Gene expression profiling (GEP) provides a complete picture of the transcriptome, which reflects the specific activation/differentiation states of a given cell population. The advent of GEP in the field of hematology has allowed significant strides to be made in defining the molecular taxonomy of leukemias and lymphomas. Tumors that are otherwise morphologically identical can now be classified according to molecular patterns predictive of distinct clinical outcomes. Success with such applications has led to the development and implementation of diagnostic and prognostic strategies based solely on microarray data.1
Our current understanding of the pathogenesis, maintenance and progression of chronic lymphocytic leukemia (CLL) has been greatly enhanced by GEP data. It is well established that CLL is a heterogeneous disease: some patients experience a slowly progressive clinical course, but most will eventually enter an advanced phase requiring repeated treatment. A significant number of CLL patients exhibit an active form of the disease from the early stages, characterized by refractoriness to treatment, infectious and autoimmune complications and a relatively rapid fatal outcome.2 One of the long-term goals of the hematologic community is to provide a molecular explanation for the marked clinical heterogeneity of CLL. A number of descriptive parameters characterizing aggressive CLL were defined in the 1980s and 1990s, but a significant breakthrough came in 1999, when two independent groups showed that patients could be placed in distinct prognostic groups according to the presence (good prognosis) or absence (poor prognosis) of somatic mutations in the immunoglobulin variable region (IgV) genes.3,4
The potential of GEP was immediately exploited to answer the long-standing questions concerning the origin of the CLL cell and its relationship with normal B lymphocytes. It has also been used to explore whether the clinical heterogeneity of the disease might depend on the genetic origin of the neoplastic cells. Recent studies on this topic by several groups, including Stamatopoulos and colleagues in this issue of the journal,5 rely on GEP to classify CLL cells according to their molecular markers and to identify the corresponding genetic signatures. Overall, the results of these studies generally concur that CLL cells are unexpectedly homogeneous in terms of their genetic signature.6,7 However, all of these studies have identified a number of genes whose differential expression distinguishes between aggressive and stable CLL.
Among the genetic markers of poor prognosis, results from Stamatopoulos5 and from our group8 have consistently pointed to genes coding for proteins involved in the control of cell movement. This highlights the significance of topographical issues in disease progression and provides convincing evidence that CLL cells with enhanced motility are associated with aggressive disease. Independent confirmation of these results stems from data generated in patients, showing that a significant proportion of the leukemic clone proliferates and that proliferation occurs predominantly in lymphoid organs.9 Furthermore, CLL cells characterized by the highest sensitivity to chemokines also express molecular markers associated with poor prognosis.10 Lastly, histological analyses indicate that proliferating CLL cells are located in specialized areas in the bone marrow (BM) and in peripheral lymphoid organs.11
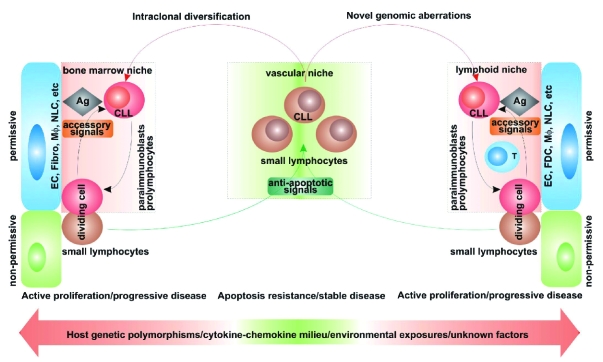
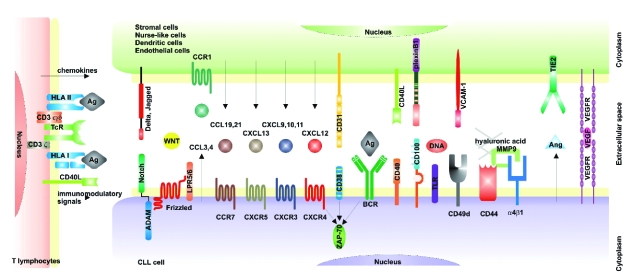
Together, these data have contributed to our current view of CLL as a disease characterized by a dynamic balance between cells circulating in the blood and cells located in permissive niches in lymphoid organs.12 The former are prevalently mature-looking small lymphocytes resistant to apoptosis, whereas the latter comprise pro-lymphocytes and paraimmunoblasts that can proliferate. It is legitimate to assume that the host microenvironment determines the balance between resistance to apoptosis and active proliferation by providing different sets of signals and conditions (Figure 1). It is therefore imperative to understand the molecular mechanisms regulating CLL trafficking between BM, blood, lymph nodes (LN) and spleen. Of the intricate network of soluble and cell-bound signals regulating interactions between CLL cells and the microenvironment, the most promising in terms of possible translational applications are briefly outlined below and schematically represented in Figure 2.
