Abstract
Background
Hepcidin is an iron regulatory peptide produced by the liver in response to inflammation and elevated systemic iron. Recent studies suggest that circulating monocytes and resident liver macrophages – Küpffer cells – may influence both basal and inflammatory expression of hepcidin.
Design and Methods
We used an in vitro co-culture model to investigate hepatocyte hepcidin regulation in the presence of activated THP1 macrophages. HuH7 hepatoma cells were co-cultured with differentiated THP1 macrophages for 24 h prior to the measurement of HuH7 hepcidin (HAMP) mRNA expression using quantitative polymerase chain reaction, and HAMP promoter activity using a luciferase reporter assay. Luciferase assays were performed using the wild type HAMP promoter, and constructs containing mutations in BMP/SMAD4, STAT3, C/EBP and E-BOX response elements. Neutralizing antibodies against interleukin-6, interleukin-1β , and the bone morphogenetic protein inhibitor noggin were used to identify the macrophage-derived cytokines involved in the regulation of HAMP expression.
Results
Co-culturing HuH7 cells with differentiated THP1 cells induced HAMP promoter activity and endogenous HAMP mRNA expression maximally after 24 h. This induction was fully neutralized in the presence of an interleukin-1β antibody, and fully attenuated by mutations of the proximal C/EBP or BMP/SMAD4 response elements.
Conclusions
Our data suggest that the interleukin-1β and bone morphogenetic protein signaling pathways are central to the regulation of HAMP expression by macrophages in this co-culture model.
Keywords: hepcidin, hepatocytes, macrophages, cytokines, inflammation
Introduction
Hepcidin was first identified as a liver expressed antimicrobial peptide,1 but has emerged as the major regulator of systemic iron homeostasis.2 An acute-phase protein and a component of the innate immune response,3 hepcidin is central to the development of the anemia of inflammation and the anemia of chronic disease by inhibiting cellular iron export into the serum through its action on macrophage ferroportin.4–7 In addition to its induction by inflammatory cytokines, hepcidin (HAMP – hepcidin antimicrobial peptide) transcription has been shown to be sensitive to changes in serum iron levels, systemic iron stores, erythropoietic factors and hypoxia.8,9 Moreover, the absence or low expression of hepcidin is implicated in the etiology of hereditary hemochromatosis as a primary cause of iron overload10 whereas its over-expression has been linked to iron deficiency anemia.11
While a number of cell types are known to express hepcidin,12–16 serum hepcidin levels are mainly determined by production and release of the peptide by hepatocytes; however, the involvement of other cell types in the regulation of hepatocyte hepcidin is unclear. Macrophages are primary candidates for such a role as they provide the majority of serum iron recycled from senescent erythrocytes and are also intrinsically involved in innate immunity and awakening of the adaptive immune response. To date, only a few studies have addressed the possibility of cytokine-secreting cells such as enterocytes or macrophages influencing hepatocyte HAMP gene expression. Studies using conditioned medium from peritoneal macrophages or THP1 monocytes have shown stimulation of hepcidin production in primary hepatocytes or HuH7 cells, respectively.17,18 Moreover, co-culturing with THP1 macrophages has been suggested to ensure an appropriate hepatocyte hepcidin response to added non-transferrin or transferrin-bound iron in vitro.19 However, this contrasts with in vivo studies in which Küpffer cells and macrophages were transiently inactivated. These studies demonstrate that hepatocytes can appropriately respond to iron challenge in isolation but that macrophages may be required for inflammatory regulation of hepcidin.20,21 Recently, there has been a report of a negative effect of Küpffer cells on hepatocyte HAMP expression and as a result a blunted hepcidin response to lipopolysaccharide treatment.15 Based on these previous studies, the precise role of macrophages in mediating or contributing towards the regulation of hepatocyte HAMP expression remains unclear. To address this issue, we developed an in vitro co-culture model utilizing human hepatoma cells (HuH7) and macrophages (THP1) to study the influence of activated macrophages on hepatic hepcidin expression.
Design and Methods
Cell culture
HuH7 human hepatoma cells were grown in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and were used for experiments at 80% confluence. THP1 cells, grown in RPMI-1640 medium containing 10% fetal bovine serum, were seeded at 1 × 106 cells per well on Transwell filters and treated overnight with phorbol myristate acetate (PMA) (100 nmol/L) to induce differentiation and attachment to the filters. Following differentiation cells were washed and incubated in fresh medium for 24 h prior to the experiments.
Co-culture
HuH7 hepatoma cells were seeded at a density of 0.5 × 106 cells per well in six-well plates and were grown for 48 h. On the day of the experiment HuH7 cells were washed and given fresh medium (containing neutralizing antibodies where necessary) and were overlaid with Transwell membranes containing either differentiated THP1 macrophages, non-activated THP1 cells (monocytes) or conditioned medium. Interleukin-6 (IL-6; R&D Systems, Abingdon, UK) and interleukin-1β (IL-1β ) neutralizing antibodies (Abcam, Cambridge, UK) and the bone morphogenetic protein (BMP) inhibitor noggin (R&D Systems) were used in some studies to identify macrophage-derived factors that might regulate hepcidin expression in HuH7 cells. In other experiments, HuH7 were exposed to THP1-conditioned medium alone (i.e. in the absence of THP1 cells). In addition, the effects of cytokines (IL-6, IL-1β and BMP2; PeproTech EC Ltd, London, UK) on HAMP expression in HuH7 monocultures was determined.
