Abstract
Cilia are cell surface organelles found on most epithelia in vertebrates. Specialized groups of cilia play critical roles in embryonic development, including left-right (LR) axis formation. Recently, cilia have been implicated as recipients of cell-cell signaling1, 2. However, little is known about cell-cell signaling pathways that control the length of cilia3. Here we provide several lines of evidence showing that fibroblast growth factor (FGF) signaling regulates cilia length and function in diverse epithelia during zebrafish and Xenopus development. Morpholino (MO) knockdown of FGF receptor 1 (FGFR1) in zebrafish cell-autonomously reduces cilia length in Kupffer’s vesicle (KV) and perturbs directional fluid flow required for LR patterning of the embryo. Expression of a dominant-negative FGFR (DN-FGFR), treatment with SU5402, a pharmacological inhibitor of FGF signaling, or genetic and morpholino reduction of redundant FGF ligands FGF8 and FGF24, reproduces this cilia length phenotype. Knockdown of FGFR1 also results in shorter tethering cilia in the otic vesicle and shorter motile cilia in the pronephric ducts. In Xenopus, expression of a DN-FGFR results in shorter monocilia in the gastrocoel roof plate (GRP) that control LR patterning4 and in shorter multicilia in external mucociliary epithelium. Together, these results suggest a fundamental and highly conserved role for FGF signaling in the regulation of cilia length in multiple tissues. Abrogation of FGFR1 signaling down-regulates expression of two ciliogenic transcription factors, foxj1 and rfx2, and the intraflagellar transport (IFT) gene, polaris, suggesting that FGF signaling mediates cilia length through an FGF8/FGF24 - FGFR1- IFT pathway. We propose that a subset of developmental defects and diseases ascribed to FGF signaling are due in part to loss of cilia function.
FGF ligands bind and activate cell surface FGF receptors (FGFR) to mediate multiple processes during embryogenesis. One ligand, FGF8, has been proposed to play divergent roles in LR patterning5-9; as a left determinant in mouse and a right determinant in chick and rabbit. Experimental manipulations of FGFR function allow cell-autonomous alterations of FGF signaling not possible with manipulations of multiple secreted ligands that activate a given receptor. Using this approach, we investigated the roles of FGFR1 in zebrafish development. To elucidate the role of FGFR1 signaling in LR development, we analyzed the expression of southpaw (spaw; zebrafish nodal homolog) the earliest known asymmetrically expressed gene10. Knockdown of FGFR1 with two distinct antisense morpholinos (MO) perturbed the normal left-sided expression of spaw in lateral plate mesoderm (LPM) (Fig. 1a-c). Ets transcription factors pea3 and erm, downstream targets of FGF signaling11, were down-regulated in fgfr1 morphants (Supplemental Fig. 1), indicating the efficacy of MO knockdown. Markers of notochord (no tail, lefty1, sonic hedgehog)12, 13 and floorplate (sonic hedgehog), were found to be normal in fgfr1 morphants (Supplemental Fig. 2), suggesting the barrier role of the embryonic midline is intact. These results indicate FGFR1 signaling is required early in LR development, preceding asymmetric expression of spaw.
Figure 1. Cell autonomous FGF signaling in Kupffer’s Vesicle controls Left-Right patterning.

(a-b) Dorsal view of left-sided spaw expression (arrow) in WT, and bilateral expression in fgfr1 MO 18-20 SS embryos. (c) Percentages of normal (left-sided), reversed, bilateral and absent spaw in WT (n=99), fgfr1 MO (n=117) and fgfr1 MO2 (n=120). (d-e) fgfr1 expression in WT 6 SS embryos. (d) Lateral view (anterior-left) showing fgfr1 expression in KV (bracket) and midbrain-hindbrain (red arrowhead). (e) Tailbud showing fgfr1 expression in KV (white arrow, dorsal view), presomitic mesoderm (red arrowhead) and lateral plate mesoderm (black arrow). (f-g) spaw expression (arrows) in DFCControl MO and DFCfgfr1 MO at 18-20 SS. (h) Percentages of spaw expression in DFC and Yolk MO injected embryos. spaw was altered in DFCfgfr1 MO (n=69) versus DFCControl MO (p<1.19e-05; n=121), with no difference between YolkControl MO (n= 57) and Yolk fgfr1 MO (p<0.90; n= 59).