Figure 1.
Hypothetical model explaining the key role played by the microenvironment in maintenance and progression of chronic lymphocytic leukemia. EC: endothelial cells; Fibro: fibroblasts; Mφ: macrophages; NLC: nurse-like cells; Ag: antigen.
Figure 2.
Main signaling pathways regulating the interactions between chronic lymphocytic leukemia cells and the microenvironment.
Chemokines and chemokine receptors
CXCR4 is expressed by most circulating CLL cells at high levels, independently from molecular markers, clinical stages or patterns of BM infiltration. BM stromal or nurse-like (NLC) cells constitutively secrete CXCL12, the ligand of CXCR4. Consequently, the CXCL12/CXCR4 axis plays a crucial role in the recruitment of neoplastic cells to growth-favorable environments.13 The functional responses induced by CXCR4 ligation, however, are highly variable among CLL patients and appear to correlate with the presence of ZAP-70 and CD38.10 A similar circuit has been proposed for the CXCL13/CXCR5 axis, where NLC and CD68+ macrophages in the BM secrete CXCL13 and CLL cells express functional CXCR5 receptors.14 CXCR3 expression by CLL cells is highly variable, although stable over time. CXCR3+ CLL patients are generally in a more advanced disease stage, present with a diffuse pattern of BM infiltration and tend to express molecular markers associated with poor prognosis. CCR7 is also expressed at high levels by CLL cells, with the intensity correlating with LN involvement.15,16 CLL cells are also markedly sensitive to the CCL19 and CCL21 ligands and show more enhanced trans-endothelial migration than normal B lymphocytes. CLL19 and CCL21 are expressed by high endothelial venules in the LN and the system is effective in recruiting CLL cells from blood to lymphoid organs.15
CLL cells can also actively secrete chemokines to modify the environment by recruiting accessory cells and creating growth-permissive conditions. Although preliminary, recent results,17,18 concur that BCR- and CD38-mediated signals effectively increase secretion of CLL3 and CLL4, recruiting supportive CD68+ macrophages and NLC.
Other soluble factors
Among the soluble factors regulating the balance between disease stability and progression are small molecules with pro-angiogenetic power. Increased angiogenesis has been consistently associated with more advanced disease phases and it is known that CLL cells themselves can secrete pro-angiogenetic factors [including fibroblast growth factor (FGF), vascular endothelial growth factor (VEGF) and angiopoietin (Ang)] and drive the construction of new blood vessels. These newly formed vessels are characterized by increased permeability, and thus contribute to disease dissemination. CLL cells can also express receptors for some of these pro-angiogenetic factors [including VEGF receptors (VEGFR) 1 and 2 and Tie-2, the Ang receptor]: the signal transduced significantly prolongs cell survival.19
Thioredoxin is a multifunctional protein, ubiquitously expressed at low levels in all cells of the body. Additionally, some types of cells have the capacity to release thioredoxin. This extracellular form of thioredoxin has cytokine and chemokine activities. It was recently shown that thioredoxin is expressed in the LN of CLL patients, where it is secreted mainly by accessory stromal cells.20 Extracellular thioredoxin can increase CLL survival, at least using in vitro models.21
Receptor/ligand pairs
CLL cells nestled in spleen, LN and BM find the ideal microenvironment for growth because the antigen is present together with the appropriate set of accessory signals. Among the latter, the best known example is represented by CD40L, abundantly expressed by T lymphocytes within the pseudofollicles and contributing to the proliferation of CD40+ CLL cells.22
We have focused on CD38, initially considered a surrogate of the absence of mutations in the IgV genes3 and now an established independent negative prognostic marker. The working hypothesis of our group is that CD38 is not merely a marker in CLL, but a cell surface receptor and adhesion molecule (with CD31 as a counter-receptor) directly involved in the delivery of growth signals.23 Prolonged CD31/CD38 contacts under static conditions significantly increase survival of the CLL clone and sustain its proliferation in vitro. The kinase ZAP-70 represents a necessary component in the molecular chain activated by CD38, providing a functional link for what were previously considered to be two independent negative prognostic markers. Furthermore, recent observations indicate that CD38+/ZAP-70+ CLL cells are characterized by the enhanced propensity to migrate in response to CXCL12, an essential chemokine in the recirculation of neoplastic cells between blood and the lymphoid organs. Thus, it appears that the CD31/CD38/ZAP-70 axis may represent a point of convergence of proliferative and migratory signals. CD38/CD31 interactions are followed by a marked upregulation of the semaphorin family member CD100, which in turn interacts with the plexin B1 ligand expressed by stromal cells and contributes to further sustain proliferation and survival of CLL cells.24,25
Adhesion molecules and MMPs
The adhesion molecule profile of CLL cells is the focus of intense investigation, since this profile can influence and determine the ability of neoplastic cells to respond to chemokines and to home where the antigen and accessory signals are abundantly present. Specific attention has been dedicated to α4β1 integrin, whose expression correlates with nodal disease and which is necessary for trans-endothelial migration.26 Together with CD44, this integrin complex serves as docking site for active form of the matrix metalloprotease-9 (MMP-9).27 The functional implications of this complex would depend on the extracellular concentration of MMP-9 and of its substrates (CXCL12 among the others) and could lead to CLL extravasation and leukemic cell arrest in a closed microenvironment.
