Abstract
Type IV secretion systems (T4SSs) are multisubunit cell-envelope-spanning structures, ancestrally related to bacterial conjugation machines, which transfer proteins and nucleoprotein complexes across membranes. T4SSs mediate horizontal gene transfer, thus contributing to genome plasticity and the evolution of pathogens through dissemination of antibiotic resistance and virulence genes. Moreover, T4SSs are also used for the delivery of bacterial effector proteins across the bacterial membrane and the plasmatic membrane of eukaryotic host cell, thus contributing directly to pathogenicity. T4SSs are usually encoded by multiple genes organized into a single functional unit. Based on a number of features, the organization of genetic determinants, shared homologies and evolutionary relationships, T4SSs have been divided into several groups. Type F and P (type IVA) T4SSs resembling the archetypal VirB/VirD4 system of Agrobacterium tumefaciens are considered to be the paradigm of type IV secretion, while type I (type IVB) T4SSs are found in intracellular bacterial pathogens, Legionella pneumophila and Coxiella burnetii. Several novel T4SSs have been identified recently and their functions await investigation. The most recently described GI type T4SSs play a key role in the horizontal transfer of a wide variety of genomic islands derived from a broad spectrum of bacterial strains.
Introduction
Many bacterial species exploit specialized secretion systems to transfer macromolecules across membranes. These secretion systems are assembled into six major groups, named types I, II, III, IV, V and VI (Thanassi and Hultgren, 2000; Henderson et al., 2004; Mougous et al., 2006). The secretion systems ancestrally related to the bacterial conjugation machinery are referred to as the type IV secretion systems (T4SSs) (Lawley et al., 2003; Christie et al., 2005). The T4SSs are unique among other bacterial secretion system types due to their ability to transfer both proteins and nucleoprotein complexes.
T4SSs are multisubunit cell-envelope-spanning structures comprising a secretion channel and often a pilus or other surface filament or protein (Lawley et al., 2003; Christie et al., 2005). Research on T4SSs began with the discovery of the F plasmid-borne conjugation system few decades ago. Conjugation systems, including the one encoded by the F plasmid, represent a large subfamily of the T4SSs and are used by bacteria in the process of the conjugative transfer of DNA from donor to recipient cells (Fig. 1). By conjugation, T4SSs mediate horizontal gene transfer, thus contributing to genome plasticity and the evolution of infectious pathogens through dissemination of antibiotic resistance and virulence genes. The T4SS of Neisseria gonorrhoeae, evolutionarily related to the conjugation machinery, mediates secretion of naked DNA into the extracellular environment instead of the donor cells (Hamilton et al., 2005). Moreover, T4SSs are also used for the delivery of bacterial effector proteins across the bacterial membrane and the plasma membrane of eukaryotic target cells, thus contributing directly to the bacterial pathogenicity (Christie et al., 2005).
Fig. 1.
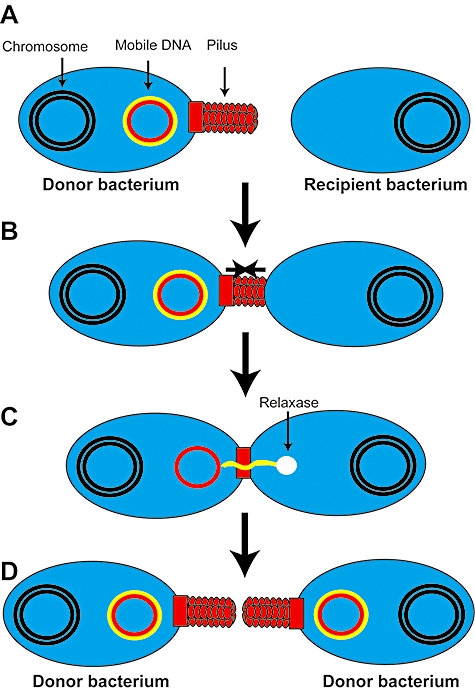
Conjugation. Conjugation systems (shown is the shot-and-pump model of conjugative DNA transfer) represent a large subfamily of the T4SSs and are used by bacteria in the process of the conjugative transfer of DNA from donor to recipient cells (A) by cell-to-cell contact usually mediated by the retraction of the pilus-like structures (B). C. ssDNA of the mobile genetic element is transferred from the donor to recipient bacteria with the help of the relaxase. D. Complementary DNA strands are synthesized in both cells and the former recipient bacterium becomes a new potential donor of the mobile DNA.
Type IV secretion systems: a challenge to simple classification
T4SSs are encoded by multiple genes organized into a single functional unit. The T4SSs gene clusters hitherto described differ significantly in a number of respects, including gene content, gene order and the number of homologues they share. Based on the number of features, including the organization of genetic determinants, shared homologies and evolutionary relationships, T4SSs have been classified into major types using two different classification schemes (Lawley et al., 2003; Christie et al., 2005).
In the original classification scheme there were initially three major types, referred to as types F, P and I, based on the incompatibility group of the representative conjugative plasmids, IncF (plasmid F), IncP (plasmid RP4) and IncI (plasmid R64) respectively (Lawley et al., 2003). In the alternative classification schemes, types F and P have been grouped together as type IVA systems, which resemble the archetypal VirB/VirD4 system of Agrobacterium tumefaciens. Type I, which varies significantly in its component modules from members of both F and P types, was named as the type IVB system. Genetic determinants of the type IVB systems are related to the archetypal Dot/Icm system of Legionella pneumophila. A third group in this classification scheme, composed of all the ‘other’ T4SS representatives that bear no or only limited homology to IVA and IVB system, has not been well characterized (Christie et al., 2005). Representatives of this group include for instance the recently identified GI lineage of T4SSs associated with broad spectrum of genomic islands in various bacteria (see below) (Juhas et al., 2007a).
