-mercaptethanol, pH 7.0) and then incubated overnight at 4°C with 200 μg of cell lysates. After extensive washing of the beads, the proteins bound to the beads were solubilized in sample buffer and subjected to SDS-polyacrylamide gel electrophoresis (PAGE).
Immunofluorescence Analysis
Immunofluorescence microscopy was performed as described previously (Tagaya et al., 1996). Cells were fixed with methanol at −20°C for 5 min for staining Man II, ZW10, RINT-1, and Rer1 or with 4% paraformaldehyde at room temperature for 20 min for other proteins.
Subcellular Fractionation
Tet-on HeLa cells (2 10-cm dishes) were washed twice with phosphate-buffered saline, collected, and suspended in homogenization buffer (10 mM Tris-HCl, 4 mM MgCl2, 120 mM NaCl, and 5 mM KCl, pH 7.2) and then homogenized with 10 strokes with a 25-gauge needle. The homogenate was centrifuged at 3000 rpm for 10 min, and then the supernatant was centrifuged again at 3000 rpm for 10 min. The postnuclear supernatant was load onto a 17.5–44% of Nycoprep gradient and centrifuged in a Beckman SW50 rotor at 80,000 × g for 3 h. After centrifugation, fractions were collected from the top, and every other fraction was subjected to SDS-PAGE after trichloroacetic acid precipitation.
Protein Transport Assay
The expression plasmid for VSVG fused with GFP was kindly donated by Dr. J. Lippincott-Schwartz (National Institutes of Health). Morphological and biochemical transport assays were performed as described previously (Iinuma et al., 2007). VSVG-GFP transported to the plasma membrane was detected using anti-VSVG (8G5) without cell permeabilization.
Retrograde Transport Assay
The plasmid encoding ts045VSVG-KDEL-R-yellow fluorescent protein (YFP) was kindly donated by Dr. Alberto Luini (Consorzio Mario Negri Sud, Santa Maria Imbaro, Italy). HeLa cells grown on 35-mm dishes were mock transfected or transfected with NAG (4160) and incubated at 37°C for 48 h. The cells were then transfected with 1 μg of the plasmid encoding VSVG-KDEL-R-YFP and incubated at 32°C for 24 h. Cycloheximide was added to the medium at a final concentration of 20 μg/ml, and the cells were incubated for another 2 h. To monitor retrograde transport, the temperature was shifted to 40°C. After 2 h, the cells were fixed and processed for immunofluorescence analysis.
Digitonin Permeabilization
HeLa cells were washed twice with permeabilization buffer (20 mM HEPES-KOH, 50 mM NaCl, 2 mM MgCl2, 250 mM sucrose, and 1 mM dithiothreitol, pH 7.4) and then incubated with 50 μg/ml digitonin (Merck, Darmstadt, Germany) at 4°C for 20 min. The cells were washed with permeabilization buffer twice and processed for immunofluorescence microscopy.
Peptide:N-glycosidase F (PNGase F) Treatment
HeLa cells depleted of NAG were solubilized with phosphate-buffered saline with 0.5% SDS and heated at 100°C for 10 min. A portion of lysates (8.2 μg) was diluted twice in reaction buffer (200 mM Tris-HCl, 2% Nonidet P-40, and 20 mM 2-mercaptoethanol, pH 8.6) and treated with PNGase F (Takara Bio, Otsu, Japan) at a final concentration of 12.5 μU/μl at 37°C for 24 h. After the reaction, the samples were subjected to SDS-PAGE.
In-Gel Digestion and Mass Spectrometry
Proteins coprecipitated with anti-p31 antibody-bound protein G-Sepharose were subjected to SDS-PAGE and visualized by silver staining. The stained bands were excised from the gel, in-gel digested with trypsin, and subjected to the direct nanoflow liquid chromatography-tandem mass spectrometry (LC-MS/MS) system (Natsume et al., 2002; Kaji et al., 2006). The peptide mixture was separated on a frit-less Mightysil-C18 (3-μm particle; Kanto Chemical, Tokyo, Japan) column (30 mm × 0.150 μm i.d.) by using a 80-min gradient of acetonitrile (0–40%) in 0.1% formic acid at a flow rate of 50 nl/min, and the eluted peptides were sprayed directly into a high resolution quadrapole time-of-flight hybrid mass spectrometer Q-TOF Ultima (Waters, Milford, MA). MS/MS spectra were acquired by data-dependent collision-induced dissociation, and the MS/MS data were analyzed using MASCOT software (Matrix Science, London, United Kingdom) for peptide assignment. The resulting data set was finally evaluated by in-house software STEM (Shinkawa et al., 2005).
RESULTS
Identification of the NAG Protein as a New Component of the Syntaxin 18 Complex
The SNARE complex is disassembled by NSF-α-SNAP in a manner coupled with NSF-mediated ATP hydrolysis (Söllner et al., 1993). We demonstrated previously that p31, ZW10, and RINT-1 remain associated after SNARE complex disassembly (Hirose et al., 2004). Our two-hybrid analysis showed that ZW10 and RINT-1 interact directly, but neither of them binds to p31, suggesting the presence of protein(s) that mediates the interaction of p31 with ZW10 and/or RINT-1.
To test this possibility, we raised mAbs against p31 and conducted an immunoprecipitation experiment using mAb 5C3 attached to protein G-Sepharose. Unexpectedly, SDS-PAGE analysis showed that many proteins were coprecipitated with the antibody-bound beads (Figure 1A, right lane). To remove nonspecific binding proteins, cell lysates were premixed with protein G-Sepharose, and then the supernatant was subjected to immunopurification. However, still many proteins were found to bind to the antibody-bound beads (data not shown). Therefore, we decided to determine p31-binding candidates and then carefully examine the specificity of binding. The proteins coprecipitated with the antibody-bound beads were separated by SDS-PAGE, digested in gels, and subjected to LC-MS/MS analysis. In addition to several proteins that are known to interact with p31 and syntaxin 18, such as RINT-1, ZW10, NSF, and Sly1, a protein of ∼270 kDa encoded by NAG was identified (Supplemental Table S1).
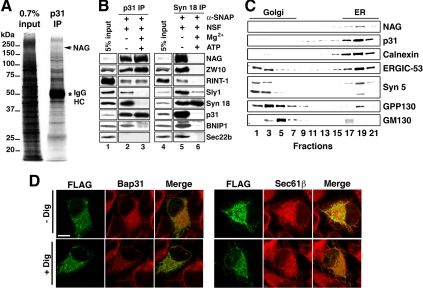
Figure 1.
Identification of the NAG protein as a component of the syntaxin 18 complex. (A) Triton X-100 extracts of 293T cells were immunoprecipitated with an anti-p31 antibody (mAb 5C3) attached to protein G-Sepharose 4B. The coprecipitated proteins were resolved by SDS-PAGE, stained with silver, and analyzed by LC-MS/MS. An asterisk denotes immunoglobulin heavy chain. (B) Triton X-100 extracts of 293T cells were incubated at 16°C for 60 min with 10 μg/ml NSF and 5 μg/ml α-SNAP in the presence or absence of 8 mM Mg2+ and 0.5 mM ATP. After incubation, the samples were immunoprecipitated with a mAb against p31 (lanes 2 and 3) or syntaxin 18 (lanes 5 and 6). The precipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. (C) Cell homogenates were subjected to Nycoprep density gradient centrifugation and analyzed by immunoblotting with the indicated antibodies. (D) HeLa cells were transfected with the plasmid encoding FLAG-NAG. At 24 h after transfection, the cells were left untreated (top row; − Dig) or treated with digitonin (bottom row; + Dig), and double-stained with antibodies against FLAG and Bap31 (left) or Sec61β (right). Bar, 20 μm.
To verify whether NAG is really associated with p31, we raised a polyclonal antibody against NAG. The NAG antibody recognized an ∼270-kDa band (Supplemental Figure S1A, lane 1), in good agreement with the calculated molecular mass of NAG. The expression of the 270-kDa protein was knocked down by siRNA [NAG (4160)] (lane 2), confirming that the recognized protein is NAG. The staining intensities of a 90-kDa and a doublet of ∼50-kDa bands were not reduced by NAG (4160), suggesting that they are proteins nonspecifically recognized by the antibody.