Conclusions
Evidence from independent experts in different fields consistently identify lymphoid organs as the carriers of the proliferative core of CLL. The host microenvironment and the resulting interplay between the genetic background and environmental influences thus play a crucial role in disease progression, as well as in resistance to treatment. The results presented by Stamatopoulos and colleagues5 further support the view that a favorable microenvironment can provide the optimal combination of antigen and co-signals for expansion of the CLL-initiating cells, leading to disease progression. Instead, under environmental conditions which do not favor growth, proliferation of the same CLL cells is impaired, and the molecular mechanisms preventing apoptosis prevail. The clinical conditions of the patient reflect the overall equilibrium between these two opposing processes. To date, several of the mechanisms described above have become the object of intense investigation for the design of new therapeutic strategies for blocking CLL cells in the circulation. The advantage of such an approach would be the increased availability of leukemic cells to associated chemotherapeutics. Relevant examples are monoclonal antibodies (mAbs) specific for CD40, CD49d and CD38 that have been proposed as potential adjuncts to conventional chemotherapeutic regimens. Alternative approaches are based on the interruption of angiogenetic or anti-apoptotic circuits. The development of these or related strategies, together or in combination, is expected to improve the outcome and quality of life of CLL patients.
Footnotes
Silvia Deaglio, M.D., Ph.D. is Assistant Professor of Medical Genetics and Fabio Malavasi, M.D. is Professor of Medical Genetics. Both work at the Laboratory of Immunogenetics in the Department of Genetics, Biology and Biochemistry of the University of Turin, Italy.
The authors’ studies examined in this perspective article have been supported by grants from Associazione Italiana Ricerca Cancro (AIRC), Compagnia di San Paolo, University of Turin, Fondazione Guido Berlucchi, PRIN/MIUR, Regione Piemonte and Fondazione Internazionale Ricerche Medicina Sperimentale (FIRMS), Italy.
References
- 1.Shaffer AL, Wright G, Yang L, Powell J, Ngo V, Lamy L, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 2.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–15. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 3.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. [PubMed] [Google Scholar]
- 4.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–54. [PubMed] [Google Scholar]
- 5.Stamatopoulos B, Haibe-Kains B, Equeter C, Meuleman N, Sorée A, de Bruyn C, et al. Gene expression profiling based on ZAP70 mRNA expression reveals differences in microenvironment interaction between patients with good and poor prognosis. Haematologica. 2009;94:790–9. doi: 10.3324/haematol.2008.002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein U, Tu Y, Stolovitzky GA, Mattioli M, Cattoretti G, Husson H, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194:1625–38. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–47. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deaglio S, Vaisitti T, Aydin S, Bergui L, D’Arena G, Bonello L, et al. CD38 and ZAP-70 are functionally linked and mark CLL cells with high migratory potential. Blood. 2007;110:4012–21. doi: 10.1182/blood-2007-06-094029. [DOI] [PubMed] [Google Scholar]
- 9.Messmer BT, Messmer D, Allen SL, Kolitz JE, Kudalkar P, Cesar D, et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J Clin Invest. 2005;115:755–64. doi: 10.1172/JCI23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson SJ, Matthews C, Catherwood MA, Alexander HD, Carey BS, Farrugia J, et al. ZAP-70 expression is associated with enhanced ability to respond to migratory and survival signals in B-cell chronic lymphocytic leukemia (B-CLL) Blood. 2006;107:3584–92. doi: 10.1182/blood-2005-04-1718. [DOI] [PubMed] [Google Scholar]
- 11.Rosati S, Kluin PM. Chronic lymphocytic leukaemia: a review of the immuno-architecture. Curr Top Microbiol Immunol. 2005;294:91–107. [PubMed] [Google Scholar]