spaw asymmetry is dependent on Kupffer’s vesicle (KV), a ciliated epithelium structure that creates directional fluid flow12-14, analogous to ‘nodal flow’ in mouse15. Fgfr1 mRNA is expressed in KV and surrounding tailbud (Fig. 1d, e). To determine whether FGF signaling functions cell-autonomously in KV cells to control spaw asymmetry, we generated chimeric DFCfgfr1 MO embryos in which fgfr1 is knocked-down in DFC/KV (dorsal forerunner cells; KV precursor cells) lineages12 but not the rest of the embryo. Similar to embryo-wide knockdown of fgfr1, DFCfgfr1 MO embryos had significant alterations in spaw expression relative to DFCControl MO (p<1.19e-05; Fig. 1c, f-h). As an important control, the effects of knockdown of FGFR1 in yolk alone (yolkfgfr1 MO) were similar to yolkcontrol MO (Fig. 1h; p<0.90). These results indicate that cell-autonomous FGFR1 signaling in DFC/KV cells is necessary for asymmetric expression of spaw in LPM.
What role does FGFR1 signaling play in DFC/KV function? Atypical Protein Kinase C (aPKC), an apical marker of polarized KV epithelial cells16, revealed that KV were of normal size and shape in fgfr1 morphants (Fig. 2a, b; n=15/15, control n=16/16), in contrast to dismorphic KV phenotypes seen in ntl or spt mutants and morphants16. Thus, morphogenesis of the KV epithelium is not dependent on FGFR1 signaling. However, KV cilia were shorter in fgfr1 MO-1 compared to Control morphants and WT embryos (Fig. 2a-c; p<1.9e-08); the number of cilia was unaltered (Fig. 2; p<0.98). Similar results were obtained from fgfr1 MO-2 (data not shown). Importantly, Xenopus fgfr1 mRNA17 rescued cilia defects induced by fgfr1 MO (Fig. 2c, p<4.70e-05), demonstrating that cilia defects in fgfr1 morphants are specific to FGFR1 knockdown.
Figure 2. FGF signaling controls cilia length and directional fluid flow in Kupffer’s Vesicle.
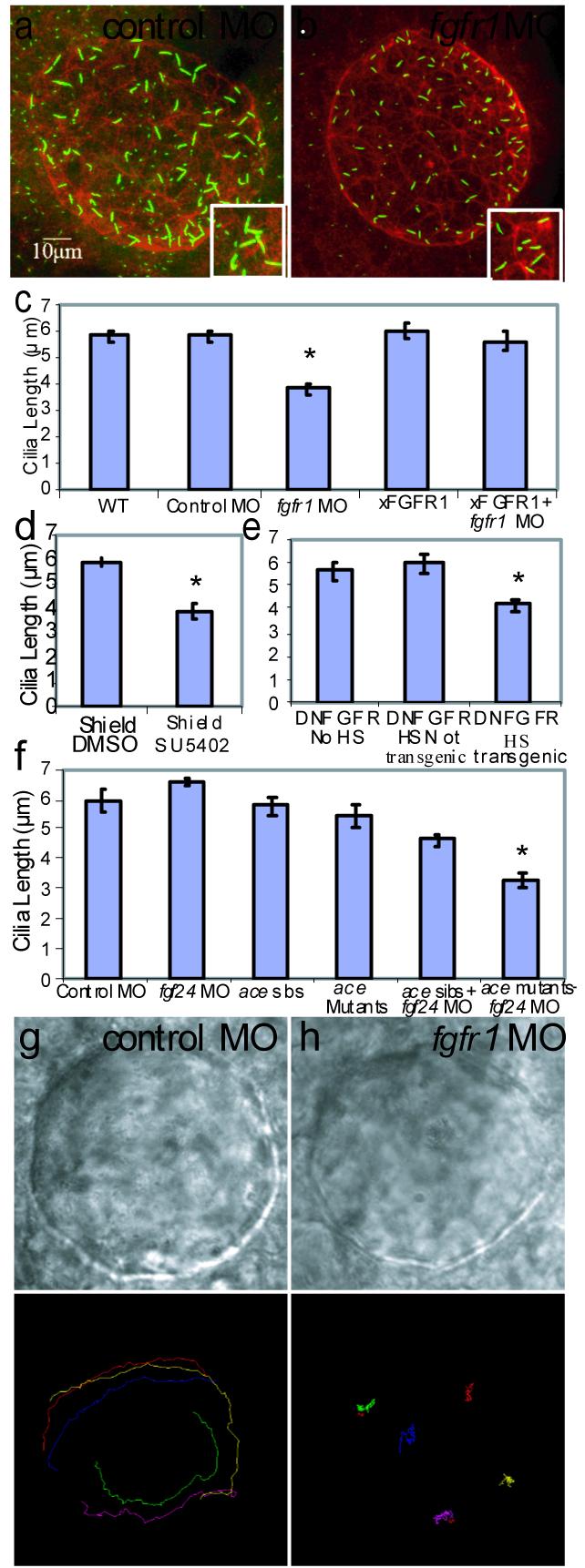
(a-b) Confocal images of 10 SS embryos, KV labeled with antibodies against aPKC (red) and acetylated tubulin (green). Control and fgfr1 morphants had similar KV structure, but cilia were shorter in fgfr1 morphants (compare insets in a and b). (c) Cilia lengths were significantly different (p< 2.88e-06) in fgfr1 morphants (688 cilia; 18 embryos) versus Control morphants (437 cilia; 9 embryos). Cilia length was similar in WT uninjected (533 cilia; 10 embryos) and Control morphant (p< 0.93), cilia numbers per KV were similar in Control and fgfr1 morphants (p< 0.26). Cilia length defects in fgfr1 morphants were rescued by Xenopus FGFR1 (xFGFR1) mRNA (p< 4.70e-05; 807 cilia; 21 embryos). Injection of xFGFR1 mRNA alone had no affect on cilia length (p<0.73; 526 cilia, 14 embryos). (d) Embryos treated with SU5402 during shield stage (248 cilia; 12 embryos) had shorter cilia compared to DMSO control