Real-time quantitative polymerase chain reaction
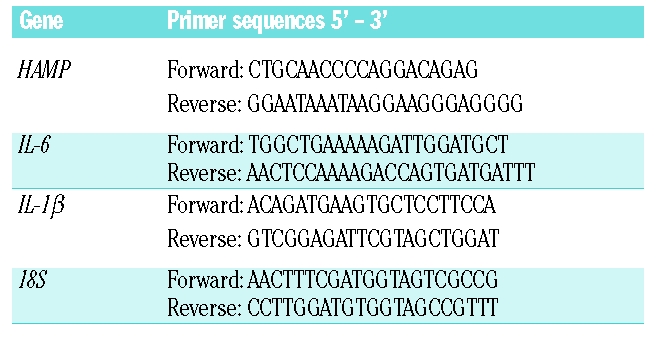
Total RNA was isolated from HuH7 cells using Trizol reagent (Invitrogen, Paisley, UK). Following first strand synthesis, expression levels of HAMP, IL-6, IL-1β and 18S (used as a housekeeper gene) mRNA were analyzed by real time quantitative polymerase chain reaction (PCR) using an ABI Prism 7000HT PCR cycler with gene-specific primers (Table 1) and a Quanti-Tect SYBR Green PCR kit (Qiagen, Crawley, UK), according to the manufacturer’s protocol. Quantitative measurements of each gene were derived from a standard curve constructed from known concentrations of PCR product.
Table 1.
Primer pairs used for quantitative real-time polymerase chain reaction analysis.
Generation of hepcidin promoter plasmid constructs
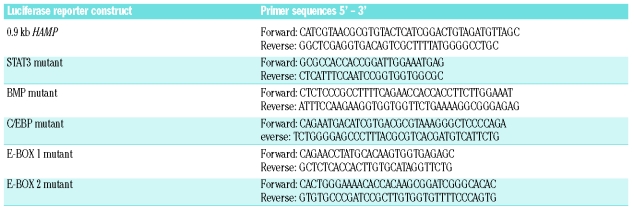
Genomic DNA was obtained from HepG2 cells and the proximal 942 bp of the human HAMP promoter was cloned into the pGL3-basic luciferase reporter vector (Promega, Southampton, UK) as described by Courselaud et al.22 Site-directed mutagenesis (Quick-change II, Stratagene, Stockport, UK) was used to insert mutations in known conserved transcription factor binding sites present in the proximal 942 bp of the HAMP promoter as detailed in Table 2. Briefly, the signal transducer and activator of transcription (STAT)3 response element was mutated according to an initial study by Wrighting and Andrews.23 The putative BMP responsive element was mutated in accordance with the observations of Verga Falzacappa et al.,24 while E-boxes, denoted 1 and 2, were inactivated as described by Bayele et al.25 The accepted proximal CCAAT enhancer binding protein (C/EBP) binding site was mutated by addition of a MluI restriction site. All constructs were sequenced prior to use in reporter assays.
Table 2.
Primer pairs used for cloning and site-directed mutagenesis of HAMP promoter to generate luciferase constructs.
Cell transfection and luciferase reporter assays
HuH7 cells were transfected with the wild type or mutant [STAT3, C/EBP, BMP-sons of mothers against decapentaplegic-4 (SMAD4) and E-BOX 1,2] HAMP reporter constructs or the empty pGL3-basic vector, using Fugene 6 (Roche, Burgess Hill, UK) according to the manufacturer’s instructions. To normalize for the transfection efficiency, an internal control - pRL-SV40 Renilla luciferase plasmid (Promega) was co-transfected alongside the HAMP constructs in a 1:50 ratio in serum-free medium. After 24 h equilibration, cells were treated for an additional 24 h and luciferase activity was determined in triplicate in at least two independent experiments using the Dual Luciferase Reporter Assay, according to the manufacturer’s instructions (Promega).
Statistical analysis
Statistical differences (p<0.05) among groups were determined using a one-way ANOVA, followed by Tukey’s post-hoc test. Student’s unpaired t test was used to compare the effects of monoculture versus co-culture within the same treatment group.
Results
The effect of co-cultured macrophages on hepcidin regulation in HuH7 hepatoma cells
THP1 cells were seeded onto Transwell inserts and incubated overnight in the presence or absence of the differentiating agent PMA. After washing, THP1 cells were incubated in fresh media for 24 h and were then overlaid onto six-well plates containing HuH7 hepatoma cells. HAMP expression in HuH7 cells co-cultured with activated THP1 macrophages was increased to 430±38% of that in the HuH7 monoculture control (p<0.001) after 24 h (Figure 1A). In contrast there was no significant difference in HAMP levels in HuH7 cells grown in the presence of non-activated THP1 cells (88±7%) compared with the levels in HuH7 cells alone. Therefore, in all subsequent experiments data are expressed relative to the monoculture control.
Figure 1.
