Abstract
Objective
The differentiation of mesenchymal stem cells (MSCs) into chondrocytes provides an attractive basis for the repair and regeneration of articular cartilage. Under clinical conditions, chondrogenesis will often need to occur in the presence of inflammatory mediators produced in response to injury or disease. Here we examine the effect of two important inflammatory cytokines, interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α), on the chondrogenic behavior of human MSCs.
Methods
Aggregate cultures of MSCs recovered from the femoral intermedullary canal were used. Chondrogenesis was assessed by the expression of relevant transcripts by quantitative RT-PCR and examination of aggregates by histology and immunohistochemistry. The possible involvement of NF-κB in mediating the effects of IL-1β was examined by delivering a luciferase reporter construct and a dominant negative inhibitor of NF-κB (srIκB), with adenovirus vectors.
Results
Both IL-1β and TNF-α inhibited chondrogenesis in a dose-dependent manner. This was associated with a marked activation of NF-κB. Delivery of srIκB abrogated the activation of NF-κB and rescued the chondrogenic response. Although expression of type X collagen followed this pattern, other markers of hypertrophic differentiation responded differently. Matrix metalloproteinase-13 was induced by IL-1β in a NF-κB dependent fashion. Alkaline phosphatase activity, in contrast, was inhibited by IL-1β regardless of srIκB delivery.
Conclusions
Cell-based repair of lesions in articular cartilage will be compromised in inflamed joints. Strategies for enabling repair under these conditions include the use of specific antagonists of individual pyrogens, such as IL-1 and TNF, or the targeting of important intracellular mediators, such as NF-κB.
Loss of articular cartilage through injury or disease presents major clinical challenges. Because cartilage has very poor regenerative capacity, various surgical techniques for cartilage repair have been devised (1). While helpful, these procedures fail to provide sustained clinical improvement in most patients, so there is much interest in the development of alternative, biological approaches to cartilage repair and regeneration (2). Among these, there is considerable research investigating the potential use of mesenchymal stem cells (MSCs) to regenerate cartilage (3).
There are two general strategies to harnessing MSCs for this purpose. In a tissue engineering approach, MSCs are recovered from the patient and used to generate a graft that is subsequently implanted into the site of cartilage damage (4). Although the graft can be developed in a bioreactor into mature cartilage, there is increasing interest in grafting immature tissue, allowing chondrogenesis to occur in situ. The second strategy, which is already in wide clinical use, supplies MSCs to the defect by penetrating the subchondral bone, thereby allowing marrow to enter the lesion. Various related surgical techniques, including microfracture and subchondral drilling, are used for this purpose. These procedures have the convenience of being performed arthroscopically in large joints (1).
Repair strategies that rely on the in situ differentiation of MSCs are attractive, but in many instances require chondrogenesis to take place within an inflamed environment. Intraarticular inflammation may result from disease, such as arthritis, or trauma, including the iatrogenic trauma of the cartilage repair surgery itself. Because interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α) are major mediators of local inflammatory processes in joints, the present study was undertaken to study their effects on chondrogenesis by MSCs derived from human bone marrow.
There is an extensive literature dating back over 20 years that describes the inhibition of cartilage matrix synthesis by chondrocytes in response to IL-1β and TNF-α (5, 6). In producing these effects, IL-1β and TNF-α activate the transcription factor NF-κB which, in turn, inhibits the synthesis of sox9, another transcription factor required for expression of the chondrocyte phenotype (7); there is evidence this occurs post-transcriptionally by destabilizing sox9 mRNA (8). However, there is surprisingly little literature concerning the influence of inflammatory mediators on the differentiation of MSCs into chondrocytes.