In both classification schemes, members within each type exhibit evidence of common ancestry. Within each type there is greater conservation of gene content and order than there is between types. Alignment of amino acid sequences shared by homologous proteins supports the division into these groups.
More recently, a novel lineage of T4SSs has been identified on the genomic island ICEHin1056, which is a vector of antibiotic resistance in Haemophilus influenzae. This T4SS is distinct from all previously described types. It represents a fourth lineage with a genetic distance as great as is observed between the F, P and I lineages. Using the alternative classification system (Christie et al., 2005), this new T4SS would be classified as ‘other’. This novel type of T4SSs is present in a wide variety of related syntenic genomic islands including pKLC102, PAPI, SPI-7 and the clc element as well as others derived from a broad variety of bacterial strains including Pseudomonas aeruginosa, Pseudomonas fluorescens, Erwinia carotovora, Salmonella enterica serovar Typhi and L. pneumophila. This lineage of T4SSs has been named GI type in order to emphasize the fact that this T4SS is found so far associated only with genomic islands (Juhas et al., 2007a).
Relationships within and among T4SS types are based mainly on three genes, named traB/virB10, traC/virB4 and traD/virD4 (named after genes on the Escherichia coli F plasmid or on the A. tumefaciens Ti plasmid respectively). As shown in our study, the high sequence variability of traD means that it is not an ideal candidate for phylogenetic analysis, whereas genes traB/virB10 and traC/virB4 encode quintessential T4SS proteins of Gram-negative bacteria that are ideal for comparative amino acid alignment and as such appear sufficient to define membership of a T4SS type (Juhas et al., 2007a). If these two genes provide the key signature that could identify other more distantly related T4SSs in genomes and, in particular, genomic islands, searching for homologues of these genes might be a way to identify other distinct T4SS lineages. However, it should be noted that, as traB/virB10 is not a component of conjugation systems of Gram-positive bacteria, traC/virB4 remains the only universally conserved T4SS component of both Gram-negative and Gram-positive bacteria.
The division of T4SSs into four groups: F, P, I, and GI seems to provide a sound framework for classifying the majority of T4SSs. It is only recently that the increased availability of genome sequence data and improved bioinformatic techniques have allowed the recognition of the novel GI type of T4SS. Thus, it is quite realistic to assume that with the use of modern computational biology techniques, other new divergent T4SS types will be identified in the forthcoming years.
F and P type IV secretion systems: paradigm of type IV secretion
The F and P type T4SSs, also referred to as type IVA systems, resemble the archetypal VirB/VirD4 system of A. tumefaciens. The T4SS of A. tumefaciens is encoded by an approximately 10 kb ‘virB operon’, comprising 11 open reading frames and a separate virD4 gene, and mediates transfer of oncogenic genes into plant cells, resulting in tumorigenesis and subsequent crown gall disease. Most of the information concerning functional properties of T4SSs has been obtained from the study of the VirB/VirD4 system of A. tumefaciens. According to the most recent views, of the 11 VirB protein determinants of the A. tumefaciens VirB/VirD4 T4SS, proteins VirB2 and VirB5 are pilus components, VirB3 and VirB7 are pilus-associated proteins, VirB4 and VirB11 are nucleoside triphosphatases that provide energy for transfer, while VirB6, VirB7, VirB8, VirB9 and VirB10 constitute components of the transmembrane channel (Fig. 2). VirB1 is a lytic transglycosylase that degrades the peptidoglycan cell wall at the site of T4SS assembly, and VirD4 is another nucleoside triphosphatase, called ‘coupling protein’, which recruits DNA to the components of the secretion system (Christie et al., 2005; Backert and Meyer, 2006; Fig. 2). Interestingly, the C-terminal part of VirB1, designated VirB1* was shown to be cleaved and secreted from cells where it was involved in the formation of the T4SS pilus (Zupan et al., 2007). Thus, VirB1 acts as a bifunctional protein that lyses peptidoglycan cell wall to facilitate insertion of the T4SS but simultaneously also promotes formation of the pilus through interaction with pilus subunits (Zupan et al., 2007). The recruitment of the transfer substrate of A. tumefaciens VirB/VirD4 T4SS consisting of T-DNA encoding oncogenic proteins and VirD2 relaxase was shown to be facilitated by the previously unrecognized recruiting protein VBP (VirD2-binding protein; Guo et al., 2007). In addition to the T-DNA-encoding oncogenic proteins bound to the VirD2 relaxase, the VirB/VirD4 T4SS of A. tumefaciens transfers several effector proteins into the host plant cells: namely VirD5, VirE2, VirE3 and VirF that increase the chance of successful infection (Vergunst et al., 2000). Recent work showed that VirE2 acts as a unique powerful ssDNA-binding molecular machine that mediates infection by actively pulling T-DNA into the host cells without the need for external energy sources (Grange et al., 2008).
Fig. 2.