embryos (p<3.26e-06; 686 cilia; 15 embryos). (e) Cilia were shorter in transgenic DN-FGFR embryos that were heat shocked at 60% epiboly (656 cilia; 19 embryos) compared to heat shocked non-transgenic siblings (p<6.94e-03; 375 cilia; 10 embryos) and non-heat-shocked siblings (p<6.99e-03; 910 cilia; 16 embryos). (f) There was no difference in cilia length (p<0.28) in fgf24 morphants (455 cilia; 10 embryos) versus Control morphants (481 cilia; 10 embryos). However, cilia were shorter when both FGF8 and FGF24 ligands were diminished (fgf24 MO in ace mutants; 12 embryos; 244 cilia), compared to single ligand knockdown (FGF8/ace mutants: p< 1.39e-04; 10 embryos; 480 cilia; fgf24 MO in ace sibs: p< 3.44e-04; 15 embryos; 643 cilia) and WT ace siblings (p<3.63e-07; 13 embryos; 626 cilia). (g-h) DIC images of bead-injected KVs in Control and fgfr1 morphants injected with fluorescent beads. (i-j) Bead paths tracked by Metamorph software. Directional KV fluid flow was absent in fgfr1 morphants (j; p< 6.4e-15; 44 beads, 9 embryos) compared to counterclockwise flow in Control morphants (i; 39 beads, 8 embryos). Error bars are standard error of the mean (s.e.m).
Additional approaches were used to assess the requirement of FGFR signaling for normal KV cilia length. Embryos treated during shield stage with a pharmacological inhibitor of FGFR activity, SU540218, 19, had shorter cilia compared to DMSO-treated controls (Fig. 2d; p<3.26e-06). Treatment at subsequent stages altered LR development but not cilia length (JMN & HJY, in prep), suggesting FGF signaling plays multiple stage-specific roles in LR development. We analyzed transgenic embryos carrying a heat-shock inducible dominant negative FGFR1 (DN-FGFR) fused to eGFP which identifies transgenic embryos from their non-transgenic siblings20. When DN-FGFR was activated at 60% epiboly, transgenic embryos had shorter cilia compared to heat-shocked non-transgenic siblings (Fig. 2e; p<6.94e-03) and non-heat-shocked siblings (Fig. 2e; p<6.99e-03), both of which had normal length cilia (Fig. 2e; p<0.61). Brief hyperactivation of FGF signaling by inducible FGFR21 avoided overexpression defects but did not increase cilia length (Supplemental Fig 3).
Which ligands signal through FGFR1 to control cilia length? FGF8 binds several FGFRs22 and FGFR1 morphants phenocopy midbrain-hindbrain defects seen in zebrafish FGF8 (acerebellar; ace) mutants5, 23. This suggests FGFR1 is a functional receptor for FGF823. ace mutants have LR defects and a minority fail to form a KV lumen5. We found fgf8 deficient embryos express KV differentiation markers (sox17, n=87/98), form an epithelium with normal apical-basal polarity (aPKC, n=10/10), and despite 33% not filling the KV lumen, develop normal numbers of cilia with normal length (Fig. 2f; p<0.53).