As shown in Figure 1B, lanes 2 and 5, NAG was coprecipitated with mAbs against p31 and syntaxin 18. Sec22b, a v/R-SNARE mainly localizes to the ER-Golgi intermediate compartment (ERGIC) (Hay et al., 1998; Zhang et al., 1999), was coprecipitated with syntaxin 18 (lane 5) but not with p31 (lane 2). This may reflect that syntaxin 18, but not p31, is the principal partner for Sec22b on the ER membrane (Aoki et al., 2008). Of note is that NAG remained associated with p31 under conditions favoring SNARE complex disassembly (lane 3), albeit its release from syntaxin 18 (lane 6). The association of NAG with p31 and syntaxin 18 was confirmed by an immunoprecipitation experiment using the NAG antibody (Supplemental Figure S1B, lane 2). These results unequivocally demonstrated that NAG is a component of the syntaxin 18 complex and forms a subcomplex with p31, ZW10, and RINT-1.
To determine the subcellular localization of NAG, cell homogenates were separated by density gradient centrifugation and analyzed by immunoblotting. As shown in Figure 1C, NAG cosedimented with p31 and an ER marker protein calnexin. To confirm the ER localization of NAG by immunofluorescence microscopy, we tried to obtain a specific anti-NAG antibody useful for immunostaining. As antigens, we tested several synthetic peptides or fragments expressed in E. coli but could not succeed to obtain good antibodies. We therefore expressed FLAG-tagged NAG in HeLa cells and examined its distribution using an anti-FLAG antibody. Although FLAG-NAG was expressed in only a few percent of the transfected cells, the expressed protein was colocalized with ER marker proteins Bap31 and Sec61β (Figure 1D). The FLAG-NAG immunostaining was not diminished by digitonin permeabilization of cells, suggesting the membrane association of the expressed protein.
The N-Terminal Region of p31 Is Required for the Interaction with NAG
In addition to the SNARE motif at the C terminus, p31 has a putative coiled-coil region at the N terminus (amino acids 3–26, predicted by the Lupus algorithm with a window size of 21 residues). Given that the N-terminal region of SNAREs is responsible for the interaction with SNARE regulators (Dietrich et al., 2003; Gerst, 2003), N-terminally truncated versions of p31 were expressed as FLAG-tag proteins in 293T cells, immunoprecipitated with an anti-FLAG antibody, and analyzed by immunoblotting. As shown in Figure 2A, NAG as well as ZW10 and RINT-1 was coprecipitated with full-length p31 and p31ΔN5 to the same extent (lanes 5 and 6), whereas no precipitation was observed with p31ΔN10 and p31ΔN15 (lanes 7 and 8). In contrast, the binding of individual mutants to syntaxin 18 was not markedly different. These results suggest that the extreme N-terminal region of p31 is required for the interaction with NAG and other accessory proteins.
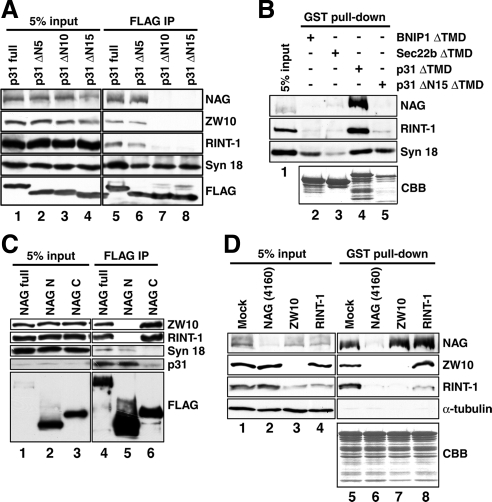
Figure 2.
NAG links between p31 and ZW10-RINT-1 through an extreme N-terminal region of p31. (A) 293T cells were transfected with the plasmids encoding full-length or N-terminally truncated versions of FLAG-p31. At 24 h after transfection, the cells were lysed, and immunoprecipitation experiments were carried out using an anti-FLAG antibody. The precipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. (B) Recombinant GST-BNIP1ΔTMD, GST-Sec22bΔTMD, GST-p31ΔTMD, and GST-p31ΔΝ15ΔTMD immobilized onto glutathione-Sepharose 4B were incubated with lysates of 293T cells at 4°C overnight. Pull-down experiments were conducted, and the proteins bound to the resin were separated by SDS-PAGE and visualized by immunoblotting or Coomassie Brilliant Blue (CBB) staining. (C) Lysates of 293T cells transfected with the plasmids encoding full-length or truncated versions of FLAG-NAG were immunoprecipitated using an anti-FLAG antibody. The precipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. (D) Recombinant GST-p31ΔTMD immobilized onto glutathione-Sepharose 4B was incubated at 4°C overnight with lysates of HeLa cells depleted of NAG, ZW10, or RINT-1. Pull-down experiments were conducted, and the proteins bound to the resin were separated by SDS-PAGE and visualized by immunoblotting or CBB staining.
To confirm this finding, we next performed pull-down experiments using recombinant p31 and ER SNAREs. Isolated recombinant GST fusion proteins with SNAREs lacking the TMD were mixed with cell lysates, pulled down with glutathione beads, and analyzed by immunoblotting (Figure 2B). Consistent with the results of the immunoprecipitation experiments, NAG and RINT-1 were pulled down with GST-p31ΔTMD (lane 4) but not with GSTp31ΔN15ΔTMD (lane 5). Essentially no binding to NAG or RINT-1 was detected for GST-BNIP1ΔTMD (lane 2) and GST-Sec22bΔTMD (lane 3). Nevertheless, GST-BNIP1ΔTMD bound syntaxin 18 to an extent comparable to that observed for GST-p31ΔTMD. These results support the idea that p31, but not BNIP1 or syntaxin 18, binds directly NAG and accessory proteins.
NAG Serves as a Link between p31 and ZW10-RINT-1
To determine which regions of NAG are responsible for the interaction with p31 and ZW10-RINT-1, NAG was divided into two fragments. As a shown in Figure 2C, lane 5, the N-terminal fragment of NAG (NAG N; 1–1035 amino acids) bound p31 and, to a much lesser extent, syntaxin 18, but not ZW10 or RINT-1. Conversely, the C-terminal fragment of NAG (NAG C; 1036–2371 amino acids) was associated with ZW10 and RINT-1 but not with p31 or syntaxin 18 (lane 6). These results raised the possibility that NAG serves as a link between p31 and ZW10-RINT-1 through its distinct regions.
To verify this hypothesis, we conducted GST-p31ΔTMD pull-down experiments using cell lysates depleted of NAG. As expected, neither ZW10 nor RINT-1 was pulled down with GST-p31ΔTMD when NAG expression was knocked down by NAG (4160) (Figure 2D, lane 6). By contrast, depletion of ZW10 or RINT-1 did not affect the binding between GST-p31ΔTMD and NAG (lanes 7 and 8). These results support the idea that NAG serves as a link between p31 and ZW10-RINT-1. Of note, ZW10 was associated with GST-p31ΔTMD even when RINT-1 was depleted. This may suggest that ZW10 is a major binding partner for NAG.
NAG Depletion Induces the Release of ZW10-RINT-1 from Membranes
To further demonstrate that NAG acts as a linker between p31 and ZW10-RINT-1, NAG expression was knocked down by NAG (4160), and cell lysates were immunoprecipitated with an antibody against syntaxin 18 (Figure 3A, lane 4) or p31 (lane 6). Clearly, the amounts of ZW10-RINT-1 coprecipitated with both antibodies were much lower than those in mock-transfected cells (lanes 3 and 5). Next, we examined whether this dissociation of ZW10-RINT-1 from p31 leads to their release from ER membranes. To this end, homogenates of NAG-depleted cells were prepared without detergent and separated into the membrane and cytosol fractions by centrifugation. As shown in Figure 3B, concomitant with NAG depletion, the amounts of membrane-bound ZW10-RINT-1 were decreased (lane 6 vs. lane 4), and accordingly, their cytosolic contents were increased (lane 9 vs. lane 7). Consistent with the idea that Sly is not a component of the NAG subcomplex (Hirose et al., 2004), no release of Sly1 from membranes was observed in NAG-depleted cells. It should be noted that NAG was almost exclusively membrane-associated and not released from membranes after p31 depletion (lanes 5 and 8). This may suggest that NAG binds to ER proteins other than p31. Alternatively, because NAG is a large protein with several hydrophobic regions (Scott et al., 2003), it may bind to ER membranes through its hydrophobic regions.
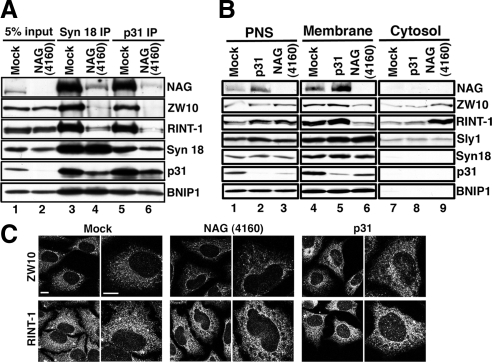
Figure 3.