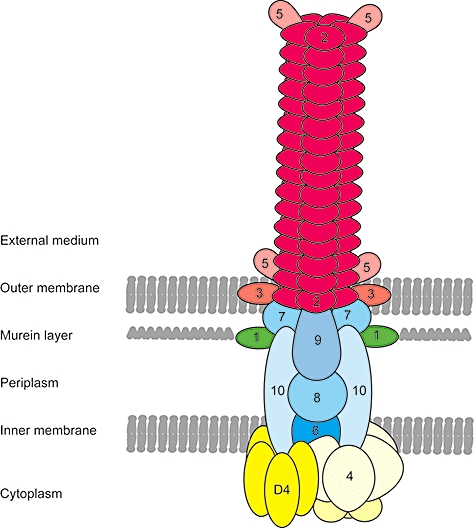
Model of the VirB/VirD4 type IV secretion machinery of Agrobacterium tumefaciens. The T4SS of A. tumefaciens is a multicomponent cell-envelope spanning structure that consists of 11 VirB proteins VirB1–VirB11 and VirD4. Colour code: yellow, nucleoside triphosphatases that provide energy for the transfer; blue, components of the transmembrane channel; red, pilus-forming components; green, lytic transglycosylase responsible for the degradation of the murein (peptidoglycan) layer at the site of assembly.
Several other T4SSs sharing a common ancestry with the VirB/VirD4 system of A. tumefaciens have been identified. Some of them contain the complete set of A. tumefaciens virB/virD4 genes, while others are chimeras of the virB/virD4 and other unrelated genes (Christie et al., 2005). One good example is from Helicobacter pylori, the causative agent of gastritis, peptic ulcer and gastric cancer in humans (Blaser and Atherton, 2004). Most virulent strains of H. pylori harbour the cag pathogenicity island encoding a VirB/VirD4-like T4SS. T4SS of H. pylori contains genes encoding proteins homologous to VirB4, VirB7, VirB9, VirB10, VirB11 and VirD4 of A. tumefaciens, in addition to other genes with unknown function. Furthermore, topological and mutational analyses suggest similar functions to A. tumefaciens proteins VirB1, VirB2, VirB6 and VirB8 for four additional non-homologous H. pylori proteins (Buhrdorf et al., 2003; Andrzejewska et al., 2006). Most recent study aiming at the elucidation of the structure of the H. pylori T4SS apparatus has shown that H. pylori T4SS contains functional analogues of all components of the VirB/VirD4 system of A. tumefaciens, except VirB5 (Kutter et al., 2008). The only effector protein of the H. pylori T4SS known to date, CagA, interacts with several host cell proteins, resulting in altered physiology of the host cells and an increased chance of successful infection (Bagnoli et al., 2005; Bourzac and Guillemin, 2005; Brandt et al., 2005; Bourzac et al., 2007; Moese et al., 2007; Zeaiter et al., 2008). The exact mechanism by which CagA is translocated by H. pylori T4SS remained elusive for a long time; however, a recent study involving fusion proteins and immunoprecipitation studies led to identification of the CagF protein, that is involved in the interaction with CagA (Couturier et al., 2006). Results from this study indicate that CagF is a protein that recognizes and delivers CagA into the T4SS channel (Couturier et al., 2006). Another protein crucial for the successful delivery of CagA into host cells is the specialized adhesin CagL. Located on the surface of the T4SS pilus, CagL triggers delivery of CagA via activation of integrin receptors on the surface of the host cells (Kwok et al., 2007).
A VirB/VirD4-like T4SS plays an important role in the pathogenesis of the intracellular Bartonella spp. (Schmid et al., 2004). Several translocated effector proteins (Beps) are involved in a wide variety of virulence associated traits, including activation of pro-inflammation, apoptosis and cytoskeleton rearrangements (Schulein et al., 2005; Backert and Meyer, 2006). A related T4SS is also exploited by the causative agent of the whooping cough, Bordetella pertussis to deliver pertussis toxin into the extracellular milieu (Rambow-Larsen and Weiss, 2004). Recently, presented model suggests that during first stage pertussis toxin interacts in the periplasm with the partially assembled secretion apparatus and only after this initial interaction, the complete T4SS is assembled and secretion of the pertussis toxin across the outer membrane can proceed (Verma and Burns, 2007).
I type IV secretion systems: protein secretion machines of intracellular pathogens
The I type T4SSs, also referred to as type IVB systems, resembling the archetypal Dot/Icm system of the IncI plasmids have been identified in two intracellular bacterial pathogens, L. pneumophila and Coxiella burnetii.