- 12.Caligaris-Cappio F, Ghia P. Novel insights in chronic lymphocytic leukemia: are we getting closer to understanding the pathogenesis of the disease? J Clin Oncol. 2008;26:4497–503. doi: 10.1200/JCO.2007.15.4393. [DOI] [PubMed] [Google Scholar]
- 13.Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–7. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- 14.Burkle A, Niedermeier M, Schmitt-Graff A, Wierda WG, Keating MJ, Burger JA. Overexpression of the CXCR5 chemokine receptor, and its ligand, CXCL13 in B-cell chronic lymphocytic leukemia. Blood. 2007;110:3316–25. doi: 10.1182/blood-2007-05-089409. [DOI] [PubMed] [Google Scholar]
- 15.Till KJ, Lin K, Zuzel M, Cawley JC. The chemokine receptor CCR7 and alpha4 integrin are important for migration of chronic lymphocytic leukemia cells into lymph nodes. Blood. 2002;99:2977–84. doi: 10.1182/blood.v99.8.2977. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Giral S, Quintana NE, Cabrerizo M, Alfonso-Perez M, Sala-Valdes M, De Soria VG, et al. Chemokine receptors that mediate B cell homing to secondary lymphoid tissues are highly expressed in B cell chronic lymphocytic leukemia and non-Hodgkin lymphomas with widespread nodular dissemination. J Leukoc Biol. 2004;76:462–71. doi: 10.1189/jlb.1203652. [DOI] [PubMed] [Google Scholar]
- 17.Burger JA, Quiroga MP, Hartmann E, Bürkle A, Wierda WG, Keating MJ, Rosenwald A. High-level expression of the T cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell co-cultures and after BCR stimulation. Blood. 2008;113:3050–8. doi: 10.1182/blood-2008-07-170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zucchetto A, Benedetti D, Tripodo C, Bomben R, Dal Bo M, Marconi D, et al. CD38/CD31, the CCL3 and CCL4 chemokines, and CD49d/VCAM-1 are interchained by sequential events sustaining chronic lymphocytic leukemia cell survival. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-08-4173. in press. [DOI] [PubMed] [Google Scholar]
- 19.Letilovic T, Vrhovac R, Verstovsek S, Jaksic B, Ferrajoli A. Role of angiogenesis in chronic lymphocytic leukemia. Cancer. 2006;107:925–34. doi: 10.1002/cncr.22086. [DOI] [PubMed] [Google Scholar]
- 20.Backman E, Bergh AC, Lagerdahl I, Rydberg B, Sundstrom C, Tobin G, et al. Thioredoxin, produced by stromal cells retrieved from the lymph node microenvironment, rescues chronic lymphocytic leukemia cells from apoptosis in vitro. Haematologica. 2007;92:1495–504. doi: 10.3324/haematol.11448. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson J, Soderberg O, Nilsson K, Rosen A. Thioredoxin prolongs survival of B-type chronic lymphocytic leukemia cells. Blood. 2000;95:1420–6. [PubMed] [Google Scholar]
- 22.von Bergwelt-Baildon M, Maecker B, Schultze J, Gribben JG. CD40 activation: potential for specific immunotherapy in B-CLL. Ann Oncol. 2004;15:853–7. doi: 10.1093/annonc/mdh213. [DOI] [PubMed] [Google Scholar]
- 23.Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, et al. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev. 2008;88:841–86. doi: 10.1152/physrev.00035.2007. [DOI] [PubMed] [Google Scholar]
- 24.Deaglio S, Vaisitti T, Aydin S, Ferrero E, Malavasi F. In-tandem insight from basic science combined with clinical research: CD38 as both marker and key component of the pathogenetic network underlying chronic lymphocytic leukemia. Blood. 2006;108:1135–44. doi: 10.1182/blood-2006-01-013003. [DOI] [PubMed] [Google Scholar]
- 25.Deaglio S, Aydin S, Vaisitti T, Bergui L, Malavasi F. CD38 at the junction between prognostic marker and therapeutic target. Trends Mol Med. 2008;14:210–8. doi: 10.1016/j.molmed.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Till KJ, Spiller DG, Harris RJ, Chen H, Zuzel M, Cawley JC. CLL, but not normal, B cells are dependent on autocrine VEGF and alpha4beta1 integrin for chemokine-induced motility on and through endothelium. Blood. 2005;105:4813–9. doi: 10.1182/blood-2004-10-4054. [DOI] [PubMed] [Google Scholar]
- 27.Redondo-Munoz J, Ugarte-Berzal E, Garcia-Marco JA, del Cerro MH, Van den Steen PE, Opdenakker G, et al. Alpha4beta1 integrin and 190-kDa CD44v constitute a cell surface docking complex for gelatinase B/MMP-9 in chronic leukemic but not in normal B cells. Blood. 2008;112:169–78. doi: 10.1182/blood-2007-08-109249. [DOI] [PubMed] [Google Scholar]