Knockdown of NAG causes the release of ZW10-RINT-1 from membranes as well as from p31. HeLa cells were mock-transfected or transfected with NAG (4160) or p31 siRNA. (A) At 72 h after transfection, cell lysates were prepared and immunoprecipitated with an antibody against syntaxin 18 (lanes 3 and 4) or p31 (lanes 5 and 6). The precipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. (B) At 72 h after transfection, the cells were homogenized by passage 40 times through a 27-gauge needle. The homogenate was centrifuged at 1100 × g for 5 min to obtain the postnuclear supernatant (PNS; lanes 1–3) and then at 135,000 × g for 30 min to separate the membrane (lanes 4–6) and cytosol (lanes 7–9). (C) At 72 h after transfection, the cells were stained with the indicated antibodies. Note that many and some RINT-1-negative large puncta were observed in NAG- and p31-depleted cells, respectively (bottom row). Bars, 10 μm.
We demonstrated previously that depletion of RINT-1 causes redistribution of ZW10 (Arasaki et al., 2006). If NAG really supports the binding of ZW10-RINT-1 to ER membranes, knockdown of NAG may cause a similar change in the distribution of ZW10. This was indeed the case (Figure 3C, top row). Depletion of NAG changed ZW10 staining from the uniform staining of the ER to a dispersed, dot-like staining (top row, middle two panels), as observed in the case of RINT-1 depletion (Arasaki et al., 2006). Such a change was not observed in the RINT-1 staining, although RINT-1-negative large puncta were observed (bottom row). These puncta correspond to patchy accumulation of ER luminal proteins, such as protein disulfide isomerase and Hsp47 (data not shown). Similar puncta were observed in cells depleted of syntaxin 18 (Iinuma et al., 2009) or p31 (Uemura et al., 2009; Figure 3C, bottom row, right two panels).
Knockdown of NAG Induces a Reduction in the Expression of p31
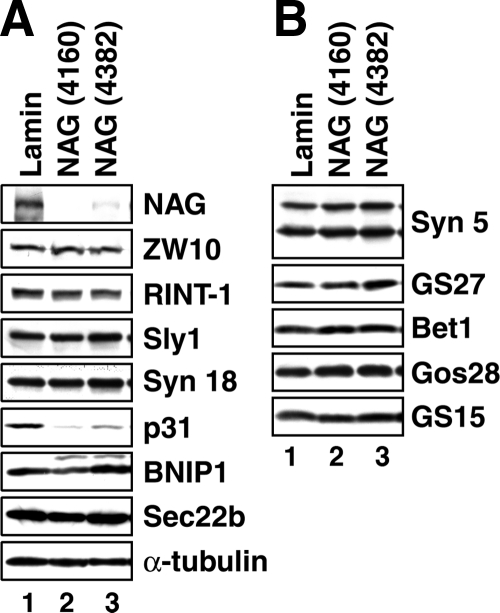
In the course of NAG knockdown experiments, we noticed that knockdown of NAG by NAG (4160) resulted in a reduction in the expression level of p31 (Figure 4A, lane 2). Another siRNAs [NAG (4382)] also induced a significant reduction in p31 expression (lane 3). The extent of p31 reduction seemed to correlate with NAG knockdown efficiency. By contrast, the expression levels of other components of the syntaxin 18 complex (Figure 4A) or Golgi-localized SNAREs (Figure 4B) were not significantly affected by knockdown of NAG. These results suggest that NAG plays a role in stabilizing p31 and confirm an intimate relationship between the two proteins.
Figure 4.
Depletion of NAG induces a reduction in the expression level of p31. HeLa cells were transfected with lamin A/C siRNA, NAG (4160), or NAG (4382). At 72 h after transfection, the cells were solubilized in phosphate-buffered saline with 0.5% SDS, and the lysates (15 μg each) were separated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies.
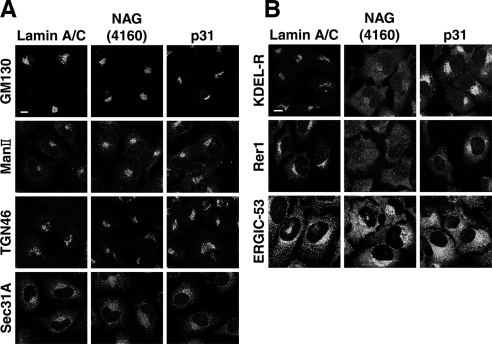
Depletion of NAG Changes the Distribution of Recycling Proteins without Substantially Affecting Golgi Morphology
To reveal the role of NAG in the early secretory pathway, we first examined the distribution of proteins in the Golgi and the ER-Golgi interface in cells transfected with NAG (4160). As shown in Figure 5A, no substantial change in the distribution of cis-, medial-, and trans-Golgi resident proteins (GM130, Man II, and TGN46, respectively) and an ER exit site marker, Sec31A, was detected at 72 h after siRNA transfection. Strikingly, knockdown of NAG caused dispersion of cis-Golgi-localized recycling proteins, KDEL-R and Rer1, and an ER-Golgi intermediate compartment marker, ERGIC-53 (Appenzeller-Herzog and Hauri, 2006) (Figure 5B, middle column). A similar phenotype was observed when NAG was knocked down with NAG (4382) (Supplemental Figure S2). The phenotype of cells transfected with NAG siRNA was more severe compared with that of cells transfected with siRNA targeting p31 (Figure 5B, middle column vs. right one), implying the contribution of NAG depletion to these changes. The p31 siRNA used could fully suppress the expression of p31 (Figure 3B, lane 2).
Figure 5.
Depletion of NAG changes the distribution of recycling proteins without affecting the Golgi apparatus. HeLa cells were transfected with lamin A/C siRNA, NAG (4160), or p31 siRNA. At 72 h after transfection, the cells were fixed and immunostained with antibodies against proteins in the Golgi or ER exit sites (A) or recycling proteins (B). Bars, 10 μm.
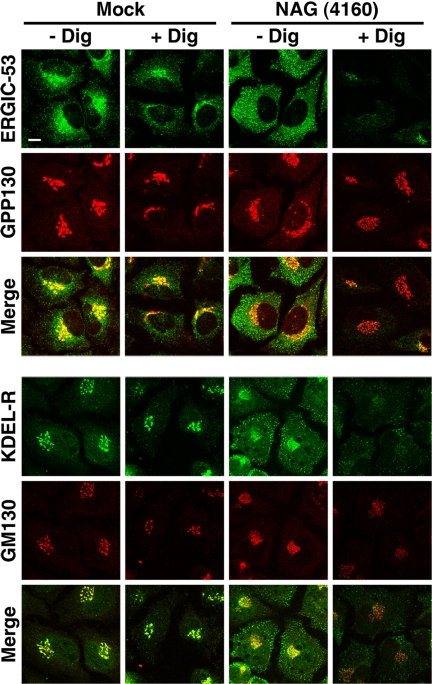
Blockage of the Tethering of Retrograde Carriers from the Golgi Apparatus in NAG-depleted Cells
To assess the state of cis-Golgi and ERGIC-localized recycling proteins in NAG-depleted cells, we permeabilized cells with digitonin. Digitonin at low concentrations is known to solubilize the plasma membrane but not other membranes including ER and Golgi membranes. Zolov and Lupashin (2005) demonstrated that nontethered Golgi vesicles in COG3-depleted cells are efficiently released from digitonin-permeabilized cells. As shown in Figure 6, the immunostaining intensity of ERGIC-53 (top row) and KDEL-R (fourth row) in NAG-depleted cells (right two columns), but not in mock-transfected cells (left two columns), was markedly reduced upon permeabilization, suggesting that ERGIC-53– and KDEL-R–containing membranes are not tightly associated with cellular structures after NAG depletion. In contrast, decreases in the immunostaining intensity of Golgi proteins, such as GPP130 (second row) and GM130 (fifth row), were not markedly different between mock-transfected and NAG-depleted cells. These results suggest that NAG depletion likely blocks the tethering step in retrograde transport from the Golgi apparatus to the ER.
Figure 6.
Digitonin sensitivity of recycling proteins in NAG-depleted cells. HeLa cells were mock transfected (left two columns) or transfected with NAG (4160) (right two columns). At 72 h after transfection, the cells were permeabilized with digitonin and double-stained with antibodies against ERGIC-53 (top row) and GPP130 (second row) or KDEL-R (fourth row) and GM130 (fifth row). Bar, 10 μm.