Legionella pneumophila is a Gram-negative facultative intracellular parasite found primarily associated with environmental amoebae and protozoa, but it is better known as the causative agent of a community acquired or nosocomial pneumonia called Legionnaires' disease. After successful infection through contaminated aerosols, L. pneumophila is phagocytosed by alveolar macrophages that do not undergo subsequent lysis due to the inhibition of the phagosome–lysosome fusion at early stages of phagosome maturation (Wiater et al., 1998). Twenty-five open reading frames whose products are crucial for the intracellular survival of the bacterium in macrophages have been identified and named icm (intracellular multiplication) and dot (defective organelle trafficking) genes. A significant number of the Dot/Icm proteins are homologous to the components of the conjugation system of the IncI plasmids such as R64 (Vogel et al., 1998; Komano et al., 2000). The Dot/Icm T4SS of L. pneumophila is absolutely required for virulence of this bacterium, as T4SS mutants are impaired in a variety of pathogenic properties. These include phagocytosis, pore formation in the host cell membranes and inhibition of phagosome–lysosome fusion. Furthermore, the Dot/Icm T4SS of L. pneumophila is involved in the recruitment of the rough endoplasmic reticulum that, together with phagosomes, forms a favourable niche for the replication of the bacterium inside the host macrophage. This T4SS also promotes apoptosis of the host cells and escape of the bacterium from the phagosome (Segal et al., 2005; Robinson and Roy, 2006). Several effector proteins secreted by the Dot/Icm secretion systems have been identified. One of them is RalF, a protein containing a Sec-7 homology domain typical of eukaryotic ARFs (ADP ribosylation factors), which is presumably used for the activation of ARF through exchange of GDP for GTP and subsequent ARF-mediated recruitment of endoplasmic reticulum vesicles (Nagai et al., 2002). Other proteins include LidA, required for the formation of the replicative vacuole, LepA and LepB, crucial for the escape of bacteria from the phagosome and numerous Sids (substrates of Icm/Dot transporter), whose function in L. pneumophila virulence is currently under intensive investigation (Conover et al., 2003; Luo and Isberg, 2004; Segal et al., 2005). SidM (DrrA) is a bifunctional enzyme involved in the activation of Rab1 through exchange of GDP to GTP and in the recruitment of Rab1 to the Legionella-containing vacuole (Ingmundson et al., 2007; Machner and Isberg, 2007). SidJ is required for efficient recruitment of endoplasmic reticulum to the bacterial phagosome (Liu and Luo, 2007) and SidF inhibits host cells from undergoing apoptosis to achieve maximal bacterial multiplication (Banga et al., 2007). Components of the Dot/Icm T4SS of L. pneumophila, together with numerous translocated effector proteins of this system were shown to be under the direct control of two regulators, PmrA and CpxR (Zusman et al., 2007; Altman and Segal, 2008). A comprehensive biochemical and genetic examination of the L. pneumophila T4SS was reported that shed more light into its structure (Vincent et al., 2006). Dot/Icm subcomplex consisting of five proteins, DotC, DotD, DotF, DotG and DotH, was identified that represents the core of the secretion complex and bridges the inner and outer membranes of L. pneumophila. This subcomplex seems to be functionally analogous to the A. tumefaciens VirB7–VirB10 subcomplex, thus suggesting a remarkable conservation of the core structure in these evolutionary distant T4SSs (Vincent et al., 2006).
Coxiella burnetii is a Gram-negative obligate intracellular bacterial pathogen of animals and the causative agent of Q fever in humans. After phagocytosis by host alveolar macrophages, C. burnetii delays phagosome–lysosome fusion, presumably to change from small-cell variants to large-cell variants, a process that allows this bacterium to thrive in the acidic environment of the phagolysosome (Zamboni et al., 2003; Parker et al., 2006). Initial sequence analysis of the C. burnetii genome revealed that it contains a majority of the L. pneumophila T4SS dot/icm genes, with the exception of icmR, dotJ and dotV; however, a protein non-homologous but functionally similar to IcmR of L. pneumophila was identified in the later study (Feldman et al., 2005). icm/dot genes of C. burnetii are clustered in a single region on the chromosome in contrast to L. pneumophila where these genes are present in two separate locations (Sexton and Vogel, 2002; Segal et al., 2005). Similarly to L. pneumophila, T4SS of C. burnetii was shown to be under the direct control of the regulatory protein PmrA (Zusman et al., 2007). Several of the C. burnetii icm/dot genes can substitute their homologues in L. pneumophila, but their direct involvement in C. burnetii virulence has not been investigated.
GI type IV secretion systems and their role in horizontal gene transfer
The horizontal gene pool contributes to the diversification and adaptation of microorganisms, thus having a significant impact on the genome plasticity of environmental bacterial species, the evolution of bacterial pathogens, and dissemination of antibiotic resistance genes and other virulence factors (Christie et al., 2005).
A major part of the horizontal gene pool consists of genomic islands which are mobile segments of bacterial genomes often inserted at tRNA genes and flanked by direct repeat sequences and whose G+C content differs from the rest of the chromosome. They often contain homologues of integrases and transposases and other genes associated with conjugative plasmids or phages. Furthermore, genomic islands often harbour a variable number of genes offering a selective advantage for host bacteria, such as metabolic, antibiotic resistance or virulence genes. According to their gene content, genomic islands are often described as pathogenicity, symbiosis, metabolic, fitness or resistance islands (Dobrindt et al., 2004).
Identification of a novel GI-like group of T4SSs brought new insight into the mechanism by which genomic islands transfer between bacteria. It was generally thought that genomic islands represent mobile elements such as conjugative plasmids that have co-integrated with the chromosome and lost their ability to further self-transfer (Dobrindt et al., 2004). This hypothesis has been challenged by experiments performed with the representatives of a family of syntenic genomic islands with deep evolutionary origin that includes ICEHin1056 of H. influenzae and the clc element, pKLC102 and PAPI of Pseudomonas spp. Several representatives of this family of genomic islands can transfer between bacterial cells after previous integration into the chromosome (Dimopoulou et al., 2002; van der Meer and Sentchilo, 2003; Klockgether et al., 2004; Mohd-Zain et al., 2004). A combination of bioinformatics and functional analyses has revealed a highly conserved set of genes encoding a novel GI-like lineage of T4SS that plays a key role in the horizontal transfer of these genomic islands (Juhas et al., 2007a).