Majumdar et al. (9) isolated CD105+ mesenchymal cells from human bone marrow, placed them in alginate culture and initiated chondrogenesis by the addition of bone morphogenetic proteins (BMPs)-2 and -9. Addition of IL-1 after 14 days of differentiation reduced the abundance of mRNAs encoding Col2A1, aggrecan and sox9. In a related study, Sitcheran et al. (8) used an established murine cell line, MC615, and placed monolayers into a chondrogenic medium. This induced sox9 mRNA, whose expression was inhibited by TNF-α through the induction of NF-κB. These data are consistent with the hypothesis that IL-1 and TNF-α inhibit chondrogenesis by MSCs, but this has not been formally demonstrated.
In the present study, human MSCs were obtained from bone marrow and placed into aggregate culture with transforming growth factor-β1 (TGF-β1) to induce chondrogenesis. IL-1β and TNF-α were found to inhibit cartilage formation very powerfully in a dose- and NF-κB-dependent fashion. These findings have important implications for the design of effective, clinically expeditious, cartilage regeneration strategies. They may also help explain why cartilage regeneration does not occur spontaneously in joints.
MATERIALS AND METHODS
Generation of adenoviral vectors
First generation (ΔE1, ΔE3), serotype 5 adenovirus encoding the cDNA for a dominant negative, ‘super-repressor’ (sr)IκB (Ad.srIκB) driven by the cytomegalovirus (CMV) immediate early promoter were generously provided by Paul Robbins (University of Pittsburgh School of Medicine, Pittsburgh, PA). Recombinant adenovirus encoding the cDNAs for firefly luciferase or green fluorescent protein and driven by the CMV promoter (Ad.CMV-Luc and Ad.GFP, respectively) were constructed by the method of Hardy et al. (10) as previously described (11). A similar vector encoding the cDNA for firefly luciferase but driven by a synthetic, NF-κB-specific promoter region (Ad.NF-κB-Luc) was acquired from Vector Biolabs (Philadelphia, PA).
To generate high-titer preparations, recombinant vectors were amplified in 293-Cre8 cells and purified over three successive CsCl gradients. Following overnight dialysis against a sucrose buffer, the preparations were aliquoted and stored in liquid nitrogen. Viral titers were estimated by optical density (260 nm) and standard plaque assay. Using this method, preparations of ∼1012 viral particles (vp)/ml were obtained with a ratio of vp: plaque forming units of less than 100:1.
MSC isolation and expansion
MSCs were isolated from intramedullary reamings collected from 4 patients (1 male, 3 female; ages 71-78) undergoing hip hemiarthroplasty at Brigham and Women’s Hospital (Boston, MA), as previously described (12), in accordance with an institutional review board-approved protocol. Briefly, material generated using a Reamer Irrigator Aspirator (Synthes, Paoli, PA) was filtered of osseous particles, and the filtrate collected in a sterile vessel. The mononuclear cell fraction within the filtrate was isolated using a Ficoll-Paque™ PLUS (StemCell Technologies, Beverly, MA) density gradient (400g for 30 min) and cultured at 5×107 nucleated cells/flask in 75-cm2 flasks with growth medium consisting of low-glucose Dulbecco’s Modified Eagles Medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT) and 1% antibiotic/antimycotic (Ab/Am) cocktail (Invitrogen). After two weeks in primary culture, adherent colonies were recovered using 0.05% trypsin, 0.5 mM EDTA buffer (Invitrogen) and expanded for 1-2 additional passages using growth medium containing 10 ng/ml recombinant human (rh)FGF-2 (PeproTech, Rocky Hill, NJ), which has been shown to maintain the chondrogenic potential of cultured human MSCs (13).
MSC transduction
Once sufficient cell numbers were obtained for differentiation cultures, MSCs were rinsed with PBS and transduced with Ad.GFP or Ad.srIκB for 2h in serum-free DMEM (14). For experiments requiring high transduction efficiency, viral transduction was enhanced by co-precipitation with lanthanum (La) phosphates (15). Briefly, lanthanum chloride anhydrous salt (Sigma, St. Louis, MO) was dissolved in deionized (DI) water to a stock concentration of 0.4 M (pH 5.5-6.0). Viral stocks were added to serum-free DMEM (phosphate source) to generate the final concentrations indicated in the figure text. To these solutions, sufficient LaCl3 stock was added to produce a final concentration of 0.2 mM La3+, and the mixtures were gently vortexed and incubated at room temp for 15 min prior to infection. After La-phosphate-adenovirus complex formation, 5 ml suspension was added to each flask, and the flasks incubated at 37°C for 30 min. An additional 5 ml DMEM + 10% FBS was added per flask, and incubation was continued for an additional 90 min. The transduction solutions were then aspirated and replaced with fresh growth medium with rhFGF-2. After 24-48 hr, cells were harvested for aggregate culture as described below.