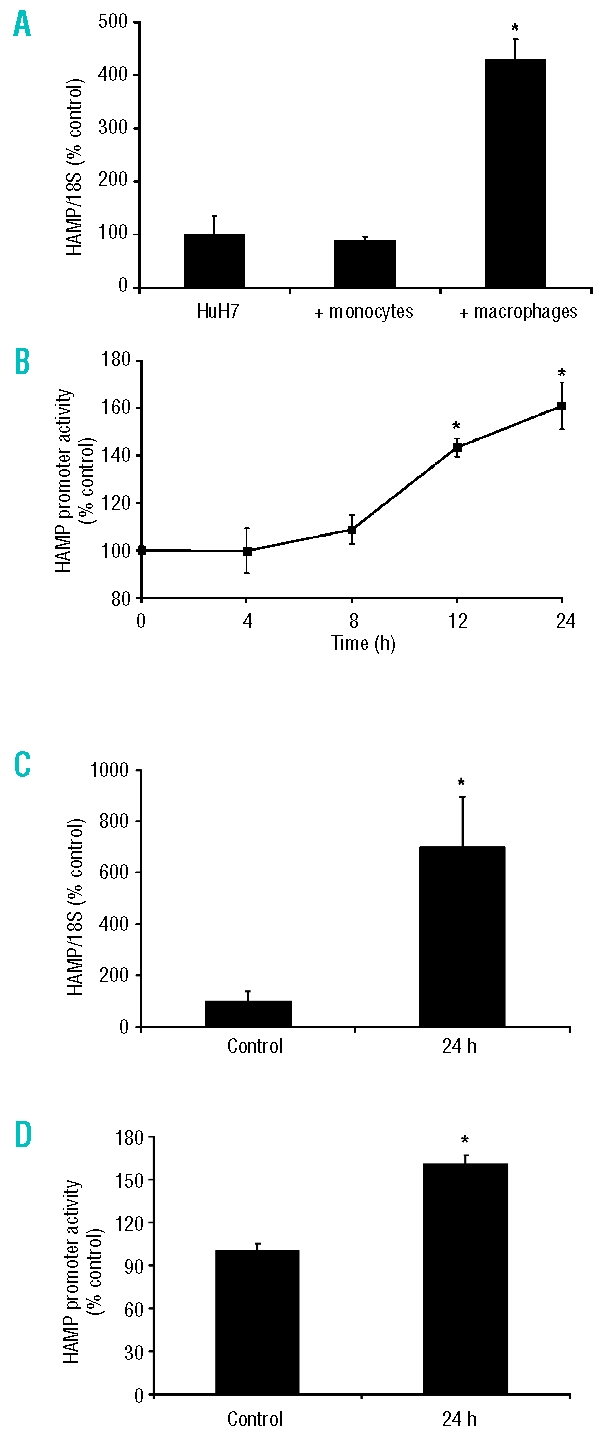
THP1 macrophages regulate HuH7 cell hepcidin levels. (A) THP1 cells (monocytes and PMA-activated macrophages) grown on Transwell inserts were co-cultured with HuH7 cells for 24 h. Hepcidin (HAMP) mRNA expression in HuH7 cells was significantly increased by co-culture with THP1 macrophages. The presence of THP1 monocytes did not alter HAMP mRNA levels. (B) HAMP promoter activity, measured in HuH7 cells using a 942 bp human HAMP -luciferase reporter construct, was also significantly increased by co-culture with THP1 macrophages. (C) HuH7 cell HAMP mRNA expression was significantly increased following incubation with conditioned-medium from THP1 macrophages. (D) HAMP promoter activity was significantly increased following incubation with conditioned-medium from THP1 macrophages. Data are presented as mean±SEM of 6–12 observations in each group and are expressed as a percentage of HuH7 monoculture data. The statistical analysis was performed using a one-way ANOVA with Tukey’s post-hoc test. *p<0.01.
In order to further examine hepcidin induction in this co-culture system a luciferase reporter construct containing 942 bp of the HAMP promoter was transiently transfected into HuH7 cells. Co-culturing HuH7 hepatoma cells with differentiated THP1 cells caused a progressive time-dependent increase in HAMP promoter activity, reaching 143±7% of the control activity after 12 h and 160±17% after 24 h (both p<0.01 compared to baseline; Figure 1B).
To demonstrate that hepcidin induction was due to macrophage-derived soluble factors, HuH7 cells were exposed to THP1 macrophage-conditioned medium for 24 h. This approach also resulted in significant induction of HAMP mRNA expression (699±196% compared with the monoculture control; p<0.01; Figure 1C), and HAMP promoter activity in reporter gene assays (161±6%; Figure 1D).
Exogenous interleukin-6, interleukin-1β and bone morphogenetic protein-2 increase the expression of hepcidin in HuH7 cells
In short-term studies (up to 4 h) a number of agents have been shown to induce hepcidin expression. Our preliminary data (Figure 2A) revealed a rapid and significant (p<0.05) increase (within 2 h) in HAMP mRNA levels in the presence of IL-6 (4.8±0.3 fold), IL-1β (2.3±0.3 fold) and BMP2 (7.0±1.1 fold). The longer term effect (24 h) of these agents on endogenous HAMP mRNA expression was subsequently determined in HuH7 cells. While modest and significant increases were still observed with IL6 and BMP-2 (3.1±0.4 and 2.7±0.4 fold, respectively; p<0.05), 24 h treatment with IL-1β caused a 43±10 fold induction of HAMP compared with untreated control cells (p<0.001; Figure 2B).
Figure 2.