First functional analysis of the GI-like T4SSs has been performed with the genomic island ICEHin1056 from H. influenzae, which confers resistance to ampicillin, chloramphenicol and tetracycline. Prior to 1972, H. influenzae was universally susceptible to ampicillin. In 1972, the first ampicillin-resistant isolate was detected and soon afterwards tetracycline, chloramphenicol, erythromycin and multiple antibiotics-resistant strains were identified that spread rapidly around the globe. Recent work has shown that a novel GI-like lineage of T4SSs plays a key role in the dissemination of the antibiotic resistance element ICEHin1056 among Haemophilus spp. Several genes of the GI T4SS of H. influenzae harboured transmembrane domains and signal peptide sequences, features typical for genes involved in T4SS (Table 1). Mutation analysis showed that inactivation of key genes of the ICEHin1056 T4SS resulted in a loss of phenotypic traits provided by a T4SS. Several mutants with a mutation in this T4SS did not produce the type IV secretion pilus and had up to 100 000-fold reduced conjugation frequencies compared with the parent strain (Juhas et al., 2007a; Table 1). In a subsequent study investigating the sequence and functional properties of various Haemophilus spp. genomic islands, the GI-like T4SS module was found to be among the most conserved parts of seven genomic islands tested, with DNA similarity ranging from 95% to 100% between islands. Furthermore, results from this study suggest that GI T4SSs of all Haemophilus genomic islands tested play a key role in the formation of the pilus and conjugative transfer of DNA (Juhas et al., 2007b).
Table 1.
The GI T4SSs.
| Gene name (tfc) |
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
| Transmembrane domains | + | − | − | − | − | − | − | + | − | + | + | − | − | − | − | − | − | + | + | + | − | − | − | − |
| Signal peptide sequence | − | + | + | + | + | − | − | − | + | − | − | + | − | + | + | − | − | − | − | − | + | + | − | − |
| Pilus formation | − | + | un | + | un | − | un | un | un | un | un | + | un | + | un | + | un | un | un | un | un | + | + | − |
| Conjugation | + | + | un | + | un | + | un | un | un | un | un | + | un | + | un | + | un | un | un | un | un | + | + | + |
Table shows characteristics (transmembrane domains, signal peptide sequences, role in the pilus formation and conjugation) of the individual gene components of the GI T4SSs. un = unknown.
This novel group of T4SSs is also harboured by a broad spectrum of other genomic islands with different properties, ranging from virulence and antibiotic resistance to biodegradation. Besides pKLC102 and PAPI of P. aeruginosa (Klockgether et al., 2007; Wurdemann and Tummler, 2007), examples include the clc element from Pseudomonas sp. strain B13 carrying the genetic information for several degradation pathways, including chlorobenzoate and chlorocatechol degradation (van der Meer and Sentchilo, 2003; Gaillard et al., 2006), or SPI-7 of S. enterica serovar Typhi encoding Vi polysaccharide antigen and SopE effector protein of the SPI-1 system (Baker et al., 2008). Furthermore, GI-like T4SSs are also harboured by genomic islands of an anaerobic aromatic-degrading denitrifying bacterium Azoarcus sp. strain EbN1 (Rabus et al., 2005), plant pathogen E. carotovora ssp. atroseptica SCRI1043 (Chen et al., 2008), methylotrophic bacterium Methylibium petroleiphilum PM1 (Kane et al., 2007), insect pathogen Photorhabdus luminescens TT01 (Held et al., 2007), plant pathogen Xylella fastidiosa (Zaini et al., 2008), human enteropathogen Yersinia enterocolitica strain 8081 (Thomson et al., 2006), Yersinia pseudotuberculosis 32777 (Collyn et al., 2006) and numerous sequenced Pseudomonas strains (Klockgether et al., 2007).
Recent findings indicate that GI-like T4SSs can be further divided into three major sublineages (Klockgether et al., 2007; Fig. 3). First, ICEHin1056 sublineage, includes T4SSs of genomic islands found in variety of Haemophilus spp. (Juhas et al., 2007b), while second, pKLC102/PAPI sublineage comprises T4SSs of metabolically very versatile group of bacteria, including different Pseudomonas spp., Azoarcus, M. petroleiphilum and X. fastidiosa (Klockgether et al., 2007). Third, SPI-7 sublineage, comprises T4SSs of genomic islands of the enteropathogen S. enterica serovar Typhi, E. carotovora and P. luminescens. GI-like T4SSs are highly conserved between genomic islands of all three sublineages (Fig. 3). The complete set of 24 genes has been identified in all H. influenzae and Haemophilus parainfluenzae genomic islands tested so far. Furthermore, almost the whole sets of the GI-like T4SSs genes have been also identified in genomic islands of the pKLC102/PAPI and SPI-7 sublineages (Fig. 3). On the other hand, only a few evolutionary distant homologues of the GI T4SS have been found in the previously described paradigmal F- and P-like T4SSs and I-like T4SSs of intracellular pathogens (Fig. 3).
Fig. 3.