To determine the range of MSC transduction using this approach, cell monolayers were transduced with three distinct levels of Ad.GFP: (i) ‘low’ = 104 vp/cell (without La3+); (ii) ‘moderate’ = 103 vp/cell + La3+; or (iii) ‘high’ = 104 vp/cell + La3+. After 6 days, cells were trypsinized and rinsed in PBS, and 20,000 per group were analyzed on a Cytomics FC 500 (Beckman Coulter) flow cytometer using a bandpass filter at 525 nm.
Aggregate chondrogenesis model
MSCs were centrifuged into cell aggregates and induced along the chondrogenic lineage as previously described (16, 17). Cells were suspended to a concentration of 1×106 cell/ml in serum-free DMEM; 200-μl aliquots (2×105 cells) per well were added to a polypropylene, v-bottom 96-well plate (Corning, Corning, NY), and the plate spun at 400g for 5 min. The supernatant was aspirated and replaced with chondrogenic inductive medium (CIM) consisting of high-glucose DMEM (containing L-glutamine and sodium pyruvate) with 1% Ab/Am cocktail, 1% ITS+Premix (BD Biosciences, San Jose, CA), 40 μg/ml proline, 100 nM dexamethasone and 50 μg/ml ascorbic acid-2-phosphate (all from Sigma). To a portion of aggregates, 10 ng/ml rhTGF-β1 (PeproTech) was added to enhance chondrogenesis and rhIL-1β or rhTNF-α (PeproTech) as inflammatory stimuli. The cell pellets formed free-floating aggregates within the first 24 hours. Media were changed every other day, and aggregates were collected at various time points for analysis, as described below.
Measurement of NF-κB activity
NF-κB activity in MSCs was assessed using the Ad.NF-κB-Luc reporter construct. Cells at 70-80% of confluence were transduced for two hours with 104 vp/cell of either Ad.NF-κB-Luc or Ad.CMV-Luc and returned to growth medium overnight. For each luciferase vector group, flasks were transduced again according to one of the following five sub-groups: (i) no virus; (ii) ‘high’ Ad.GFP (see “MSC transduction” above); (iii) ‘low’ Ad.srIκB; (iv) ‘moderate’ Ad.srIκB; or (v) ‘high’ Ad.srIκB.
The next day, cells were formed into aggregates and cultured for 5 days in CIM with 10 ng/ml TGF-β1. On day 5, IL-1β (10 ng/ml) was added to a portion of cultures. After an additional 5 hours, aggregates were collected in 200 μl GLO Lysis buffer (Promega, Madison, WI) and luciferase activities were determined by Bright-Glo™ Luciferase Assay System (Promega). Measurements from Ad.NF-κB-Luc groups were normalized by those from matched Ad.CMV-Luc groups to control for any effects the various treatments might have on general uptake and expression of recombinant adenoviral vector (i.e., independent of NF-κB activity).
Immunoblotting
For Western blot analysis, aggregates were collected in Laemmli buffer after 6 days of culture in CIM with TGF-β1. Protein concentrations were determined by the Lowry method using a RC/DC protein assay (BioRad, Hercules, CA). Equal amounts of proteins (20-40 μg) were resolved by SDS-PAGE on a 10% polyacrylamide gel and transferred to a PVDF membrane. Immunodetection was performed with rabbit antibodies against human IκBα, phospho-IκBα, and β-actin (all Santa Cruz Biotechnology, Santa Cruz, CA) and a HRP-conjugated goat anti-rabbit IgG (Chemicon, Temecula, CA). Bands were visualized using a Western Lightning™ Chemiluminescence System (PerkinElmer, Waltham, MA) and a Kodak Image Station 2000MM (Eastman Kodak, Rochester, NY).