Another FGF ligand, fgf24, has overlapping expression with fgf8 in and around DFC/KV cells24. fgf24 mutants (ikarus; ika)25 and siblings had normal length KV cilia (average cilia length=6.2 μm; 498 cilia; 12 embryos). To test for redundant function of FGF8 and FGF24, we injected fgf24 MO into ace mutants to reduce the amount of FGF8/FGF24 activity. ace heterozygotes injected with fgf24 MO had shorter KV cilia than uninjected ace heterozygotes (Fig. 2f; p<0.015), and ace homozygotes injected with fgf24 MO had KV cilia lengths comparable to fgfr1 morphants (Fig.2f; p<3.63e-07). Similarly, ika mutants injected with fgf8 MO had shorter cilia (Supplemental Figure 4). WT, ika mutants and siblings injected with fgf24 MO had normal length cilia (Fig. 2f; p<0.28), arguing against off-target MO effects. These results indicate that FGF8 and FGF24 ligands function, likely through FGFR1, to control cilia length. Thus, results from MO against FGFR1, pharmacological inhibitors of FGFRs, transgenic expression of DN-FGFR, and mutants and MO of multiple FGF ligands indicate that FGF signaling is necessary to control KV cilia length.
To assess whether cilia-driven directional fluid flow in KV was altered by the cilia defects in fgfr1 morphants, we tracked movement of fluorescent beads injected into the lumen of KV13. In control morphants, fluorescent beads exhibited a persistent counter-clockwise directional flow (Fig. 2i, Supplemental Movie 1). In contrast, beads in fgfr1 morphants had no persistent directional flow (Fig. 2j, Supplemental Movie 2) indicating FGF signaling controls LR patterning by regulating cilia length and KV fluid flow prior to initiation of asymmetric spaw expression.
The discovery that FGF signaling plays a role in LR patterning by regulating cilia suggests other developmental roles attributed to FGF signaling might be due to cilia defects. To determine whether FGF-dependent regulation of cilia length is a more general developmental mechanism, we examined cilia in two epithelia that express FGFR1, the pronephric ducts and ear (otic vesicle; Supplemental Fig. 5b, c). Pronephric ducts are primitive excretory organs containing motile cilia14. Inhibition of FGF signaling during Xenopus embryogenesis inhibits pronephric development26, but no mechanism has been elucidated. Pronephric duct cilia at 26 somite stage (SS) were shorter in fgfr1 morphants compared to WT embryos (Fig. 3a, b, e; p<4.24e-04). Consistent with pronephric cilia defects, fgfr1 morphants develop cystic kidneys (Supplemental Fig. 6). In the zebrafish ear, two types of cilia are required for otolith formation: tethering cilia and motile cilia. Tethering cilia attract seeding granules and when reduced in number or length, granules are not organized correctly for otolith formation19. In zebrafish FGF8 or FGFR1 knock-down perturbs otic vesicle and otolith formation23, and otic vesicle cilia number is altered when FGF signaling is pharmacologically inhibited18. Here, fgfr1 morphants had shorter tethering cilia and otolith defects (Fig. 3c-d, d, Supplementary Fig. 6d-e; p< 1.1e-07), suggesting the otic vesicle and otolith defects seen in fgfr1 MO are due to defects in cilia length. Thus FGF signaling controls cilia length and function in multiple tissues during zebrafish development.
Figure 3. Cilia length in pronephric ducts, otic vesicles, gastrocoel roof plate epithelia and mucociliary epithelia is controlled by FGF signaling.

(a-b) Pronephric duct cilia were shorter and disorganized in fgfr1 morphants (p<4.24×10-4; 528 cilia; 10 embryos,) compared to WT (517 cilia; 10 embryos) 26 SS embryos. (c-d) Otic vesicle tethering cilia (arrows, and inset) were shorter (p< 1.10e-07) in fgfr1 morphants (325 cilia; 10 embryos) compared to WT embryos (322 cilia; 8 embryos) at 24 hpf. (g-j, m). GRP cilia in Xenopus embryos were normal length in cells expressing GFP alone (green cells in g, outlined in h; 316 cilia 18 embryos, p<0.11), neighboring cells (outside boundaries in h; 653, 18 embryos), and cells neighboring DN-FGFR+GFP expression (outside boundaries in j; 652 cilia 15 embryos, p<0.99). In contrast, GRP cilia were shorter in cells expressing DN-FGFR+GFP (i, inside boundaries in j; 155 cilia, 15 embryos) compared to neighboring cells (p<6.1e-03) and cells expressing GFP alone (p<2.7e-03). (k, l) Z plane rendering of mucociliary epithelia (scale bar 20 um), showing shorter cilia in cells expressing DN-FGFR+GFP (13 cells, 7 embryos) compared to controls expressing GFP alone (14 cells, 4 embryos). (n) Multicilia area is reduced in cells expressing DN-FGFR+GFP (p<0.019). Error bars are s.e.m.