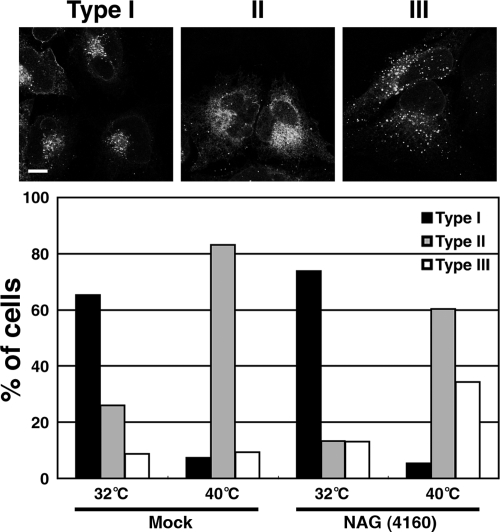
To verify that retrograde transport to the ER is inhibited after NAG knockdown, we carried out a retrograde transport assay by using ts045 VSVG-KDEL-R chimera. At 32°C (permissive temperature), this chimera is localized in the cis-Golgi but redistributes to the ER through the retrograde pathway upon shift to 40°C (nonpermissive temperature) (Cole et al., 1998; Yang et al., 2005, 2008). When expressed at the permissive temperature, VSVG-KDEL-R-YFP was localized in the perinuclear region in many NAG-depleted cells as well as mock-transfected cells (Figure 7, type I). In some cells, however, VSVG-KDEL-R-YFP was localized in the ER with some in punctate structures (type II) or distributed as punctate structures throughout the cytoplasm (type III). When the temperature was shifted to 40°C, VSVG-KDEL-R-YFP redistributed to the ER (type II) in most mock-transfected cells, whereas redistribution of VSVG-KDEL-R-YFP to the ER was substantially inhibited in NAG-depleted cells (Figure 7, bottom). These results suggest that the retrograde transport of VSVG-KDEL-R-YFP to the ER is inhibited by NAG depletion.
Figure 7.
Retrograde transport of VSVG-KDEL-R is impaired in NAG-depleted cells. The experiments were conducted as described in Materials and Methods. Expressed VSVG-KDEL-R-YFP was predominantly in the perinuclear region (type I), in the ER (type II), or in diffuse distribution (type III). Bottom, quantitative data. The average of three independent experiments is shown. For each sample, >200 cells were evaluated. Bar, 10 μm.
Depletion of NAG Causes a Defect in the Glycosylation of Secretory and Lysosomal Proteins
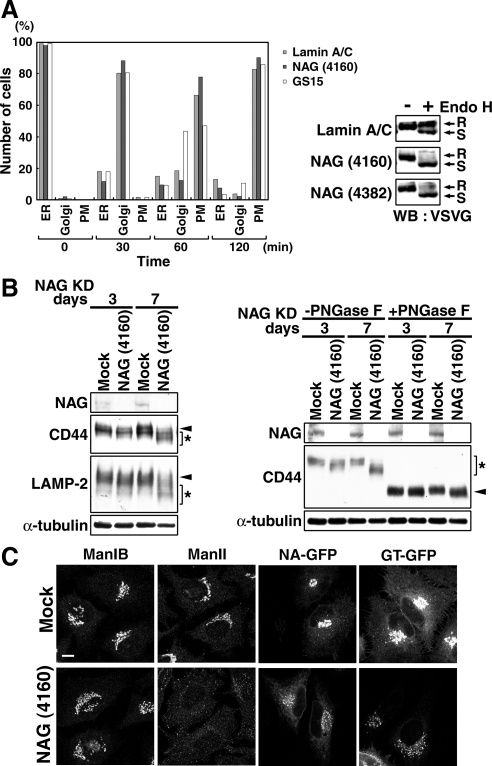
Because Golgi morphology was not markedly disrupted in cells treated with NAG siRNA for 72 h, we examined whether protein transport from the ER to the Golgi is unaffected in NAG-depleted cells. To this end, NAG (4160) and the plasmid encoding VSVG-GFP were sequentially transfected into HeLa cells, and the cells were incubated at the nonpermissive temperature to allow the accumulation of VSVG-GFP at the ER. The transport of VSVG-GFP was monitored by immunofluorescence microscopy after the cells were shifted to the permissive temperature. As shown in Figure 8A, left, no significant retardation of VSVG-GFP transport was observed in NAG-depleted cells, whereas a marked delay was detected in GS15-depleted cells (a positive control). However, when Endo H resistance of VSVG-GFP was analyzed, a large fraction of VSVG-GFP remained Endo H sensitive (Figure 8A, right). A similar result was obtained when NAG (4382) was used to suppress NAG expression. These results suggest that the processing of glycosylation is defective in NAG-depleted cells. To confirm this, we examined the glycosylation of endogenous secretory and lysosomal glycoproteins in NAG-depleted cells. As shown in Figure 8B, left, the gel mobility of both CD44, a plasma membrane-localized protein, and LAMP-2, a lysosomal resident, on SDS-PAGE was increased after 72 h of NAG knockdown, implying the production of underglycosylated protein species. The mobility shifts of the two proteins were more enhanced after a prolonged incubation (7 d). To verify that the increased gel mobility of CD44 was due to defective glycosylation, lysates of NAG-depleted HeLa cells were subjected to PNGase F treatment to remove N-linked oligosaccharides. The molecular weight of the deglycosylated form of CD44 in NAG-depleted cells was found to be almost identical with that in mock transfected cells (Figure 8B, right), indicating that the increased gel mobility of CD44 after NAG knockdown is mainly due to glycosylation defect. Because glycosylation defect was enhanced after a prolong NAG knockdown, we examined the distribution of Golgi glycosylation enzymes at 7 d after transfection of cells with NAG siRNA. In contrast to the phenotype observed at 3-d knockdown of NAG, glycosylation enzymes were detected in centrally, disconnected elements (mannosidase IB, N-acetylglucosaminyltransferase I-GFP, and β-1,4-galactosyltransferase 1-GFP) or in puncta distributed throughout the cytoplasm (Man II) (Figure 8C).
Figure 8.
Depletion of NAG causes a defect in glycosylation of secretory and lysosomal proteins. (A) HeLa cells were successively transfected with lamin A/C siRNA, NAG (4160), or GS15 siRNA and then with the plasmid for VSVG-GFP. Transport of VSVG-GFP was monitored as described in Materials and Methods. The number of cells in which VSVG-GFP was localized in the ER, the Golgi apparatus, and plasma membrane (PM) were counted. Alternatively, lysates of cells double transfected were prepared after 120-min incubation at the permissive temperature and treated with endoglycosidase H (Endo H). The samples were subjected to SDS-PAGE and analyzed by immunoblotting with an anti-VSVG antibody. R, Endo H-resistant bands; S, Endo H-sensitive bands. (B, left) HeLa cells were mock treated or treated with NAG (4160) for 3 or 7 d, solubilized in phosphate-buffered saline with 0.5% SDS, separated by SDS-PAGE, and analyzed by immunoblotting with the indicated antibodies. Arrowheads and asterisks indicate fully glycosylated and underglycosylated proteins, respectively. (B, right) Lysates of HeLa cells depleted of NAG for 3 or 7 d were treated with PNGase F. Samples were loaded on SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. Asterisk and arrowheads indicate the positions of glycosylated and deglycosylated forms of CD44, respectively. (C) Distribution of glycosylation enzymes in HeLa cells treated with NAG (4160) for 7 d. In the expression of N-acetylglucosaminyltransferase I-GFP (NA-GFP) and β-1,4-galactosyltransferase 1-GFP (GT-GFP), the plasmids encoding GFP fusion proteins were transfected into HeLa cells at 6 d after transfection with NAG (4160). Bar, 10 μm.
DISUCUSSION
The NAG gene was first identified as a gene coamplified with the MYCN gene in neuroblastoma (Wimmer et al., 1999). The function of NAG is not known, and there is some controversy concerning whether NAG is associated with tumor progression and prognosis (Scott et al., 2003; Weber et al., 2004; Kaneko et al., 2007). Here, we show that NAG is a subunit of the syntaxin 18 complex and serves as a link between p31 and ZW10-RINT-1 (Figure 9). The N-terminal region (amino acids 1-1035) and C-terminal region (amino acids 1036-2371) of NAG bind p31 and ZW10-RINT-1, respectively. The latter region corresponds to that encoded by a 4.5-kb transcript, which was originally assumed to code for the entire protein (Wimmer et al., 1999). However, a later study demonstrated that a 7.3-kb mRNA encoding 2371 amino acids is the major transcript (Scott et al., 2003). Our immunoblotting analysis showed the presence of the major ∼270-kDa protein, but not the ∼150-kDa protein encoded by the 4.5-kb transcript, suggesting that the 4.5-kb product is not efficiently translated. We confirmed that the major NAG species in IMR-32, a neuroblastoma cell line where NAG was coamplified with MYCN (Scott et al., 2003), is also an ∼270-kDa protein (data not shown). The finding that NAG is encoded by the 7.5-kb transcript makes sense because the N-terminal region that is absent in the product of the 4.5-kb transcript is required for the important function of NAG, i.e., the interaction with p31.