Pellet size
After six weeks in culture, pictures were taken of the PBS-washed aggregates while still in wells. These images were analyzed with ImageJ software (http://rsb.info.nih.gov/ij/), measuring aggregate cross-sectional areas. Area measurements were converted from pixel2 to mm2 using a reference object of known size.
DNA and glycosaminoglycan content
The DNA content of aggregates was determined using the Hoechst Dye 33258 method (18). Samples were digested overnight at 65°C using 100 μg/ml proteinase K in 30 mM Tris (pH 7.8), 50 mM NaCl, 10 mM MgCl2. Pellet digests were taken through three freeze/thaw cycles, and aliquots were added to 100 ng/ml Hoechst Dye 33258 (Sigma) in 10 mM Tris (pH 7.4), 1 mM Na2EDTA, 100 mM NaCl. The fluorescence intensity was measured immediately on a DQ300 Fluorometer (Hoefer Scientific Instruments, San Francisco, CA), and DNA concentration was determined from a standard curve of calf thymus DNA (Sigma).
Proteinase K digests were also analyzed for glycosaminoglycan (GAG) content using the dimethylmethylene blue (DMMB) dye binding assay (19). Briefly, aliquots of pellet digest (or serial dilutions) were combined with DMMB solution, and samples were measured at 595 nm. GAG concentrations were interpolated from a standard curve of shark chondroitin-6-sulphate (Sigma), and results were normalized by DNA content.
Histology and immunohistochemistry
Aggregates were fixed for 30 min in 4% paraformaldehyde, encapsulated in 0.5% agarose gels (for better handling), embedded in paraffin and sectioned at 5-μm thickness. Sections were mounted onto glass slides, deparaffinized with three xylene washes (5 min each) and re-hydrated in graded alcohol solutions. For detection of matrix proteoglycan, representative sections were stained with 1.0% toluidine blue (Sigma-Aldrich), pH 3.0 for 30 min. Slides were rinsed in DI water, dehydrated in graded alcohol, rinsed three times in xylene and cover slipped with Cytoseal™ XYL mounting medium (Richard-Allan Scientific, Kalamazoo, USA).
For immunohistochemistry, endogenous peroxidases were quenched in hydrogen peroxide solution, and aggregate sections were digested with 0.1 U/ml chondroitinase ABC (Sigma-Aldrich) in PBS with 1% bovine serum albumin (Sigma-Aldrich) for 1 hr at 37°C. After blocking with 1% BSA, 10% normal donkey serum in PBS for 1 hr, slides were incubated overnight (4°C) with polyclonal goat anti-human collagen type I or type II antibodies (both from Santa Cruz Biotechnology, Santa Cruz, CA) or goat anti-collagen type X (a generous gift from Gary Gibson, Henry Ford Hospital, Detroit, MI) in blocking buffer. Antigens were visualized using a biotin-conjugated, donkey anti-goat secondary antibody (Santa Cruz Biotechnology) and a streptavidin-horseradish peroxidase labeling kit (Dako, Carpinteria, CA) using 3,3′-diaminobenzidine (DAB). Slides were rinsed, counterstained with hematoxylin, dehydrated, cover slipped and imaged on a Leica DM LB microscope (Leica Microsystems, Wetzlar, Germany).
Quantitative RT-PCR
Total RNA was extracted from pooled MSC aggregates (3 per group) following homogenization in a pyrex tissue grinder (Wheaton, Millville, NJ) and lysis in RLT buffer (Qiagen, Valencia, CA). RNA was purified using RNeasy microkit reagents and protocol (Qiagen). For cDNA synthesis, 0.2 μg of total RNA for each group was reverse transcribed using oligo (dT)18 primers and SuperScript III reverse transcriptase (Invitrogen). The cDNA product was diluted 1:3 and two microliters were used for subsequent amplification by quantitative PCR.