To explore whether control of cilia length by FGF signaling is conserved in vertebrates, two types of epithelial cilia were examined in Xenopus laevis: monocilia on gastrocoel roof plate (GRP) implicated in LR patterning4, and mucociliary epithelial cilia that move fluid across the external epidermis3. Since DN-FGFR causes gastrulation defects when expressed ubiquitously during early embryogenesis, we co-injected DN-FGFR and GFP mRNA into cell lineages that contribute to either the GRP or mucociliary epithelium (Supplemental Fig. 1d-f). GRP cells co-expressing GFP and DN-FGFR had shorter cilia compared to neighboring GRP cells in the same embryo (p<6.0e-03; Fig. 3i-j, m) and GRP cells in embryos expressing GFP alone (p<2.7e-03; Fig. 3g-h, m). In mucociliary epithelial, cells co-expressing GFP and DN-FGFR had shorter cilia compared to cells expressing GFP alone (p<0.019; Fig. 3k-i, n). These results indicate that FGF signaling controls cilia length in diverse epithelia, and suggests that the regulation of cilia length by FGF signaling is evolutionarily conserved.
How does FGFR1 regulate cilia length? To address this, we analyzed cell differentiation, epithelial cell polarization and cilia formation of KV cells in zebrafish16. In fgfr1 morphants, two markers of the DFC/KV cell lineage, sox1712 and dnah913, showed similar expression in WT and fgfr1 morphants, indicating correct DFC/KV cell differentiation (Fig. 4a-d, i). Apical membrane marker aPKC and tight junction marker ZO-1 revealed apical-basal polarity in KV cells was intact in fgfr1 morphants compared to WT controls (Supplemental Fig. 7a-d). Further, cilia in fgfr1 morphants were correctly positioned at the apical surface facing the KV lumen (Supplemental Fig. 7e, f). In contrast to the apparent normal differentiation and polarization of KV cells in fgfr1 morphants, two members of transcription factor families implicated in ciliogenesis27, 28, foxJ1 and rfx2 (BWB and HJY, in preparation) were down-regulated in these embryos (Fig. 4e, f, i). Correspondingly, expression of polaris, an intraflagellar transport gene (Ift88) required for normal length cilia in zebrafish29, was diminished in fgfr1 morphants (Fig. 4g-i). Reduced polaris expression is consistent with IFT-defective phenotypes seen in fgfr1 morphants, including curved body axis, kidney cysts and shortened cilia (Fig. 2a-f, Supplemental Fig. 6). From these results, we propose that FGF8 and FGF24 activate FGFR1 cell-autonomously in KV cells to maintain a transcriptional network that allows normal expression of IFT proteins required for normal length cilia (Fig. 4j).
Figure 4. FGF signaling controls ciliogenic genes in DFC/KV cells.
(a, b) sox17 expression in DFC/KV (and endoderm cells in a different focal plane) in 90% epiboly embryos was normal in fgfr1 morphants and WT embryos. (c, d) Expression of dnah9 in 95 % epiboly embryos was normal in fgfr1 morphants and WT embryos. (e, f) In contrast, foxJ1 was down-regulated in fgfr1 morphants versus WT embryos at 90% epiboly. (g, h) Similarly, polaris was down-regulated in fgfr1 morphants versus WT embryos at tailbud stage. (i) Comparison of percentage of embryos with WT expression levels of each gene indicated. (j) Proposed mechanism by which FGF signaling controls length of motile cilia: FGF ligands bind to FGFR1 activating downstream transcription factors (TF) including foxj1 and rfx2, these TF activate IFT genes (e.g. polaris) to maintain motile cilia length on epithelial cells.
Monocilia are found on almost all cells and have been implicated as sites for receiving or modulating cell-cell signaling pathways such as Hedgehog1, PDGF1 and Wnt2. Interactions among signaling pathways are of great interest in understanding how cells integrate diverse signals. Extrapolating from our discovery of a link between FGF signaling and cilia function in zebrafish and Xenopus, we propose that (1) some of the apparent interactions between FGF signaling and other cell signaling pathways might be due to FGF-dependent changes in cilia, which then influence the ability of cells to receive and integrate other cell-cell signals, and (2) a spectrum of developmental defects and human diseases due to defects in FGF signaling might be due to defects in cilia length or function.