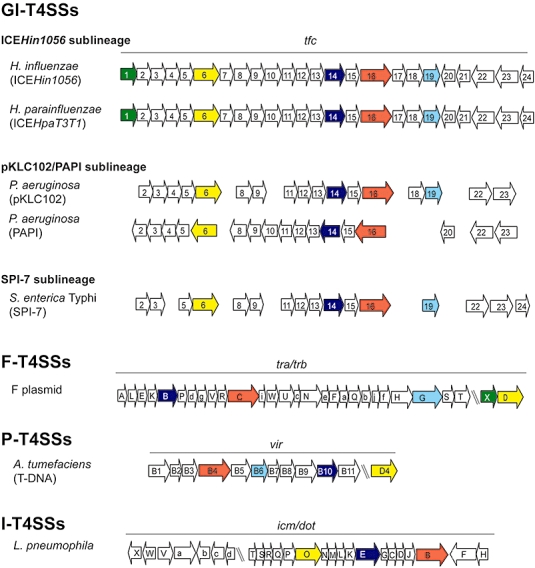
The variable T4SSs. The picture shows genetic organization of the GI T4SS in three known sublineages (ICEHin1056, pKLC102/PAPI, SPI-7) and its homologues in paradigmal F and P-like T4SSs and I-like T4SSs. GI T4SSs are well conserved in genomic islands from different bacterial species and share only limited homology to few components of the F- and P- and I-like T4SSs. Genes homologous across T4SS groups are highlighted with the same colour. Upper case gene names = Tra, Icm and lower case gene names = Trb, Dot for F plasmid and L. pneumophila respectively.
However, despite strong conservation of this T4SS, it should be noted that there are significant differences in mobility and expression of GI T4SS-harbouring genomic islands. For example, T4SS-dependent conjugative transfer of ICEHin1056 between two H. influenzae strains proceeds at various frequencies ranging from 10−1 to 10−9 (Juhas et al., 2007b). The clc element of Pseudomonas sp. strain B13 is transferable at frequencies of around 10−1−10−2 to Pseudomonas putida and P. aeruginosa (Gaillard et al., 2006; Gaillard et al., 2008). pKLC102 and PAPI-1 of P. aeruginosa are capable of self-transfer at frequencies similar to the clc element, but no conjugative transfer has been demonstrated for other members of this sublineage, like PAGI-2 and PAGI-3 (Qiu et al., 2006; Klockgether et al., 2007). Similarly, no conjugative transfer has been shown for the SPI-7 of S. enterica serovar Typhi (Baker et al., 2008). Variations in the conjugation frequencies of the more distant genomic islands could be attributed to the slight differences in the gene content of their T4SS modules; however, this explanation would not suffice for the closely related islands with the same set of 24 GI T4SS genes like ICEHin1056 and ICEHpaT3T1 (Fig. 3). Rather, this could be related to the differences in the host strain. Recent findings indicate that host strain background has a tremendous impact on the conjugal transfer efficiency and expression of genomic islands (Juhas et al., 2007b; Gaillard et al., 2008).
In conclusion, GI-like T4SSs allow genomic islands to mobilize and spread through a bacterial population, thus playing a key role in bacterial virulence, evolution and adaptation to variable environments. What remains to be seen is whether the many and diverse genomic islands not related to ICEHin1056, pKLC102/PAPI and SPI-7 have a similar secretion machinery that enables them to propagate rapidly through horizontal gene transfer.
Conclusions
Due to the contribution of a number of researchers worldwide, type IV secretion is one of the most rapidly advancing fields of research in microbiology. T4SSs mediate the horizontal transfer of genes, thus contributing to the plasticity of bacterial genomes and transmission of antibiotic resistance genes and other fitness factors between bacterial species. Furthermore, T4SSs are used for the delivery of bacterial effector proteins into the eukaryotic host cells. Since the discovery of the transfer region of the conjugative F plasmid and Ti plasmid of A. tumefaciens, similar systems have been discovered in a wide variety of other bacteria and it is reasonable to predict that many more will be discovered with the increasing number of bacterial genomes being sequenced. Functional studies and detailed characterization of the components of T4SSs diverse from the archetypal T4SSs will enhance our understanding of the function and diversity of bacterial secretion machines.
Acknowledgments
We cordially thank Professor E. Richard Moxon for critical reviews and comments on the manuscript. We would also like to thank all scientists who contributed with their results and ideas to the field of type IV secretion and apologize to those whose work was not cited due to space limitation. This work was supported by a grant from the Medical Research Council UK. Dr M. Juhas and Dr D. Crook are funded by NIHR Biomedical Research Programme, Oxford Infection Theme.