Real-time quantitative PCR was performed using TaqMan technology and ABI PRISM 7300 apparatus (Applied Biosystems, Foster City, CA). PCR reaction parameters were as follows: (a) reaction mix: 2 μl cDNA and 7 μl dH2O mixed with 10 μl Master Mix 2X Buffer (Applied Biosystems) and 1 μl TaqMan primer in a final volume of 20 μl. (b) PCR cycles: 10 minutes at 95°C followed by 40 amplification cycles (95°C for 15 seconds and 60°C for 60 seconds). To minimize the effects of unequal quantities of starting RNA and to eliminate potential sources of inconsistency, relative expression levels of each gene was normalized to GAPDH using the -ΔΔCT method (20). Pre-designed real-time PCR primers for all gene targets were obtained from Applied Biosystems.
Alkaline phosphatase (ALP) activity
For measurement of ALP activity, pellets were homogenized (as above) and resuspended in 0.25 ml Triton X-100. ALP activity was assayed using p-nitrophenyl phosphate (Sigma) as the substrate in a buffer containing 0.15 M Tris-HCl (pH 9.0), 0.1 mM MgCl2, and 0.1 mM ZnCl2 according to the method of Teixeira et al. (21). Hydrolysis of p-nitrophenyl phosphate was monitored as a change in absorbance at 410 nm. ALP activity was expressed as nmol of product (p-nitrophenol) per min, where 1 absorbance unit = 64 nmol of product. Activity values were normalized by the total protein content for each sample, as determined using the DC protein assay kit (BioRad).
Statistical Analysis
Aggregate experiments were repeated at least three times using cells from different donors. Representative data sets are shown for all results except for qRT-PCR and ALP assays, where the averages from multiple donor populations are presented. Statistical significance between treatment groups was determined by two-tailed, unpaired Student’s t-tests, with p values < 0.05 considered significant.
RESULTS
IL-1β and TNF-α suppress TGF-β-induced chondrogenesis of hMSCs
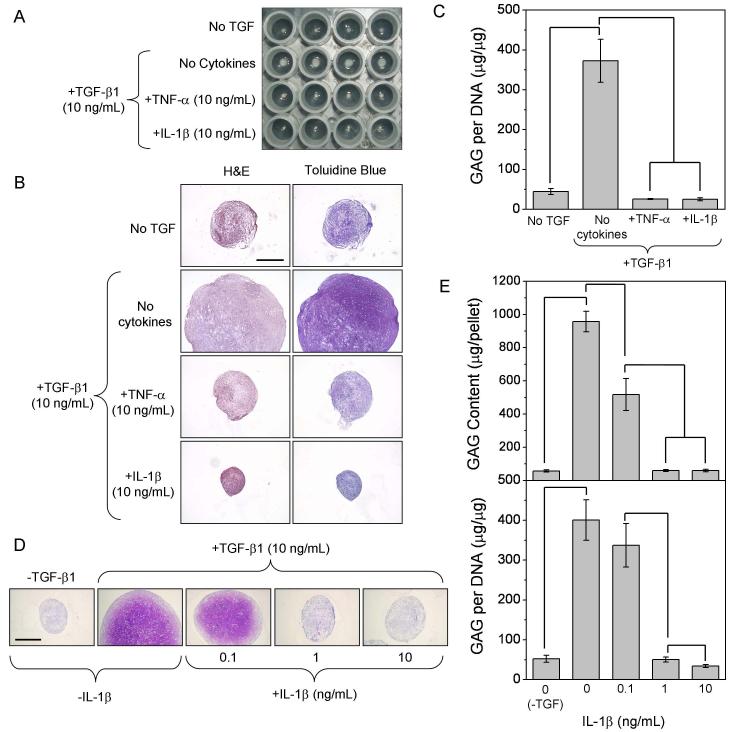