Methods Summary
Xenopus mRNA Injections
For Xenopus GRP monocilia analysis, embryos were injected with 200 pg GFP mRNA alone (lineage tracer) or co-injected with 400 pg DN-FGFR mRNA into two dorsal cells of 32-cell embryo. For Xenopus epithelial cell analysis, embryos were injected with 200 pg GFP mRNA alone or co-injected with 600 pg dnFGFR mRNA into a single ventral cell of a 16-cell embryo.
Statistics
Cilia measurements were analyzed using a two-tailed student’s T-test, and analysis of spaw proportions were conducted using Fisher’s exact test. In a given embryo each cilium was measured in the tissue of interest and the average cilia length per embryo was determined. Averages for controls and experimentals were compared within each clutch of embryos. Outcomes were the same using a second analytical approach in which all cilia lengths were pooled and compared across all series of experiments. Analysis was done by R-Commander software package within the R Statistical Software platform30. Results are considered significant when p< 0.05 and results are expressed as mean±standard error of the mean.
Supplementary Material
Acknowledgements
We thank A. Moon and M. Condic for critical discussions on the manuscript; M. Karthikeyan, J. Shen, D. Coombs, and E. Martini for technical help, S. Miyagawa-Tomita, K. Poss, and H. Issacs for reagents. This work was supported by American Heart Association predoctoral fellowship to JMN, NRSA Postdoctoral fellowship to JDA and grants from NHLBI, NICHD, and Primary Children’s Medical Foundation to HJY.
Appendix
Methods
Zebrafish and Xenopus embryo culture
Oregon AB WT zebrafish (Danio rerio) were collected from natural matings, and were injected, raised and staged as described previously13. Heterozygote crosses with aceti282a, fgf24t22030, and hsp70:dn-fgfr1were used to produce ace and fgf24 mutant embryos and hsp:dn-fgfr1 transgenic embryos respectively5, 20, 23, 25. Hsp70:dnfgfr1 embryos from heterozygote crosses were incubated at 28°C (no heat-shock activation) or at 60% epiboly for one hour at 37 °C (heat-shock activation) and then returned to 28°C until collected for IHC. Xenopus embryos were obtained using standard methods as previously described3.
Morpholino and mRNA injections
Antisense morpholino oligonucleotides (MO) were obtained from Gene Tools, LLC and Open Biosystems. Fluorescently labeled MOs against FGFR1 were designed using previously described sequences: translation blocking 3-carboxyfluorescein-labeled fgfr1 MO-1 (5′- GCAGCAGCGTGGTCTTCATTATCAT-3′)23, 31, translation blocking 3-carboxyfluorescein-labeled fgfr1 MO-2 (5′-CAAAGATCCTCTACATCTGAACTCC-3′)31. The fgf24 MO (5′-AGGAGACTCCCGTACCGTACTTGCC-3′) and the 3-lissamine-labeled fgf8 MO (5′-TAGGATGCTCTTACCATGAACGTCG-3′) have also been previously described24, 32. Fluorescein-labeled standard negative control (5′-CCTCTTACCTCAGTTACAATTTATA-3′) from Gene Tools, LLC was used in control injections. MO was injected into 1-4 cell zebrafish embryos for whole embryo protein knock-down experiments 13. A volume of 1 nl was delivered containing 5 ng of fgf24 MO, 4 ng of fgfr1 MO-1, 8 ng of fgfr1 MO-2, or 4 ng of Control MO. For DFCMO experiments, fluorescent MO was injected into the yolk of embryos at 500-1000 cell stage and embryos were selected by fluorescent microscopy for MO accumulation in DFC as previously described12. To control for activity of the protein of interest in the yolk alone we used yolkMO control injections: fluorescent MO was injected into dome-30% epiboly embryos, and embryos were selected by fluorescent microscopy for MO diffusion throughout the yolk. For DFCMO and yolkMO injections 1 nl was delivered containing 2 ng of fgfr1 MO-1 or 2 ng of Control MO. Capped xFGFR1, DN-FGFR, iFGFR21 and GFP mRNAs were made from linearized plasmid using the using the mMessage machine SP6 transcription kit (Ambion)17. For MO rescue experiments, 100 pg of xFGFR1 was injected alone or co-injected with 5 ng of fgfr1 MO-1 into 1-4 cell stage zebrafish embryos. For iFGFR experiments, 2.5 pg of iFGFR was injected into 1-4 cell stage zebrafish embryos.