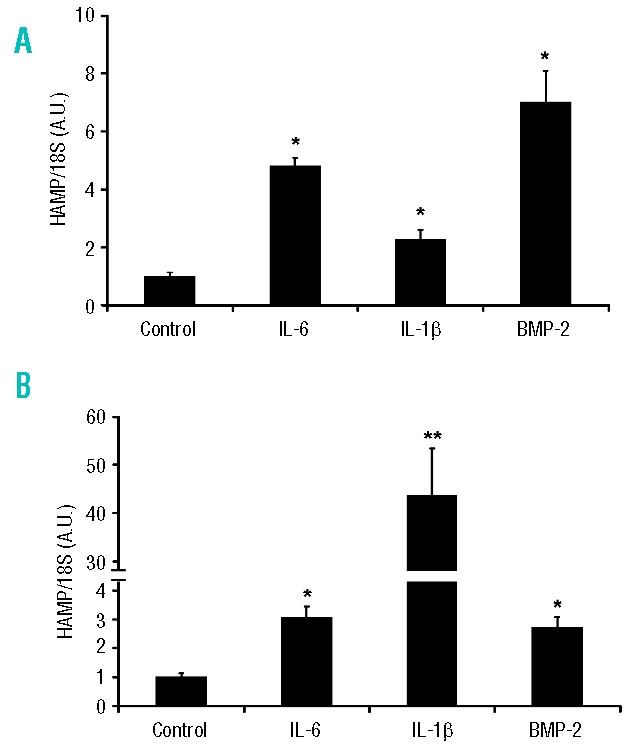
Effect of recombinant cytokines on HAMP mRNA expression in HuH7 cells. Cells were treated with IL-6 (10 ng/mL), IL-1β (10 ng/mL) or BMP-2 (25 ng/mL) for (A) 2 or (B) 24 h and HAMP mRNA expression was determined using quantitative real-time-PCR. Data are mean±SEM of four to six observations in each group and are expressed as a ratio of 18S mRNA in arbitrary units (A.U.). The statistical analysis was performed using a one-way ANOVA with Tukey’s post-hoc test. *p<0.05; **p<0.001.
The effect of anti-interleukin-1β , anti-interleukin-6, or noggin on the induction of hepcidin by macrophages
Both IL-6 and IL-1β are released by macrophages during inflammation and increase hepatic HAMP expression. In our cell system, activation of THP1 macrophages with PMA resulted in a 52±9-fold increase in IL-6 (p<0.001) and a 192±25-fold increase in IL-1β mRNA expression (p<0.001), compared with untreated cells. Following co-culture with THP-1 macrophages, HuH7 IL-6 mRNA levels were increased 12.5±1.4-fold (p<0.001), whereas IL-1β mRNA levels were induced 23.9±5.0-fold (p<0.001) compared with untreated controls. The relative levels of these cytokines in THP-1 macrophages were significantly higher than those observed in co-cultured HuH7 (IL-6, 7.4±1.3-fold, p<0.001; IL-1β , 1046±135-fold, p<0.001). This marked induction of IL-1β suggests that this cytokine may play a significant role in regulating HuH7 HAMP levels in the co-culture system.
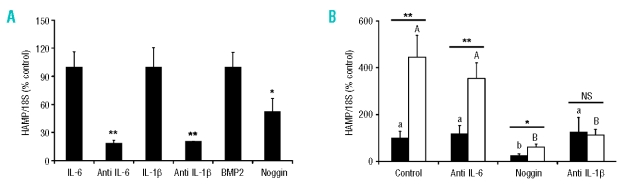
To further distinguish between the contributions of IL-6 and IL-1β to the regulation of HAMP expression, THP1 macrophages/HuH7 hepatoma cells were co-cultured in the presence or absence of either anti-IL-6 or anti-IL-1β neutralizing antibodies. The effects of the BMP antagonist noggin were also investigated. In preliminary studies to validate the use of these agents we observed that the induction of HAMP by IL-6, IL-1β , and BMP2 could be blocked in the presence the anti-IL-6 antibody (−82% of the IL-6 response), the anti-IL-1β antibody (−79% of the IL-1β response) and noggin (−48% of the BMP2 response), respectively (Figure 3A). While the presence of anti-IL-6 had no significant effect on co-culture-induced hepatic HAMP expression after 24 h, the anti-IL-1β neutralizing antibody abolished the induction of hepcidin under these conditions. The BMP antagonist noggin significantly reduced basal HAMP expression in HuH7 cell monocultures to 25±7% (p<0.001) of the control level (Figure 3B) but could not prevent a 2.4-fold induction in HAMP expression from this level in the presence of THP1 macrophages.
Figure 3.
The effects of neutralizing antibodies or noggin on macrophage-dependent induction of hepatoma cell HAMP expression. (A) Neutralizing antibodies, anti-IL-6 (0.1 μL/mL), anti-IL-1β (5 μL/mL), and noggin (0.1 μg/mL), were validated by measuring their ability to block HAMP mRNA induction by IL-6 (10 ng/mL), IL-1β (10 ng/mL) or BMP-2 (25 ng/mL). The effect of each cytokine was normalized to 100%. Data are mean±SEM of six observations in each group and are expressed as a ratio of 18S mRNA (% control) (B) HuH7 cells were cultured in the absence (filled bars) or presence (open bars) of activated THP1 macrophages, in the presence of neutralizing antibodies or noggin for 24 h. Anti-IL-6 antibody had no significant effect on HAMP mRNA expression, however the IL-1β antibody prevented the macrophage-dependent induction of HAMP. Noggin decreased basal expression HAMP expression but HAMP mRNA was significantly increased in the presence of THP1 macrophages. Data are mean±SEM of four to six observations in each group and are expressed as a ratio of 18S mRNA (% control). The statistical analysis to determine differences between groups was performed using a one-way ANOVA with Tukey’s post-hoc test. Different letters above data bars (UPPERCASE – THP1/HuH7 co-cultures; lowercase – HuH7 monocultures) indicate that these groups are significantly different (p<0.05). To compare differences between co-culture and monoculture within the same treatment group we employed Student’s unpaired t test. *p<0.05; **p<0.01.