We first examined the effects of IL-1β and TNF-α on the chondrogenic differentiation of hMSCs induced by TGF-β1. When stimulated with 10 ng/ml TGF-β1, cell aggregate size increased substantially over six weeks (Figure 1A). Cells within these aggregates were surrounded by a GAG-rich extracellular matrix, as demonstrated by purple staining with toluidine blue (Figure 1B). In the presence of IL-1β or TNF-α, aggregate size and GAG staining did not exceed those of cultures lacking TGF-β1. Quantitative analysis of GAG content from pellet digests confirmed that both cytokines completely blocked the TGF-β1-induced increase in proteoglycan synthesis (Figure 1C). GAG levels in pellet-conditioned media paralleled that of corresponding digest samples, increasing from 13.4 ± 3.0 to 112.0 ± 26.7 μg/pellet/24-hr (n=6 pellets from two independent experiments) with addition of TGF-β1 but decreasing to 4.0 ± 2.4 μg/pellet/24-hr (p<0.0001) with IL-1β co-stimulation. This suggests that the pro-inflammatory cytokines inhibited proteoglycan biosynthesis rather than curtailing their deposition (i.e., via increased proteolytic activity).
Figure 1.
Testing the dose-dependence of inhibition, we found that IL-1β significantly reduced pellet size (Figure 1D) and GAG content at levels as low as 0.1 ng/ml. Quantitative analysis of GAG content confirmed histological results (Figure 1E, upper panel); however, on a per-cell basis, GAG synthesis was only significantly reduced at concentrations >0.1 ng/ml IL-1β (lower panel). TNF-α trends were similar to those of IL-1β (data not shown).
NF-κB activation and inhibition within hMSC aggregates
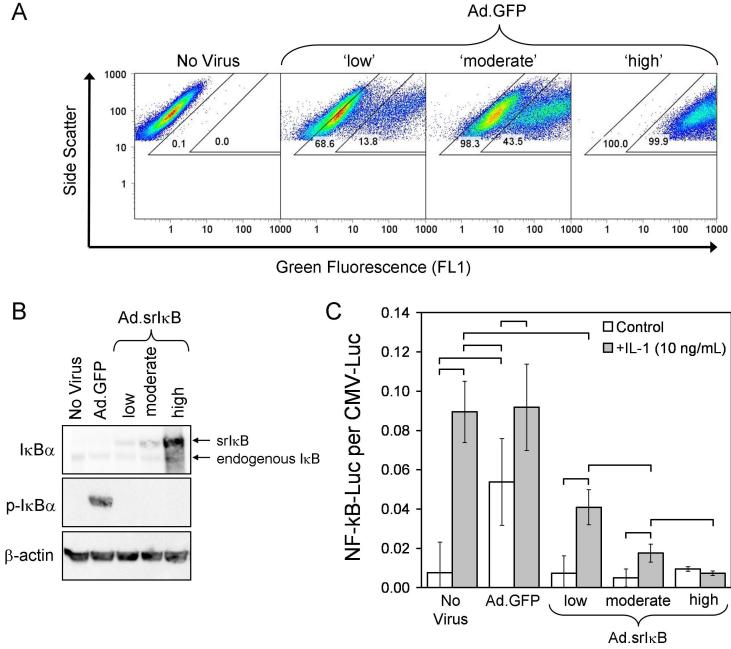
To evaluate the possible role of NF-κB signaling in mediating IL-1β effects, we used an adenoviral construct encoding a dominant negative, ‘super-repressor’ (sr)IκB. In this mutant form of IkBα, serines 32 and 36 are replaced by alanine residues (S32A/S36A), preventing phosphorylation and subsequent degradation of the transgene product (22, 23). Because IκB acts intracellularly, comprehensive effects require its efficient delivery to nearly all cells in culture. Thus we first studied a similar adenoviral construct encoding the GFP reporter (Ad.GFP) to determine transduction efficiency under different conditions, using the lanthanum method (15) to overcome the otherwise modest susceptibility of MSCs to infection by adenovirus. As shown in Figure 2A, the three conditions selected produced between 60-100% GFP+ cells (outer gates), with 10-100% of those cells fluorescing more intensely (inner gate).