In Situ Hybridization
Digoxigenin RNA probes were generated using a Roche DIG RNA labeling kit. cDNA templates used include spaw10, shh13, ntl12, fgfr123, sox1712, pea311, erm11, lefty113, dnah913, polaris29, foxJ1 (BWB unpublished), rfx2 (BWB unpublished). In situ hybridization were performed as previously described13, with automated wash and antibody incubation using a Biolane HTI machine (Huller and Huttner HG). After post-fixation, embryos were cleared in 100% EtOH for imaging. Embryos were stored in 70% glycerol and images were obtained and processed using a Nikon Coolpix5000 camera and Photoshop Software (Adobe).
Immunofluorescence Microscopy
For zebrafish immunohistochemistry, embryos were fixed in 4% paraformaldehyde at 4°C, dehydrated in a MeOH series, stored in 100% MeOH, rehydrated, boiled in 1 mM EDTA for five minutes (except IHC for pronephric cilia), and subsequently blocked for 1 hour in PBS containing 5% sheep serum, 1% BSA, 1% DMSO, and 0.1% Triton-X. Embryos were incubated in primary antibody including mouse anti- acetylated Tubulin (1:300, Sigma T-6793), rabbit anti-atypical Protein Kinase C ζ (1:100; Santa Cruz sc-216), and mouse anti-ZO-1 (1:150; Zymed 33-9100). After washes with PBS/0.1% Triton-X/1% DMSO/1% BSA embryos were blocked for 1 hour and incubated in secondary antibody, including goat anti-rabbit Alexa Fluor 647 and goat anti-mouse Alexa Fluor 488. Embryos were cleared and mounted in Slow Fade Reagent (Molecular Probes). Images were acquired using an Olympus Fluoview FV300 laser scanning confocal microscope and assembled using ImageJ (NIH) and Photoshop (Adobe) software. Confocal z-series images were assembled to present the sum of the focal planes; cilia length was measured using Metamorph software (Universal Imaging Corp). For GRP monocilia imaging, injected Xenopus embryos were collected at stage 1733, the vitelline membrane removed, fixed overnight in 4% PFA in PBS, dehydrated in methanol and stored at -20C. Embryos were dissected following rehydration to expose GRP cilia according previous methods4. For epithelial cilia analysis, injected embryos were collected at stage 26 and kept whole3. Embryos were blocked in 10% lamb serum in PBS/0.1% Triton-X (PBST), with PBST only washes. Cilia were labeled as for zebrafish and injected cells were visualized using a polyclonal GFP antibody (1:400; Torrey Pines Biolabs). Anti-mouse Alexa fluor 568 and anti-rabbit Alexa fluor 488 secondary antibodies were used. Samples were mounted in PBST and imaged using an Olympus Fluoview FV300 confocal microscope. To measure epithelial cilia length, images were processed using Fluoview software to render the cilia in the x-z plane and then images and cilia length for both epithelial and GRP cilia were measured as for zebrafish.
KV Flow Analysis
Embryos were dechorionated at 6-8 SS and mounted in 1% low melt agarose. Fluorescent beads (0.5-2 μm; Polysciences, Inc.) were injected into KV and imaged on a Leica DMRA compound microscope using a 40x Plan Apo objective using a Coolsnap HQ digital camera (Photometrics), Metamorph (Universal Imaging Corp) to track individual beads and calculate velocity and Quicktime (Apple) to display movies.
Pharmacological Treatments
Shield-stage embryos were incubated in 24 well tissue culture dishes (25-30 embryos per well) in either SU5402 (Calbiochem)18, 19 resuspended in DMSO or AP20187 (Ariad) resuspended in EtOH, diluted into embryo water to a concentration of 20-25μM for SU5402 (concentration dependant on drug lot) or 1.25 μM for AP20187. For a vehicle control, an equivalent volume of DMSO or EtOH was added to embryo water. At after 1 hour, embryos were washed with embryo water and incubated in the 24 well dishes until fixed for IHC.