Mutations in the SMAD4 and C/EBP binding sites of the HAMP promoter abolish sensitivity to macrophages
After transfecting HuH7 hepatoma cells with a luciferase reporter containing 942 bp of human HAMP promoter, co-culture with activated THP-1 macrophages resulted in a 165±11% induction relative to the level in the monoculture control (p<0.05; Figure 4). Mutations of the STAT3 response element resulted in a 30±3% (p<0.05) decrease in basal activity, but interestingly did not prevent the induction of promoter activity due to the presence of macrophages. Moreover, the mutation of both E-boxes resulted in significant de-repression of basal promoter activity (140±5% of control; p<0.05), suggestive of promoter occupancy by repressor proteins. Interestingly, HAMP promoter activity was not significantly further induced in the presence of macrophages. In contrast, mutation of the BMP response element decreased basal expression to 26±7% of the wild type control level and prevented the induction by macrophages. Finally, when the proximal C/EBP response element was mutated, there was no effect on basal promoter activity; however, the induction of HAMP promoter following exposure to macrophages was fully abrogated.
Figure 4.
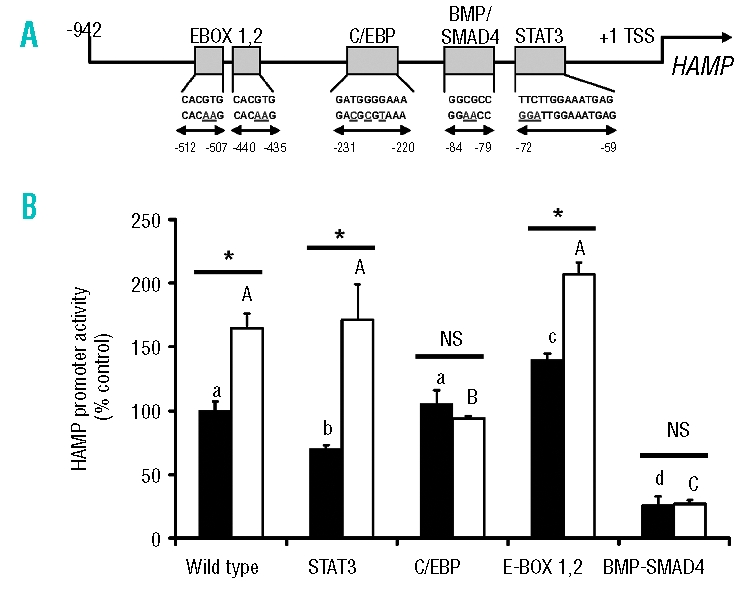
Mutations in the SMAD4 and C/EBP binding sites of the HAMP promoter abolish sensitivity to macrophages. (A) Schematic representation of the proximal 942 bp of the human HAMP promoter. Positions of particular consensus elements and engineered mutations are indicated relative to the transcriptional start site (TSS). (B) HuH7 cells were transfected with luciferase-reporter constructs and 24 h later were co-cultured with activated THP1 cells. Luciferase activity was measured in monoculture (filled bars) and co-culture (open bars) after 24 h. Data are means±SEM of six observations in each group and are expressed as a percentage of the monoculture wild type control. The statistical analysis to determine differences between groups was performed using a one-way ANOVA with Tukey’s post-hoc test. Different letters above data bars (UPPERCASE – THP1/HuH7 co-cultures; lowercase – HuH7 monocultures) indicate that these groups are significantly different (p<0.05). To compare differences between co-culture and monoculture within the same treatment group we employed Student’s unpaired t test. *p<0.001.
Discussion
In this study we developed an in vitro co-culture system to investigate the cross-talk between hepatocytes and macrophages in mediating hepcidin regulation. Similar models have been employed previously to study the effects of monocyte/macrophage factors on hepatocyte HAMP expression. A strong induction of HAMP was observed in primary hepatocytes following the addition of peritoneal macrophage conditioned medium.17 Moreover, Jacolot and colleagues demonstrated that PMA-differentiated THP1 cells alter hepatoma cell iron sensing.19 In agreement with these studies,17,19 we have shown that both co-culture of HuH7 cells with THP1 macrophages, and the addition of conditioned medium from THP1 macrophages exert a positive effect on endogenous HAMP expression and HAMP promoter activity through soluble factors found in the medium. Interestingly, Verga Falzacappa and colleagues demonstrated a time-dependent increase in HAMP expression in HuH7 cells following the addition of conditioned medium from THP1 monocytes.18 However, we could not replicate this finding when co-culturing THP1 monocytes with HuH7 cells. This suggests that in our co-culture system THP1 macrophages must be activated to elicit a stimulatory effect on HAMP expression.