References
- Altman E, Segal G. The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J Bacteriol. 2008;190:1985–1996. doi: 10.1128/JB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrzejewska J, Lee SK, Olbermann P, Lotzing N, Katzowitsch E, Linz B, et al. Characterization of the pilin ortholog of the Helicobacter pylori type IV cag pathogenicity apparatus, a surface-associated protein expressed during infection. J Bacteriol. 2006;188:5865–5877. doi: 10.1128/JB.00060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Bagnoli F, Buti L, Tompkins L, Covacci A, Amieva MR. Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc Natl Acad Sci USA. 2005;102:16339–16344. doi: 10.1073/pnas.0502598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S, Pickard D, Whitehead S, Farrar J, Dougan G. Mobilization of the incQ plasmid R300B with a chromosomal conjugation system in salmonella typhi. J Bacteriol. 2008;190:4084–4087. doi: 10.1128/JB.00065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga S, Gao P, Shen X, Fiscus V, Zong WX, Chen L, Luo ZQ. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc Natl Acad Sci USA. 2007;104:5121–5126. doi: 10.1073/pnas.0611030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. J Clin Invest. 2004;113:321–333. doi: 10.1172/JCI20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourzac KM, Botham CM, Guillemin K. Helicobacter pylori CagA induces AGS cell elongation through a cell retraction defect that is independent of Cdc42, Rac1, and Arp2/3. Infect Immun. 2007;75:1203–1213. doi: 10.1128/IAI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourzac KM, Guillemin K. Helicobacter pylori–host cell interactions mediated by type IV secretion. Cell Microbiol. 2005;7:911–919. doi: 10.1111/j.1462-5822.2005.00541.x. [DOI] [PubMed] [Google Scholar]
- Brandt S, Kwok T, Hartig R, Konig W, Backert S. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc Natl Acad Sci USA. 2005;102:9300–9305. doi: 10.1073/pnas.0409873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhrdorf R, Forster C, Haas R, Fischer W. Topological analysis of a putative virB8 homologue essential for the cag type IV secretion system in Helicobacter pylori. Int J Med Microbiol. 2003;293:213–217. doi: 10.1078/1438-4221-00260. [DOI] [PubMed] [Google Scholar]
- Chen LL, Ma BG, Gao N. Reannotation of hypothetical ORFs in plant pathogen Erwinia carotovora subsp. atroseptica SCRI1043. FEBS J. 2008;275:198–206. doi: 10.1111/j.1742-4658.2007.06190.x. [DOI] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collyn F, Guy L, Marceau M, Simonet M, Roten CA. Describing ancient horizontal gene transfers at the nucleotide and gene levels by comparative pathogenicity island genometrics. Bioinformatics. 2006;22:1072–1079. doi: 10.1093/bioinformatics/bti793. [DOI] [PubMed] [Google Scholar]
- Conover GM, Derre I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- Couturier MR, Tasca E, Montecucco C, Stein M. Interaction with CagF is required for translocation of CagA into the host via the Helicobacter pylori type IV secretion system. Infect Immun. 2006;74:273–281. doi: 10.1128/IAI.74.1.273-281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulou ID, Russell JE, Mohd-Zain Z, Herbert R, Crook DW. Site-specific recombination with the chromosomal tRNA (Leu) gene by the large conjugative Haemophilus resistance plasmid. Antimicrob Agents Chemother. 2002;46:1602–1603. doi: 10.1128/AAC.46.5.1602-1603.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- Feldman M, Zusman T, Hagag S, Segal G. Coevolution between nonhomologous but functionally similar proteins and their conserved partners in the Legionella pathogenesis system. Proc Natl Acad Sci USA. 2005;102:12206–12211. doi: 10.1073/pnas.0501850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard M, Vallaeys T, Vorholter FJ, Minoia M, Werlen C, Sentchilo V, et al. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J Bacteriol. 2006;188:1999–2013. doi: 10.1128/JB.188.5.1999-2013.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard M, Pernet N, Vogne C, Hagenbuchle O, van der Meer JR. Host and invader impact of transfer of the clc genomic island into Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci USA. 2008;105:7058–7063. doi: 10.1073/pnas.0801269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange W, Duckely M, Husale S, Jacob S, Engel A, Hegner M. VirE2: a unique ssDNA-compacting molecular machine. PLoS Biol. 2008;6:e44. doi: 10.1371/journal.pbio.0060044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Jin S, Sun D, Hew CL, Pan SQ. Recruitment of conjugative DNA transfer substrate to Agrobacterium type IV secretion apparatus. Proc Natl Acad Sci USA. 2007;104:20019–20024. doi: 10.1073/pnas.0701738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol. 2005;55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- Held KG, LaRock CN, D'Argenio DA, Berg CA, Collins CM. A metalloprotease secreted by the insect pathogen Photorhabdus luminescens induces melanization. Appl Environ Microbiol. 2007;73:7622–7628. doi: 10.1128/AEM.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- Juhas M, Crook DW, Dimopoulou ID, Lunter G, Harding RM, Ferguson DJ, Hood DW. Novel type IV secretion system involved in propagation of genomic islands. J Bacteriol. 2007a;189:761–771. doi: 10.1128/JB.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Power PM, Harding RM, Ferguson DJ, Dimopoulou ID, Elamin AR, et al. Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 2007b;8:R237. doi: 10.1186/gb-2007-8-11-r237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane SR, Chakicherla AY, Chain PS, Schmidt R, Shin MW, Legler TC, et al. Whole-genome analysis of the methyl tert-butyl ether-degrading beta-proteobacterium Methylibium petroleiphilum PM1. J Bacteriol. 2007;189:1931–1945. doi: 10.1128/JB.01259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klockgether J, Reva O, Larbig K, Tummler B. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J Bacteriol. 2004;186:518–534. doi: 10.1128/JB.186.2.518-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klockgether J, Wurdemann D, Reva O, Wiehlmann L, Tummler B. Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa. J Bacteriol. 2007;189:2443–2459. doi: 10.1128/JB.01688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T, Yoshida T, Narahara K, Furuya N. The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol Microbiol. 2000;35:1348–1359. doi: 10.1046/j.1365-2958.2000.01769.x. [DOI] [PubMed] [Google Scholar]