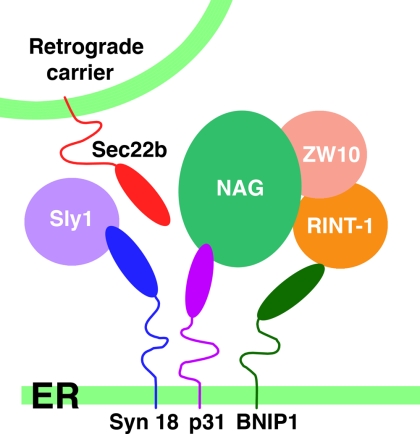
Figure 9.
Schematic representation of the syntaxin 18 complex. NAG associates with p31 and ZW10-RINT-1 through its N-terminal and C-terminal regions, respectively. RINT-1 may weakly interact with BNIP1.
Sequence comparison revealed that the central region of NAG (amino acids 577-1707) shares 17% amino acid identity with yeast 82-kDa Dsl3p/Sec39p (Mnaimneh et al., 2004; Kraynack et al., 2005). NAG seems to be the orthologue of yeast Dsl3p/Sec39p, although their molecular sizes are remarkably different. The finding that the C-terminal region of NAG (amino acids 1036-2371) binds ZW10-RINT-1 is in line with the observation of Kraynack et al. (2005) that the C-terminal region of Dsl3p/Sec39p (amino acids 548-675) interacts with Dsl1p, the yeast ZW10 orthologue (Andag and Schmitt, 2003; Hirose et al., 2004), in a yeast two-hybrid assay. In yeast, Dsl3p/Sec39p as well as Dsl1p greatly facilitates the recruitment of Use1p/Slt1p (yeast p31) to the ternary complex consisting of the Ufe1p (syntaxin 18), Sec20p (BNIP1), and Tip20p (RINT-1), whereas Tip20p plays a central role within the complex (Kraynack et al., 2005). In mammalian cells, NAG is a critical factor to link between p31 and accessory proteins but does not seem to be important for SNARE core complex assembly for the following reasons. First, BNIP1 was fully associated with syntaxin 18 in the absence of NAG (Figure 3A). Second, p31 lacking the N-terminal NAG-binding region (p31ΔN15ΔTMD), as well as full-length p31, was found to bind syntaxin 18 (Figure 2, A and B).
A knockdown experiment demonstrated that NAG contributes to stabilize p31. A similar down-regulation was reported for Use1p/Slt1p in a yeast dsl3 mutant (dsl3-1) (Kraynack et al., 2005). Several syntaxin family members are known to be stabilized by SM proteins (Bryant and James, 2001; Toonen et al., 2005; Braun and Jentsch, 2007; Carpp et al., 2007). Braun and Jentsch (2007) recently demonstrated that Ufe1p is ubiquitinated and degraded by an ERAD-like mechanism in the absence of an SM protein, Sly1p. Our preliminary data also showed that p31 can be ubiquitinated (unpublished data). Further studies will clarify the mechanism of p31 down-regulation and its implication in the formation of the syntaxin 18 complex.
ERGIC-53, KDEL-R, and Rer1 are known to cycle between either the ERGIC or Golgi and the ER in a COPI-dependent, Rab6-independent manner (Sato et al., 1995, 2001; Füllekrug et al., 1997; Kappeler et al., 1997; Orci et al., 1997; Tisdale et al., 1997; Majoul et al., 2001; del Nery et al., 2006). In cells treated with NAG (4160) for 72 h, these proteins showed a diffuse distribution, whereas Golgi-resident proteins, which can be transported to the ER in a Rab6-dependent manner (Girod et al., 1999; White et al., 1999; Malsam et al., 2005; Young et al., 2005), remained in a ribbon-like structure (Figure 5), suggesting that NAG is involved in the COPI-dependent pathway. Indeed, Golgi-to-ER retrograde transport of KDEL-R, a protein that uses a COPI route (Girod et al., 1999), was impaired in NAG-depleted cells. The diffuse distribution of ERGIC and Golgi recycling proteins in NAG-depleted cells may represent nontethered vesicles or tubules. In support of this idea, these vesicles or tubules were found to be easily released from digitonin-permeabilized cells. In contrast, ERGIC-53 and Golgi recycling protein were not released from digitonin-permeabilized control cells. Although Okumura et al. (2006) reported no change in the localization of KDEL-R upon p31 depletion, our results showed redistribution of recycling proteins, including KDEL-R upon knockdown of p31. The difference between their result and ours may be in part due to the fact that the distribution of KDEL-R was analyzed at 24 h after transfection of siRNA by Okumura et al. (2006), but 72 h in our case. Alternatively, the difference may be derived from the use of different cell lines; NIH3T3 cells by Okumura et al. (2006) and HeLa cells by us. It is also possible that Okumura et al. (2006) might not notice a distributional change of KDEL-R in p31-depleted cells because the change was probably more subtle compared with the phenotype of NAG-depleted cells, as shown in this study.
Despite the intimate relationship between NAG and ZW10, the relatively intact Golgi phenotype in NAG-depleted cells is significantly different from that in cells where ZW10 is knocked down (Hirose et al., 2004; Varma et al., 2006; Sun et al., 2007). There are several possible explanations. The first is that the two proteins may control different Golgi-to-ER retrograde trafficking pathways by interacting with other unidentified proteins. Although our data unequivocally demonstrated that NAG and ZW10 are components of the syntaxin 18 complex, it is possible that they interact with other sets of proteins in the ER. Some fractions of ZW10-RINT1 remained associated with membranes in the absence of NAG (Figure 3B). Another possibility is that ZW10 plays a role not only in Golgi-to-ER retrograde transport but also in ER-to-Golgi anterograde transport, as proposed previously (Hirose et al., 2004), whereas NAG is exclusive for the retrograde pathway. ZW10 is a dynamitin-interacting protein and helps the recruitment of the microtubule minus-end–directed motor dynein-dynactin to kinetochores (Starr et al., 1998) and perhaps to other cellular structures, including the ER and Golgi. Dynein-dynactin is known to mediate ER-to-Golgi anterograde transport (Burkhardt et al., 1997, Presley et al., 1997). In certain cells, ZW10 is localized not only in the ER but also in the Golgi apparatus (Varma et al., 2006; Arasaki et al., 2007), near where the minus-end of microtubules is located. Golgi localization of ZW10 may be a result of its movement to the minus-end of microtubules accompanied by anterograde transport.
NAG depletion did not significantly inhibit anterograde transport of VSVG-GFP from the ER to the plasma membrane through the Golgi apparatus, but it affected glycosylation of VSVG-GFP and endogenous proteins. A prolonged depletion of NAG caused a severe glycosylation defect. Shestakova et al. (2006) reported that Golgi modification of CD44 and LAMP-2 is defective in a prolonged (6–9 d) knockdown of COG3, a subunit of the conserved oligomeric Golgi (COG) complex most likely implicated in retrograde transport within the Golgi apparatus (Ungar et al., 2006; Smith and Lupashin, 2008). Glycosylation defect in COG3-depleted cells is likely due to the relocation of Golgi enzymes to COG complex-dependent vesicles (Shestakova et al., 2006), which likely mediate retrograde transport within the Golgi apparatus under normal conditions (Zolov and Lupashin, 2005). Although the distribution of Golgi enzymes was not markedly altered in cells treated with NAG siRNA for 72 h (Figure 5; data not shown), a prolonged knockdown induced their redistribution (Figure 8C). One simple explanation for impaired glycosylation of VSVG-GFP in cells treated with NAG siRNA for 72 h is that glycosylation enzymes could not fully gain access to VSVG-GFP, although they remained in a ribbon-like structure at the level of immunofluorescence microscopy. Because a large amount of VSVG-GFP passed through the Golgi apparatus in a short time, a small change in the distribution of glycosylation enzymes may largely affect glycosylation efficiency of secretory proteins. Alternatively, recycling of oligosaccharide processing enzymes in the ER–Golgi interface may be impaired. It has been reported that yeast α-mannosidase I interacts with Rer1 (Massaad and Herscovics, 2001). In mammals, UDP-glucose:glycoprotein glucosyltransferase, a protein folding sensor and glucosyltransferase, is enriched in the ER-Golgi interface, in addition to the presence in the ER (Zuber et al., 2001). It is possible that perturbation of retrograde transport of proteins, such as Rer1 and KDEL-R, responsible for recycling of oligosaccharide processing enzymes causes a defect in glycosylation.