Figure 2.
Using the same viral doses, we next confirmed expression of the srIκB construct within the chondrogenesis model. Western blot analysis of aggregates collected six days post-transduction verified viral-dose-dependent expression of the super-repressor, which has a higher molecular weight than endogenous IκBα due to the addition of a hemaglutinin tag (Figure 2B). Employing a NF-κB-driven firefly luciferase reporter, we then demonstrated quantitatively that the increase in NF-κB activity in response to IL-1β could be inhibited by the srIκB construct in a dose-dependent fashion (Figure 2C). As suggested by the phoshorylated IκB band visible in Figure 2B, Ad.GFP administered at the ‘high’ dose induced NF-κB activity to levels near those produced by IL-1β (Figure 2C). This is consistent with the ability of recombinant adenovirus vectors to activate NF-κB (24), and helps explain our previous observation (25) that high doses of empty adenovirus vector inhibit chondrogenesis in aggregate culture. The absence of a phosphorylated IκB band in cells transduced with Ad.srIκB confirms the biological activity of the srIκB encoded by this vector. Based on these results, we chose the lower two doses of vector (designated ‘low’ and ‘moderate’; precise transduction conditions are described in the Material and Methods) for subsequent chondrogenesis experiments.
NF-κB inhibition reverses IL-1 suppression of hMSC chondrogenesis
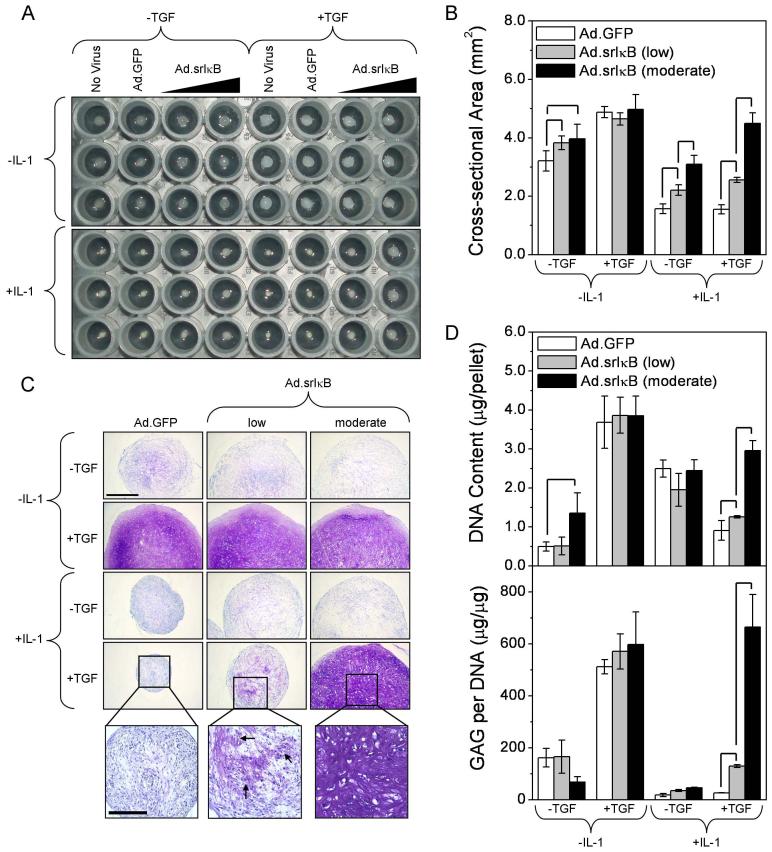
When srIκB was delivered to hMSCs prior to addition of IL-1β, aggregate growth in response to TGF-β1 was completely restored by Ad.srIκB in a dose-dependent manner (Figure 3A-B). Toluidine blue staining demonstrated dose-dependent recovery of proteoglycan synthesis with srIκB delivery (Figure 3C). Interestingly, isolated regions of GAG deposition existed within IL-1-treated pellets given the ‘low’ dose of Ad.srIκB, which is expected to transduce only a fraction of cells. We suggest that these regions contain cells which received sufficient copies of the srIκB gene, and that neighboring, non-chondrogenic cells did not. The dose-dependent increase in proteoglycan content in response to Ad.srIκB was verified by quantitative assays (Figure 3D, lower panel). No significant difference was seen between the IL-1β positive and negative groups at the ‘moderate’ dose of Ad.srIκB. In some treatment groups, DNA content increased with the level of srIκB expression (Figure 3D, upper panel), suggesting a mitogenic response to IL-1β in the absence of a chondrogenic one. This may partially explain corresponding differences in aggregate cross-sectional area (Figure 3B). Of note, the extent of DNA content and aggregate size responses varied among donor cell populations (data not shown). No differences in aggregate size or proteoglycan content were observed between Ad.GFP and untransduced controls.