- 31.Thummel R, et al. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006;235:336–346. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- 32.Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- 33.Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) : a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. Garland Pub.; New York: 1994. [Google Scholar]
References
- 1.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerdes JM, et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- 3.Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet. 2008;40:871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schweickert A, et al. Cilia-driven leftward flow determines laterality in Xenopus. Curr Biol. 2007;17:60–66. doi: 10.1016/j.cub.2006.10.067. [DOI] [PubMed] [Google Scholar]
- 5.Albertson RC, Yelick PC. Roles for fgf8 signaling in left-right patterning of the visceral organs and craniofacial skeleton. Developmental biology. 2005;283:310–321. doi: 10.1016/j.ydbio.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 6.Boettger T, Wittler L, Kessel M. FGF8 functions in the specification of the right body side of the chick. Curr Biol. 1999;9:277–280. doi: 10.1016/s0960-9822(99)80119-5. [DOI] [PubMed] [Google Scholar]
- 7.Fischer A, Viebahn C, Blum M. FGF8 acts as a right determinant during establishment of the left-right axis in the rabbit. Curr Biol. 2002;12:1807–1816. doi: 10.1016/s0960-9822(02)01222-8. [DOI] [PubMed] [Google Scholar]
- 8.Meyers EN, Martin GR. Differences in left-right axis pathways in mouse and chick: functions of FGF8 and SHH. Science. 1999;285:403–406. doi: 10.1126/science.285.5426.403. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka Y, Okada Y, Hirokawa N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 2005;435:172–177. doi: 10.1038/nature03494. [DOI] [PubMed] [Google Scholar]
- 10.Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- 11.Roehl H, Nusslein-Volhard C. Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr Biol. 2001;11:503–507. doi: 10.1016/s0960-9822(01)00143-9. [DOI] [PubMed] [Google Scholar]
- 12.Amack JD, Yost HJ. The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Curr Biol. 2004;14:685–690. doi: 10.1016/j.cub.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 14.Kramer-Zucker AG, et al. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development. 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- 15.Nonaka S, et al. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 16.Amack JD, Wang X, Yost HJ. Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer’s vesicle in zebrafish. Developmental biology. 2007;310:196–210. doi: 10.1016/j.ydbio.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 17.Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- 18.Millimaki BB, Sweet EM, Dhason MS, Riley BB. Zebrafish atoh1 genes: classic proneural activity in the inner ear and regulation by Fgf and Notch. Development. 2007;134:295–305. doi: 10.1242/dev.02734. [DOI] [PubMed] [Google Scholar]
- 19.Riley BB, Zhu C, Janetopoulos C, Aufderheide KJ. A critical period of ear development controlled by distinct populations of ciliated cells in the zebrafish. Developmental biology. 1997;191:191–201. doi: 10.1006/dbio.1997.8736. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- 21.Pownall ME, et al. An inducible system for the study of FGF signalling in early amphibian development. Developmental biology. 2003;256:89–99. doi: 10.1016/s0012-1606(02)00120-3. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, et al. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scholpp S, Groth C, Lohs C, Lardelli M, Brand M. Zebrafish fgfr1 is a member of the fgf8 synexpression group and is required for fgf8 signalling at the midbrain-hindbrain boundary. Dev Genes Evol. 2004;214:285–295. doi: 10.1007/s00427-004-0409-1. [DOI] [PubMed] [Google Scholar]
- 24.Draper BW, Stock DW, Kimmel CB. Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development. 2003;130:4639–4654. doi: 10.1242/dev.00671. [DOI] [PubMed] [Google Scholar]
- 25.Fischer S, Draper BW, Neumann CJ. The zebrafish fgf24 mutant identifies an additional level of Fgf signaling involved in vertebrate forelimb initiation. Development. 2003;130:3515–3524. doi: 10.1242/dev.00537. [DOI] [PubMed] [Google Scholar]
- 26.Urban AE, et al. FGF is essential for both condensation and mesenchymal-epithelial transition stages of pronephric kidney tubule development. Developmental biology. 2006;297:103–117. doi: 10.1016/j.ydbio.2006.04.469. [DOI] [PubMed] [Google Scholar]
- 27.Brody SL, Yan XH, Wuerffel MK, Song SK, Shapiro SD. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am J Respir Cell Mol Biol. 2000;23:45–51. doi: 10.1165/ajrcmb.23.1.4070. [DOI] [PubMed] [Google Scholar]
- 28.Bonnafe E, et al. The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol Cell Biol. 2004;24:4417–4427. doi: 10.1128/MCB.24.10.4417-4427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bisgrove BW, Snarr BS, Emrazian A, Yost HJ. Polaris and Polycystin-2 in dorsal forerunner cells and Kupffer’s vesicle are required for specification of the zebrafish left-right axis. Developmental biology. 2005;287:274–288. doi: 10.1016/j.ydbio.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 30.Team, R.D.C. R Foundation for Statistical ComputingVienna. Austria: 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