In future studies, it is important to examine whether NAG plays a role in determining tumor behavior, and if so, what is the mechanism? It should be noted that NAG partners, ZW10 and RINT-1, are spindle and G2/M checkpoint proteins, respectively (Williams et al., 1992; Chan et al., 2000; Xiao et al., 2001). ZW10 in a complex with ROD and Zwilch plays a role in turning off of the spindle checkpoint (Williams et al., 2003). It has been reported that ∼81% of the RINT-1 heterozygotes succumb to multiple tumor formation with haploinsufficiency during their average life span of 24 mo (Lin et al., 2007). Moreover, together with the p130 member of the Rb family, RINT-1 is known to control telomere length in a telomerase-independent manner (Kong et al., 2006). It is tempting to speculate that overexpression of NAG perturbs the quantitative balance of complexes involved in cell cycle-related events and membrane traffic, leading to uncontrolled cell proliferation. Alternatively, overexpression of NAG may affect the attachment of cells to matrix by perturbing cell–matrix binding molecules. It has been reported that MYCN amplification down-regulates CD44 or may affect the glycosylation of CD44 (Gross et al., 1997, 2000). CD44 is involved in cell–cell and cell–matrix interactions and its glycosylation regulates the binding to hyaluronan (Skelton et al., 1998).
In conclusion, our results disclose that the product of a gene coamplified with MYCN is a subunit of the ER SNARE complex and participates in Golgi-to-ER retrograde membrane traffic.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. M. G. Waters, H.-P. Hauri, J. Lippincott-Schwartz, H. D. Söling, N. Nakamura, A. Luini, and B. Mayer for gifts of reagents. We are grateful to K. Suzuki, Y. Aikawa, H. Inaba, K. Miyazaki, and R. Kato for technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research 18370081, 18050036, 18657044, and 19036032 from the Ministry of Education, Science, Sports and Culture of Japan.
Abbreviations used:
- ER
endoplasmic reticulum
- ERGIC
ER-Golgi intermediate compartment
- GFP
green fluorescent protein
- KDEL-R
KDEL receptor
- mAb
monoclonal antibody
- Man II
mannosidase II
- NAG
neuroblastoma-amplified gene
- PNGase F
peptide:N-glycosidase F
- siRNA
short interfering RNA
- SNARE
N-ethylmaleimide-sensitive factor attachment protein receptor
- TMD
transmembrane domain
- VSVG
vesicular stomatitis virus-encoded glycoprotein
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-11-1104) on April 15, 2009.
REFERENCES
- Andag U., Neumann T., Schmitt H. D. The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J. Biol. Chem. 2001;276:39150–39160. doi: 10.1074/jbc.M105833200. [DOI] [PubMed] [Google Scholar]
- Andag U., Schmitt H. D. Dsl1p, an essential component of the Golgi-endoplasmic reticulum retrieval system in yeast, uses the same sequence motif to interact with different subunits of the COPI vesicle coat. J. Biol. Chem. 2003;278:51722–51734. doi: 10.1074/jbc.M308740200. [DOI] [PubMed] [Google Scholar]
- Aoki T., Kojima M., Tani K., Tagaya M. Sec22b-dependent assembly of endoplasmic reticulum Q-SNARE proteins. Biochem. J. 2008;410:93–100. doi: 10.1042/BJ20071304. [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Hauri H. P. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J. Cell Sci. 2006;119:2173–2183. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- Arasaki K., Taniguchi M., Tani K., Tagaya M. RINT-1 regulates the localization and entry of ZW10 to the syntaxin 18 complex. Mol. Biol. Cell. 2006;17:2780–2788. doi: 10.1091/mbc.E05-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasaki K., Uemura T., Tani K., Tagaya M. Correlation of Golgi localization of ZW10 and centrosomal accumulation of dynactin. Biochem. Biophys. Res. Commun. 2007;359:811–816. doi: 10.1016/j.bbrc.2007.05.188. [DOI] [PubMed] [Google Scholar]
- Belgareh-Touze N., Corral-Debrinski M., Launhardt H., Galan J. M., Munder T., Le Panse S., Haguenauer-Tsapis R. Yeast functional analysis: identification of two essential genes involved in ER to Golgi trafficking. Traffic. 2003;4:607–617. doi: 10.1034/j.1600-0854.2003.00116.x. [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Glick B. S. The mechanism of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Braun S., Jentsch S. SM-protein-controlled ER-associated degradation discriminates between different SNAREs. EMBO Rep. 2007;8:1176–1182. doi: 10.1038/sj.embor.7401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant N. J., James D. E. Vps45p stabilizes the syntaxin homologue Tlg2p and positively regulates SNARE complex formation. EMBO J. 2001;20:3380–3388. doi: 10.1093/emboj/20.13.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt J. K., Echeverri C. J., Nilsson T., Vallee R. B. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt P., Hattendorf D. A., Weis W. I., Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27:923–933. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri L., Varlamov O., Doege C. A., Hofmann K., Beilharz T., Rothman J. E., Söllner T. H., Lithgow T. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2003;100:9873–9877. doi: 10.1073/pnas.1734000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpp L. N., Shanks S. G., Struthers M. S., Bryant N. J. Cellular levels of the syntaxin Tlg2p are regulated by a single mode of binding to Vps45p. Biochem. Biophys. Res. Commun. 2007;363:857–860. doi: 10.1016/j.bbrc.2007.09.067. [DOI] [PubMed] [Google Scholar]
- Chan G.K.T., Jablonski S. A., Starr D. A., Goldberg M. L., Yen T. J. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2000;2:944–947. doi: 10.1038/35046598. [DOI] [PubMed] [Google Scholar]
- Cole N. B., Ellenberg J., Song J., DiEuliis D., Lippincott-Schwartz J. Retrograde transport of Golgi-localized proteins to the ER. J. Cell Biol. 1998;140:1–15. doi: 10.1083/jcb.140.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Nery E., Miserey-Lenkei S., Falguières T., Nizak C., Johannes L., Perez F., Goud B. Rab6A and Rab6A' GTPases play non-overlapping roles in membrane trafficking. Traffic. 2006;7:394–407. doi: 10.1111/j.1600-0854.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- Dietrich L. E., Boeddinghaus C., LaGrassa T. J., Ungermann C. Control of eukaryotic membrane fusion by N-terminal domains of SNARE proteins. Biochim. Biophys. Acta. 2003;1641:111–119. doi: 10.1016/s0167-4889(03)00094-6. [DOI] [PubMed] [Google Scholar]
- Dilcher M., Veith B., Chidambaram S., Hartmann E., Schmitt H. D., Fischer von Mollard G. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 2003;22:3664–3674. doi: 10.1093/emboj/cdg339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., Sutton R. B., Brunger A. T., Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllekrug J., Boehm J., Röttger S., Nilsson T., Mieskes G., Schmitt H. D. Human Rer1 is localized to the Golgi apparatus and complements the deletion of the homologous Rer1 protein of Saccharomyces cerevisiae. Eur. J. Cell Biol. 1997;74:31–40. [PubMed] [Google Scholar]
- Gerst J. E. SNARE regulators: matchmakers and matchbreakers. Biochem. Biophys. Acta. 2003;1641:99–110. doi: 10.1016/s0167-4889(03)00096-x. [DOI] [PubMed] [Google Scholar]
- Girod A., Storrie B., Simpson J. C., Johannes L., Goud B., Roberts L. M., Lord J. M., Nilsson T., Pepperkok R. Evidence for a COP-I-independent transport route from the Golgi complex to the endoplasmic reticulum. Nat. Cell Biol. 1999;1:423–430. doi: 10.1038/15658. [DOI] [PubMed] [Google Scholar]
- Gross N., Balmas K., Brognara C. B. Absence of functional CD44 hyaluronan receptor on human NMYC-amplified neuroblastoma cells. Cancer Res. 1997;57:1387–1393. [PubMed] [Google Scholar]
- Gross N., Balmas–Bourloud K., Brognara C. B. MYCN-related suppression of functional CD44 expression enhances tumorigenic properties of human neuroblastoma cells. Exp. Cell Res. 2000;260:396–403. doi: 10.1006/excr.2000.5007. [DOI] [PubMed] [Google Scholar]