Various studies have described an array of cytokines that can induce hepcidin expression, including IL-6, IL-1 and a number of BMP.17,27–30 Most of these cytokines are known secreted products of macrophages and monocytes. Our data revealed a substantial induction of both IL-6 and IL-1β mRNA in THP1 cells following activation with PMA. To distinguish between the effects of these cytokines we utilized neutralizing antibodies to IL-1β or IL-6, and noggin, a BMP inhibitor. Preliminary studies showed that the antibodies and noggin were able to suppress the increase in HAMP elicited by their respective cytokines. While the IL-1β antibody blocked macrophage-induced HAMP production there was no effect of the IL-6 neutralizing antibody on this response. Interestingly, a microarray study showed an 84-fold increase in IL-1β expression following addition of PMA to THP1 cells,31 suggesting that it was the predominant cytokine produced by activated THP1 macrophages. In support of this hypothesis, Lee et al. suggested that IL-1α and β but not IL-6 were the main cytokines found in the conditioned medium from peritoneal macrophage preparations. Furthermore, HAMP expression in primary hepatocyte preparations from IL-6−/− mice also responded appropriately to the addition of conditioned medium.17
Next we attempted to address the molecular basis for the induction of hepcidin by macrophage-derived factors. A number of consensus elements within the proximal 0.9 kb human HAMP promoter have been mapped and functionally assessed. These include binding elements for transcription factors of the CCAAT enhancer family, STAT3, SMAD4, and transcription factors of the β helix-loop-helix (β HLH) family (HIF, USF, c-myc, c-max). Using a series of HAMP promoter constructs we explored the role of these transcriptional regulatory sites on the induction of hepcidin by activated macrophages.
A conserved STAT3 binding site (−72 to −59 bp, relative to the transcriptional start site), has been identified and has been shown to be essential for the classical IL-6-gp130 induction of HAMP promoter via the JAK/STAT3 pathway.18,23 Interestingly, we have recently shown that the adipokine leptin, through its cognate Ob-Rb receptor, can also induce this pathway and this observation may be pertinent to the understanding of obesity-associated hypoferremia.32 Mutation of the STAT3 binding site resulted in a significant decrease in basal HAMP promoter activity. These findings are in keeping with previous observations18 and confirm the importance of STAT3 in the regulation of HAMP expression. Interestingly, mutation of the STAT3 binding site in the HAMP promoter did not influence luciferase activity in the presence of activated macrophages, suggesting that it does not play a major role in controlling hepcidin regulation in our co-culture model. Importantly, this hypothesis is further supported by our data from the studies using the IL-6 neutralizing antibody.
Two E- box elements, labeled 1 and 2 in this study (−440 to −435 and −512 to −507 bp), have been shown to bind β HLH factors.25 In particular, the region containing both of these elements was also demonstrated, in vivo, to be occupied by hypoxia inducible factors and thus could be important in conferring sensitivity of HAMP expression to hypoxia.33 Under the conditions of this study mutation of the E-box sequences resulted in a significant increase in basal HAMP promoter activity. Taken together with the findings of Bayele et al.,25 these data indicate that E boxes may be important in both positive and negative regulation of HAMP promoter activity depending on the relative abundance of the transcription factors binding to this region.
In close proximity to the STAT3 site lies a conserved SMAD4/BMP responsive element (−84 to −79 bp), which is regulated by BMP and other members of the transforming growth factor-β superfamily. This region has been shown to confer sensitivity to a range of BMP in vitro, and to mediate, in part, the regulation of hepatocyte hepcidin in response to changes in serum iron.24 Inhibition of BMP signaling by the mutation of the SMAD4/BMP response element in the proximal HAMP promoter completely abolished the induction of hepcidin by macrophages and dramatically decreased basal promoter activity. These data indicate that a functional SMAD4/BMP response element may be essential for the activity of upstream response elements involved in the induction of hepcidin by macrophages in this system. In support of this hypothesis, the BMP antagonist noggin significantly decreased basal HAMP mRNA expression in HuH7 monocultures. However, in the presence of a functional SMAD4/BMP response element macrophage-derived factors were still able to induce HAMP by approximately 2.4 fold in noggin-treated cells.
The molecular actions of IL-1 have been studied in great detail. Both IL1-αand β operate through type I IL-1 receptor (IL-1RI) and mainly trans-activate the transcription factor nuclear factor κ B (NF-κ B), the mitogen-activated protein kinase (MAPK/ERK1,2) signaling cascade or C/EBP proteins.34 To date, the involvement of NF-κ B or MAPK/ERK signaling has not been reported in hepcidin regulation. However, the C/EBP proteins (α and β ) have been implicated in iron-mediated regulation of the HAMP promoter.22 A conserved C/EBP binding site (−231 to −222 bp) has also been implicated in maintaining basal HAMP promoter activity.18,22 Interestingly, in our studies mutation of the C/EBP binding site did not alter basal HAMP promoter activity. This is likely to reflect differences in the promoter constructs used – we mutated the C/EBP binding site by addition of a MluI restriction site, whereas in previous studies18,22 the C/EBP binding site was deleted by truncation of the promoter construct. The involvement of C/EBP in the acute phase response has been demonstrated in a range of different tissues and principally involves C/EBP-α, β and Δ.35 The C/EBP proteins are known to augment IL-6 transcription (C/EBP-β is also termed NF-IL6 as it can directly affect IL-6 transcription), affect STAT3 signaling or independently trans-activate the target gene. Our data suggest that STAT3 is not necessary for the activation of hepcidin in the co-culture model and that C/EBP factors play a major role in the induction of hepcidin by macrophages.