- Kutter S, Buhrdorf R, Haas J, Schneider-Brachert W, Haas R, Fischer W. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J Bacteriol. 2008;190:2161–2171. doi: 10.1128/JB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- Liu Y, Luo ZQ. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun. 2007;75:592–603. doi: 10.1128/IAI.01278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- van der Meer JR, Sentchilo V. Genomic islands and the evolution of catabolic pathways in bacteria. Curr Opin Biotechnol. 2003;14:248–254. doi: 10.1016/s0958-1669(03)00058-2. [DOI] [PubMed] [Google Scholar]
- Moese S, Selbach M, Brinkmann V, Karlas A, Haimovich B, Backert S, Meyer TF. The Helicobacter pylori CagA protein disrupts matrix adhesion of gastric epithelial cells by dephosphorylation of vinculin. Cell Microbiol. 2007;9:1148–1161. doi: 10.1111/j.1462-5822.2006.00856.x. [DOI] [PubMed] [Google Scholar]
- Mohd-Zain Z, Turner SL, Cerdeno-Tarraga AM, Lilley AK, Inzana TJ, Duncan AJ, et al. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J Bacteriol. 2004;186:8114–8122. doi: 10.1128/JB.186.23.8114-8122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- Parker NR, Barralet JH, Bell AM. Q fever. Lancet. 2006;367:679–688. doi: 10.1016/S0140-6736(06)68266-4. [DOI] [PubMed] [Google Scholar]
- Qiu X, Gurkar AU, Lory S. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2006;103:19830–19835. doi: 10.1073/pnas.0606810104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabus R, Kube M, Heider J, Beck A, Heitmann K, Widdel F, Reinhardt R. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch Microbiol. 2005;183:27–36. doi: 10.1007/s00203-004-0742-9. [DOI] [PubMed] [Google Scholar]
- Rambow-Larsen AA, Weiss AA. Temporal expression of pertussis toxin and Ptl secretion proteins by Bordetella pertussis. J Bacteriol. 2004;186:43–50. doi: 10.1128/JB.186.1.43-50.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CG, Roy CR. Attachment and fusion of endoplasmic reticulum with vacuoles containing Legionella pneumophila. Cell Microbiol. 2006;8:793–805. doi: 10.1111/j.1462-5822.2005.00666.x. [DOI] [PubMed] [Google Scholar]
- Schmid MC, Schulein R, Dehio M, Denecker G, Carena I, Dehio C. The VirB type IV secretion system of Bartonella henselae mediates invasion, proinflammatory activation and antiapoptotic protection of endothelial cells. Mol Microbiol. 2004;52:81–92. doi: 10.1111/j.1365-2958.2003.03964.x. [DOI] [PubMed] [Google Scholar]
- Schulein R, Guye P, Rhomberg TA, Schmid MC, Schroder G, Vergunst AC, et al. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci USA. 2005;102:856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Feldman M, Zusman T. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol Rev. 2005;29:65–81. doi: 10.1016/j.femsre.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Sexton JA, Vogel JP. Type IVB secretion by intracellular pathogens. Traffic. 2002;3:178–185. doi: 10.1034/j.1600-0854.2002.030303.x. [DOI] [PubMed] [Google Scholar]
- Thanassi DG, Hultgren SJ. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr Opin Cell Biol. 2000;12:420–430. doi: 10.1016/s0955-0674(00)00111-3. [DOI] [PubMed] [Google Scholar]
- Thomson NR, Howard S, Wren BW, Holden MT, Crossman L, Challis GL, et al. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2006;2:e206. doi: 10.1371/journal.pgen.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuink TJ, Hooykaas PJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- Verma A, Burns DL. Requirements for assembly of PtlH with the pertussis toxin transporter apparatus of Bordetella pertussis. Infect Immun. 2007;75:2297–2306. doi: 10.1128/IAI.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent CD, Friedman JR, Jeong KC, Buford EC, Miller JL, Vogel JP. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol. 2006;62:1278–1291. doi: 10.1111/j.1365-2958.2006.05446.x. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- Wiater LA, Dunn K, Maxfield FR, Shuman HA. Early events in phagosome establishment are required for intracellular survival of Legionella pneumophila. Infect Immun. 1998;66:4450–4460. doi: 10.1128/iai.66.9.4450-4460.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdemann D, Tummler B. In silico comparison of pKLC102-like genomic islands of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2007;275:244–249. doi: 10.1111/j.1574-6968.2007.00891.x. [DOI] [PubMed] [Google Scholar]
- Zaini PA, Fogaca AC, Lupo FG, Nakaya HI, Vencio RZ, da Silva AM. The iron stimulon of Xylella fastidiosa includes genes for type IV pilus and colicin V-like bacteriocins. J Bacteriol. 2008;190:2368–2378. doi: 10.1128/JB.01495-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni DS, McGrath S, Rabinovitch M, Roy CR. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol Microbiol. 2003;49:965–976. doi: 10.1046/j.1365-2958.2003.03626.x. [DOI] [PubMed] [Google Scholar]
- Zeaiter Z, Cohen D, Musch A, Bagnoli F, Covacci A, Stein M. Analysis of detergent-resistant membranes of Helicobacter pylori infected gastric adenocarcinoma cells reveals a role for MARK2/Par1b in CagA-mediated disruption of cellular polarity. Cell Microbiol. 2008;10:781–794. doi: 10.1111/j.1462-5822.2007.01084.x. [DOI] [PubMed] [Google Scholar]
- Zupan J, Hackworth CA, Aguilar J, Ward D, Zambryski P. VirB1* promotes T-pilus formation in the vir-Type IV secretion system of Agrobacterium tumefaciens. J Bacteriol. 2007;189:6551–6563. doi: 10.1128/JB.00480-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol. 2007;63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]