- Hatsuzawa K., Hirose H., Tani K., Yamamoto A., Scheller R. H., Tagaya M. Syntaxin 18, a SNAP receptor that functions in the endoplasmic reticulum, intermediate compartment, and cis-Golgi vesicle trafficking. J. Biol. Chem. 2000;275:13713–13720. doi: 10.1074/jbc.275.18.13713. [DOI] [PubMed] [Google Scholar]
- Hatsuzawa K., Tamura T., Hashimoto H., Hashimoto H., Yokoya S., Miura M., Nagaya H., Wada I. Involvement of syntaxin 18, an endoplasmic reticulum (ER)-localized SNARE protein, in ER-mediated phagocytosis. Mol. Biol. Cell. 2006;17:3964–3977. doi: 10.1091/mbc.E05-12-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay J. C., Klumperman J., Oorschot V., Steegmaier M., Kuo C. S., Scheller R. H. Localization, dynamics, and protein interactions reveal distinct roles for ER and Golgi SNAREs. J. Cell Biol. 1998;141:1489–1502. doi: 10.1083/jcb.141.7.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose H., Arasaki K., Dohmae N., Takio K., Hatsuzawa K., Nagahama M., Tani K., Yamamoto A., Tohyama M., Tagaya M. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 2004;23:1267–1278. doi: 10.1038/sj.emboj.7600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W. SNAREs and traffic. Biochim. Biophys. Acta. 2005;1744:120–144. doi: 10.1016/j.bbamcr.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Iinuma T., Shiga A., Nakamoto K., O'brien M. B., Aridor M., Arimitsu N., Tagaya M., Tani K. Mammalian Sec16/p250 plays a role in membrane traffic from the endoplasmic reticulum. J. Biol. Chem. 2007;282:17632–17639. doi: 10.1074/jbc.M611237200. [DOI] [PubMed] [Google Scholar]
- Iinuma T., Aoki T., Arasaki K., Hirose H., Yamamoto A., Samata R., Hauri H. P., Arimitsu N., Tagaya M., Tani K. Role of syntaxin 18 in the organization of endoplasmic reticulum subdomains. J. Cell Sci. 2009;122:1680–1690. doi: 10.1242/jcs.036103. [DOI] [PubMed] [Google Scholar]
- Inoue M., Arasaki K., Ueda A., Aoki T., Tagaya M. N-terminal region of ZW10 serves not only as a determinant for localization but also as a link with dynein function. Genes Cells. 2008;13:905–914. doi: 10.1111/j.1365-2443.2008.01215.x. [DOI] [PubMed] [Google Scholar]
- Jahn R., Scheller R. H. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Kaji H., Yamauchi Y., Takahashi N., Isobe T. Mass spectrometric identification of N-linked glycopeptides using lectin-mediated affinity capture and glycosylation site-specific stable isotope tagging. Nat. Protoc. 2006;1:3019–3302. doi: 10.1038/nprot.2006.444. [DOI] [PubMed] [Google Scholar]
- Kaneko S., Ohira M., Nakamura Y., Isogai E., Nakagawara A., Kaneko M. Relationship of DDX1 and NAG gene amplification/overexpression to the prognosis of patients with MYCN-amplified neuroblastoma. J. Cancer Res. Clin. Oncol. 2007;133:185–192. doi: 10.1007/s00432-006-0156-y. [DOI] [PubMed] [Google Scholar]
- Kappeler F., Klopfenstein D. R., Foguet M., Paccaud J.-P., Hauri H. P. The recycling of ERGIC-53 in the early secretory pathway. ERGIC-53 carries a cytosolic endoplasmic reticulum-exit determinant interacting with COP II. J. Biol. Chem. 1997;272:31801–31808. doi: 10.1074/jbc.272.50.31801. [DOI] [PubMed] [Google Scholar]
- Kong L. J., Meloni A. R., Nevins J. R. The Rb-related p130 protein controls telomere lengthening through an interaction with a Rad50-interacting protein, RINT-1. Mol. Cell. 2006;22:63–71. doi: 10.1016/j.molcel.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Kraynack B. A., Chan A., Rosenthal E., Essid M., Umansky B., Waters M. G., Schmitt H. D. Dsl1p, Tip20p, and the novel Dsl3 (Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast. Mol. Biol. Cell. 2005;16:3963–3977. doi: 10.1091/mbc.E05-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M., Pelham H.R.B. SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell. 1996;85:205–215. doi: 10.1016/s0092-8674(00)81097-1. [DOI] [PubMed] [Google Scholar]
- Lewis M. J., Rayner J. C., Pelham H. R. A novel SNARE complex implicated in vesicle fusion with the endoplasmic reticulum. EMBO J. 1997;16:3017–3024. doi: 10.1093/emboj/16.11.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Liu C. C., Gao Q., Zhang X., Wu G., Lee W. H. RINT-1 serves as a tumor suppressor and maintains Golgi dynamics and centrosome integrity for cell survival. Mol. Cell. Biol. 2007;27:4905–4916. doi: 10.1128/MCB.02396-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malsam J., Satoh A., Pelletier L., Warren G. Golgin tethers define subpopulations of COPI vesicles. Science. 2005;307:1095–1098. doi: 10.1126/science.1108061. [DOI] [PubMed] [Google Scholar]
- Majoul I., Straub M., Hell S. W., Duden R., Söling H. D. KDEL-cargo regulates interactions between proteins involved in COPI vesicle traffic: measurements in living cells using FRET. Dev. Cell. 2001;1:139–153. doi: 10.1016/s1534-5807(01)00004-1. [DOI] [PubMed] [Google Scholar]
- Massaad M. J., Herscovics A. Interaction of the endoplasmic reticulum α-1,2-mannosidase Mns1p with Rer1p using the split-ubiquitin system. J. Cell Sci. 2001;114:4629–4635. doi: 10.1242/jcs.114.24.4629. [DOI] [PubMed] [Google Scholar]
- McNew J. A., Parlati F., Fukuda R., Johnston R. J., Paz K., Paumet F., Söllner T. H., Rothman J. E. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- Mnaimneh S., et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–34. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Hirose H., Taniguchi M., Kurashina H., Arasaki K., Nagahama M., Tani K., Yamamoto A., Tagaya M. Involvement of BNIP1 in apoptosis and endoplasmic reticulum membrane fusion. EMBO J. 2004;23:3216–3226. doi: 10.1038/sj.emboj.7600333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T., Yamauchi Y., Nakayama H., Shinkawa T., Yanagida M., Takahashi N., Isobe T. A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal. Chem. 2002;74:4725–4733. doi: 10.1021/ac020018n. [DOI] [PubMed] [Google Scholar]
- Okumura A. J., Hatsuzawa K., Tamura T., Nagaya H., Saeki K., Okumura F., Nagao K., Nishikawa M., Yoshimura A., Wada I. Involvement of a novel Q-SNARE, D12, in quality control of the endomembrane system. J. Biol. Chem. 2006;281:4495–4506. doi: 10.1074/jbc.M509715200. [DOI] [PubMed] [Google Scholar]
- Orci L., Stamnes M, Ravazzola M., Amherdt M., Perrelet A, Söllner T. H., Rothman J. E. Bidirectional transport by distinct populations of COPI-coated vesicles. Cell. 1997;90:335–349. doi: 10.1016/s0092-8674(00)80341-4. [DOI] [PubMed] [Google Scholar]
- Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- Patel S. K., Indig F. E., Olivieri N., Levine N. D., Latterich M. Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell. 1998;92:611–620. doi: 10.1016/s0092-8674(00)81129-0. [DOI] [PubMed] [Google Scholar]
- Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J., Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Reilly B. A., Kraynack B. A., VanRheenen S. M., Waters M. G. Golgi-to-endoplasmic reticulum (ER) retrograde traffic in yeast requires Dsl1p, a component of the ER target site that interacts with a COPI coat subunit. Mol. Biol. Cell. 2001;12:3783–3796. doi: 10.1091/mbc.12.12.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R., Saraste J., Goud B. Retrograde traffic in the biosynthetic-secretory route: pathways and machinery. Curr. Opin. Cell Biol. 2003;15:438–445. doi: 10.1016/s0955-0674(03)00077-2. [DOI] [PubMed] [Google Scholar]
- Sato K., Nishikawa S., Nakano A. Membrane protein retrieval from the Golgi apparatus to the endoplasmic reticulum (ER): characterization of the RER1 gene product as a component involved in ER localization of Sec12p. Mol. Biol. Cell. 1995;6:1459–1477. doi: 10.1091/mbc.6.11.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Sato M., Nakano A. Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J. Cell Biol. 2001;152:935–944. doi: 10.1083/jcb.152.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. K., Board J. R., Lu X., Pearson A. D., Kenyon R. M., Lunec J. The neuroblastoma amplified gene, NAG: genomic structure and characterisation of the 7.3 kb transcript predominantly expressed in neuroblastoma. Gene. 2003;307:1–11. doi: 10.1016/s0378-1119(03)00459-1. [DOI] [PubMed] [Google Scholar]