It is tempting to speculate that the stimulation of C/EBP proteins by IL-1β results in an increase in HAMP expression. This hypothesis is supported by recent studies in hepatoma and human primary hepatocytes, showing that hepcidin was induced by IL-1β directly and does not require any cross-talk with the IL6/STAT3 pathway.36 However, it is also possible that IL-1β may be working indirectly, perhaps through the modulation and release of BMP. Future studies will explore this possibility.
In summary, using a co-culture model we have shown that activated macrophages induce hepatic HAMP expression. Our data suggest that IL-1β is the predominant macrophage factor involved in mediating this response. These findings provide novel insights into the cross-talk between macrophages and hepatocytes and add evidence to the suggestion that during inflammation these two cell types act in concert to regulate hepcidin production.
Footnotes
Authorship and Disclosures
PM and TBC performed experiments, analyzed data and co-wrote the paper; BC performed experiments and analysed data; ATM and SKS co-wrote the paper; PAS analyzed data and co-wrote the paper. PM and TBC made equal contributions.
The authors declare that they have no conflicts of interest.
Funding: this work was funded by the Biotechnology & Biological Sciences Research Council. Pavle Matak was funded by a studentship from the Medical Research Council.
References
- 1.Krause A, Neitz S, Magert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, et al. LEAP-1, a novel highly disul-fide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000;480:147–50. doi: 10.1016/s0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 2.Nemeth E, Ganz T. Regulation of iron metabolism by hepcidin. Annu Rev Nutr. 2006;26:323–42. doi: 10.1146/annurev.nutr.26.061505.111303. [DOI] [PubMed] [Google Scholar]
- 3.Nemeth E, Valore EV, Territo M, Schiller G, Lichtenstein A, Ganz T. Hepcidin, a putative mediator of anemia of inflammation, is a type II acute-phase protein. Blood. 2003;101:2461–3. doi: 10.1182/blood-2002-10-3235. [DOI] [PubMed] [Google Scholar]
- 4.Chaston T, Chung B, Mascarenhas M, Marks J, Patel B, Srai SK, et al. Evidence for differential effects of hepcidin in macrophages and intestinal epithelial cells. Gut. 2008;57:374–82. doi: 10.1136/gut.2007.131722. [DOI] [PubMed] [Google Scholar]
- 5.Delaby C, Pilard N, Goncalves AS, Beaumont C, Canonne-Hergaux F. Presence of the iron exporter ferroportin at the plasma membrane of macrophages is enhanced by iron loading and down-regulated by hepcidin. Blood. 2005;106:3979–84. doi: 10.1182/blood-2005-06-2398. [DOI] [PubMed] [Google Scholar]
- 6.Knutson MD, Oukka M, Koss LM, Aydemir F, Wessling-Resnick M. Iron release from macrophages after erythrophagocytosis is up-regulated by ferroportin 1 overexpression and down-regulated by hepcidin. Proc Natl Acad Sci USA. 2005;102:1324–8. doi: 10.1073/pnas.0409409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivera S, Nemeth E, Gabayan V, Lopez MA, Farshidi D, Ganz T. Synthetic hepcidin causes rapid dose-dependent hypoferremia and is concentrated in ferroportin-containing organs. Blood. 2005;106:2196–9. doi: 10.1182/blood-2005-04-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemeth E. Iron regulation and erythropoiesis. Curr Opin Hematol. 2008;15:169–75. doi: 10.1097/MOH.0b013e3282f73335. [DOI] [PubMed] [Google Scholar]
- 9.Pinto JP, Ribeiro S, Pontes H, Thowfeequ S, Tosh D, Carvalho F, et al. Erythropoietin mediates hepcidin expression in hepatocytes through EPOR signaling and regulation of C/EBPα. Blood. 2008;111:5727–33. doi: 10.1182/blood-2007-08-106195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolas G, Andrews NC, Kahn A, Vaulont S. Hepcidin, a candidate modifier of the hemochromatosis phenotype in mice. Blood. 2004;103:2841–3. doi: 10.1182/blood-2003-09-3358. [DOI] [PubMed] [Google Scholar]
- 11.Nicolas G, Bennoun M, Porteu A, Mativet S, Beaumont C, Grand-champ B, et al. Severe iron deficiency anemia in transgenic mice expressing liver hepcidin. Proc Natl Acad Sci USA. 2002;99:4596–601. doi: 10.1073/pnas.072632499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bekri S, Gual P, Anty R, Luciani N, Dahman M, Ramesh B, et al. Increased adipose tissue expression of hepcidin in severe obesity is independent from diabetes and NASH. Gastroenterology. 2006;131:788–96. doi: 10.1053/j.gastro.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Gnana-Prakasam JP, Martin PM, Mysona BA, Roon P, Smith SB, Ganapathy V. Hepcidin expression in mouse retina and its regulation via lipopolysaccharide/Toll-like receptor-4 pathway independent of Hfe. Biochem J. 2008;411:79–88. doi: 10.1042/BJ20071377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen NB, Callaghan KD, Ghio AJ, Haile DJ, Yang F. Hepcidin expression and iron transport in alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2006;291:L417–25. doi: 10.1152/ajplung.00484.2005. [DOI] [PubMed] [Google Scholar]