- Shestakova A., Zolov S., Lupashin V. COG complex-mediated recycling of Golgi glycosyltransferases is essential for normal protein glycosylation. Traffic. 2006;7:191–204. doi: 10.1111/j.1600-0854.2005.00376.x. [DOI] [PubMed] [Google Scholar]
- Shimoi W., Ezawa I., Nakamoto K., Uesaki S., Gabreski G., Aridor M., Yamamoto A., Nagahama M., Tagaya M., Tani K. p125 is localized in endoplasmic reticulum exit sites and involved in their organization. J. Biol. Chem. 2005;280:10141–10148. doi: 10.1074/jbc.M409673200. [DOI] [PubMed] [Google Scholar]
- Shinkawa T., Taoka M., Yamauchi Y., Ichimura T., Kaji H., Takahashi N., Isobe T. STEM: a software tool for large-scale proteomic data analyses. J. Proteome Res. 2005;4:1826–1831. doi: 10.1021/pr050167x. [DOI] [PubMed] [Google Scholar]
- Skelton T. P., Zeng C., Nocks A., Stamenkovic I. Glycosylation provides both stimulatory and inhibitory effects on cell surface and soluble CD44 binding to hyaluronan. J. Cell Biol. 1998;140:431–446. doi: 10.1083/jcb.140.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. D., Lupashin V. V. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr. Res. 2008;343:2024–2031. doi: 10.1016/j.carres.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T., Whiteheart S. W., Brunner M., Erdjument-Bromage H., Geromanos S., Tempst P., Rothman J. E. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Starr D. A., Williams B. C., Hays T. S., Goldberg M. L. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Shestakova A., Hunt L., Sehgal S., Lupashin V., Storrie B. Rab6 regulates both ZW10/RINT-1- and conserved oligomeric Golgi complex-dependent Golgi trafficking and homeostasis. Mol. Biol. Cell. 2007;18:4129–4142. doi: 10.1091/mbc.E07-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Sweet D. J., Pelham H. R. The Saccharomyces cerevisiae SEC20 gene encodes a membrane glycoprotein which is sorted by the HDEL retrieval system. EMBO J. 1992;11:423–432. doi: 10.1002/j.1460-2075.1992.tb05071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet D. J., Pelham H. R. The TIP1 gene of Saccharomyces cerevisiae encodes an 80 kDa cytoplasmic protein that interacts with the cytoplasmic domain of Sec20p. EMBO J. 1993;12:2831–2840. doi: 10.1002/j.1460-2075.1993.tb05944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagaya M., Furuno A., Mizushima S. SNAP prevents Mg2+-ATP-induced release of N-ethylmaleimide-sensitive factor from the Golgi apparatus in digitonin-permeabilized PC12 cells. J. Biol. Chem. 1996;271:466–470. doi: 10.1074/jbc.271.1.466. [DOI] [PubMed] [Google Scholar]
- Tani K., Shibata M., Kawase K., Kawashima H., Hatsuzawa K., Nagahama M., Tagaya M. Mapping of functional domains of γ-SNAP. J. Biol. Chem. 2003;278:13531–13538. doi: 10.1074/jbc.M213205200. [DOI] [PubMed] [Google Scholar]
- Tisdale E. J., Plutner H., Matteson J., Balch W. E. p53/58 binds COPI and is required for selective transport through the early secretory pathway. J. Cell Biol. 1997;137:581–593. doi: 10.1083/jcb.137.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonen R. F., Verhage M. Vesicle trafficking: pleasure and pain from SM genes. Trends Cell Biol. 2003;13:177–186. doi: 10.1016/s0962-8924(03)00031-x. [DOI] [PubMed] [Google Scholar]
- Toonen R. F., de Vries K. J., Zalm R., Südhof T. C., Verhage M. Munc18-1 stabilizes syntaxin 1, but is not essential for syntaxin 1 targeting and SNARE complex formation. J. Neurochem. 2005;93:1393–1400. doi: 10.1111/j.1471-4159.2005.03128.x. [DOI] [PubMed] [Google Scholar]
- Uemura T., Sato T., Aoki T., Yamamoto A., Okada T., Hirai R., Harada R., Mori K., Tagaya M., Harada A. p31 deficiency influences endoplasmic reticulum tubular morphology and cell survival. Mol. Cell. Biol. 2009;29:1869–1881. doi: 10.1128/MCB.01089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D., Oka T., Krieger M., Hughson F. M. Retrograde transport on the COG railway. Trends Cell Biol. 2006;16:113–120. doi: 10.1016/j.tcb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Varma D., Dujardin D. L., Stehman S. A., Vallee R. B. Role of the kinetochore/cell cycle checkpoint protein ZW10 in interphase cytoplasmic dynein function. J. Cell Biol. 2006;172:655–662. doi: 10.1083/jcb.200510120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen S. M., Reilly B. A., Chamberlain S. J., Waters M. G. Dsl1p, an essential protein required for membrane traffic at the endoplasmic reticulum/Golgi interface in yeast. Traffic. 2001;2:212–231. doi: 10.1034/j.1600-0854.2001.020307.x. [DOI] [PubMed] [Google Scholar]
- Wakana Y., Takai S., Nakajima K., Tani K., Yamamoto A., Watson P., Stephens D. J., Hauri H. P., Tagaya M. Bap31 is an itinerant protein that moves between the peripheral endoplasmic reticulum (ER) and a juxtanuclear compartment related to ER-associated degradation. Mol. Biol. Cell. 2008;19:1825–1836. doi: 10.1091/mbc.E07-08-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Söllner T. H., Rothman J. E. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weber A., Imisch P., Bergmann E., Christiansen H. Coamplification of DDX1 correlates with an improved survival probability in children with MYCN-amplified human neuroblastoma. J. Clin. Oncol. 2004;22:2681–2690. doi: 10.1200/JCO.2004.07.192. [DOI] [PubMed] [Google Scholar]
- Weimbs T., Low S. H., Chapin S. J., Mostov K. E., Bucher P., Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc. Natl. Acad. Sci. USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J., et al. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J. Cell Biol. 1999;147:743–760. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. C., Karr T. L., Montgomery J. M., Goldberg M. L. The Drosophila l(1) zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J. Cell Biol. 1992;118:759–773. doi: 10.1083/jcb.118.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. C., Li Z.-X., Liu S., Williams E. V., Leung G., Yen T. J., Goldberg M. L. Zwilch, a new component of the ZW10/ROD complex required for kinetochore functions. Mol. Biol. Cell. 2003;14:1379–1391. doi: 10.1091/mbc.E02-09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer K., Zhu X. X., Lamb B. J., Kuick R., Ambros P. F., Kovar H., Thoraval D., Motyka S., Alberts J. R., Hanash S. M. Co-amplification of a novel gene, NAG, with the N-myc gene in neuroblastoma. Oncogene. 1999;18:233–238. doi: 10.1038/sj.onc.1202287. [DOI] [PubMed] [Google Scholar]
- Xiao J., Liu C.-C., Chen P.-L., Lee W.-H. RINT-1, a novel rad50-interacting protein, participates in radiation induced G2/M checkpoint control. J. Biol. Chem. 2001;276:6105–6111. doi: 10.1074/jbc.M008893200. [DOI] [PubMed] [Google Scholar]
- Yang J. S., Lee S. Y., Spanò S., Gad H., Zhang L., Nie Z., Bonazzi M., Corda D., Luini A., Hsu V. W. A role for BARS at the fission step of COPI vesicle formation from Golgi membrane. EMBO J. 2005;24:4133–4143. doi: 10.1038/sj.emboj.7600873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. S., et al. A role for phosphatidic acid in COPI vesicle fission yields insights into Golgi maintenance. Nat. Cell Biol. 2008;10:1146–1153. doi: 10.1038/ncb1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J., Stauber T., del Nery E., Vernos I., Pepperkok R., Nilsson T. Regulation of microtubule-dependent recycling at the trans-Golgi network by Rab6A and Rab6A'. Mol. Biol. Cell. 2005;16:162–177. doi: 10.1091/mbc.E04-03-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wong S. H., Tang B. L., Xu Y., Hong W. Morphological and functional association of Sec22b/ERS-24 with the pre-Golgi intermediate compartment. Mol. Biol. Cell. 1999;10:435–453. doi: 10.1091/mbc.10.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolov S. N., Lupashin V. V. Cog3p depletion blocks vesicle mediated Golgi retrograde trafficking in HeLa cells. J. Cell Biol. 2005;168:747–759. doi: 10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber C., Fan J. Y., Guhl B., Parodi A., Fessler J. H., Parker C., Roth J. Immunolocalization of UDP-glucose:glycoprotein glucosyltransferase indicates involvement of pre-Golgi intermediates in protein quality control. Proc. Natl. Acad. Sci .USA. 2001;98:10710–10715. doi: 10.1073/pnas.191359198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.