- 15.Theurl M, Theurl I, Hochegger K, Obrist P, Subramaniam N, van Rooijen N, et al. Kupffer cells modulate iron homeostasis in mice via regulation of hepcidin expression. J Mol Med. 2008;86:825–35. doi: 10.1007/s00109-008-0346-y. [DOI] [PubMed] [Google Scholar]
- 16.Zhang AS, Xiong S, Tsukamoto H, Enns CA. Localization of iron metabolism-related mRNAs in rat liver indicate that HFE is expressed predominantly in hepatocytes. Blood. 2004;103:1509–14. doi: 10.1182/blood-2003-07-2378. [DOI] [PubMed] [Google Scholar]
- 17.Lee P, Peng H, Gelbart T, Wang L, Beutler E. Regulation of hepcidin transcription by interleukin-1 and interleukin-6. Proc Natl Acad Sci USA. 2005;102:1906–10. doi: 10.1073/pnas.0409808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its i nflammatory stimulation. Blood. 2007;109:353–8. doi: 10.1182/blood-2006-07-033969. [DOI] [PubMed] [Google Scholar]
- 19.Jacolot S, Ferec C, Mura C. Iron responses in hepatic, intestinal and macrophage/monocyte cell lines under different culture conditions. Blood Cells Mol Dis. 2008;41:100–8. doi: 10.1016/j.bcmd.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Lou DQ, Lesbordes JC, Nicolas G, Viatte L, Bennoun M, Van Rooijen N, et al. Iron- and inflammation-induced hepcidin gene expression in mice is not mediated by Kupffer cells in vivo. Hepatology. 2005;41:1056–64. doi: 10.1002/hep.20663. [DOI] [PubMed] [Google Scholar]
- 21.Montosi G, Corradini E, Garuti C, Barelli S, Recalcati S, Cairo G, et al. Kupffer cells and macrophages are not required for hepatic hepcidin activation during iron overload. Hepatology. 2005;41:545–52. doi: 10.1002/hep.20620. [DOI] [PubMed] [Google Scholar]
- 22.Courselaud B, Pigeon C, Inoue Y, Inoue J, Gonzalez FJ, Leroyer P, et al. C/EBPα regulates hepatic transcription of hepcidin, an antimicrobial peptide and regulator of iron metabolism. Cross-talk between C/EBP pathway and iron metabolism. J Biol Chem. 2002;277:41163–70. doi: 10.1074/jbc.M202653200. [DOI] [PubMed] [Google Scholar]
- 23.Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood. 2006;108:3204–9. doi: 10.1182/blood-2006-06-027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verga Falzacappa MV, Casanovas G, Hentze MW, Muckenthaler MU. A bone morphogenetic protein (BMP)-responsive element in the hepcidin promoter controls HFE2-mediated hepatic hepcidin expression and its response to IL-6 in cultured cells. J Mol Med. 2008;86:531–40. doi: 10.1007/s00109-008-0313-7. [DOI] [PubMed] [Google Scholar]
- 25.Bayele HK, McArdle H, Srai SK. Cis and trans regulation of hepcidin expression by upstream stimulatory factor. Blood. 2006;108:4237–45. doi: 10.1182/blood-2005-07-027037. [DOI] [PubMed] [Google Scholar]
- 26.Andriopoulos B, Pantopoulos K. Hepcidin generated by hepatoma cells inhibits iron export from co-cultured THP1 monocytes. J Hepatol. 2006;44:1125–31. doi: 10.1016/j.jhep.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 27.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–9. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 28.Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, et al. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113:1271–6. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietrangelo A, Dierssen U, Valli L, Garuti C, Rump A, Corradini E, et al. STAT3 is required for IL–6-gp130-dependent activation of hepcidin in vivo. Gastroenterology. 2007;132:294–300. doi: 10.1053/j.gastro.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Truksa J, Peng H, Lee P, Beutler E. Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1 expression independently of Hfe, transferrin receptor 2 (Tfr2), and IL-6. Proc Natl Acad Sci USA. 2006;103:10289–93. doi: 10.1073/pnas.0603124103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohro T, Tanaka T, Murakami T, Wada Y, Aburatani H, Hamakubo T, et al. A comparison of differences in the gene expression profiles of phorbol 12-myristate 13-acetate differentiated THP-1 cells and human monocyte-derived macrophage. J Atheroscler Thromb. 2004;11:88–97. doi: 10.5551/jat.11.88. [DOI] [PubMed] [Google Scholar]
- 32.Chung B, Matak P, McKie AT, Sharp P. Leptin increases the expression of the iron regulatory hormone hepcidin in HuH7 human hepatoma cells. J Nutr. 2007;137:2366–70. doi: 10.1093/jn/137.11.2366. [DOI] [PubMed] [Google Scholar]
- 33.Peyssonnaux C, Zinkernagel AS, Schuepbach RA, Rankin E, Vaulont S, Haase VH, et al. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J Clin Invest. 2007;117:1926–32. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunne A, O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE. 2003;2003:re3. doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 35.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–75. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer F, Torzewski J, Kamenz J, Veit K, Hombach V, Dedio J, et al. Interleukin-1β stimulates acute phase response and C-reactive protein synthesis by inducing an NFκB-and C/EBPβ-dependent autocrine interleukin-6 loop. Mol Immunol. 2008;45:2678–89. doi: 10.1016/j.molimm.2007.12.017. [DOI] [PubMed] [Google Scholar]