Figure 3.
The protective effects of NF-κB inhibition on chondrogenesis were further examined by molecular analysis of the 6-week aggregates. Quantitative RT-PCR showed that expression of collagen type II and aggrecan core protein was induced by TGF-β1, inhibited by IL-1β, and restored by Ad.srIκB, but not Ad.GFP. (Figure 4A). Levels of collagen type X mRNA generally followed collagen type II trends, which is common for this in vitro model (26). MMP-13 expression was only substantially upregulated in the presence of both TGF-β1 and IL-1β. In this treatment group, MMP-13 expression was downregulated approximately 100-fold when Ad.GFP was substituted with Ad.srIκB. The large variance in fold-induction measurements can be partially attributed to the pooling of intra-experiment aggregates and comparison of expression levels among multiple donor populations. However, statistically significant differences among groups were determined by comparison of ΔCT values, as shown in Table 1.
Figure 4.
Table 1. ΔCT Values (CT[target] - CT[GAPDH]).
| Vector | Treatment | ACP | COL II | COL X | MMP-13 |
|---|---|---|---|---|---|
| Ad.GFP | -IL-1, -TGF | 7.9 (1.5) | 11.2 (0.2) | 9.2 (3.2) | 3.9 (3.6) |
| Ad.GFP | -IL-1, +TGF | 0.1 (0.3) | -1.5 (0.80) | -0.5 (1.0) | 2.8 (1.4) |
| Ad.GFP | +IL-1, -TGF | 10.0 (3.5) | 10.3 (4.1) | 10.1 (3.9) | 2.4 (2.5) |
| Ad.GFP | +IL-1, +TGF | 6.3 (1.8) | 5.7 (2.7) | 4.3 (0.6) | -2.7 (2.4) |
| Ad.srIκB | -IL-1, -TGF | 7.0 (3.2) | 9.3 (4.2) | 7.8 (2.4) | 6.1 (3.2) |
| Ad.srIκB | -IL-1, +TGF | 0.6 (0.8) | -0.7 (1.7) | 0.5 (2.5) | 3.6 (1.6) |
| Ad.srIκB | +IL-1, -TGF | 9.7 (3.7) | 11.7 (5.3) | 12.8 (5.6) | 3.8 (3.7) |
| Ad.srIκB | +IL-1, +TGF | 2.2 (2.3) | 0.8 (3.8) | 3.9 (4.4) | 0.3 (4.7) |
| p-values (Student’s t-test) | |||||
|---|---|---|---|---|---|
| Ad.GFP | +TGF, -IL-1 vs. +IL-1 | 0.004 | 0.012 | 0.002 | 0.024 |
| Ad.srIκB | +TGF, -IL-1 vs. +IL-1 | 0.315 | 0.561 | 0.311 | 0.303 |
ΔCT values presented as Mean (Standard Deviation)
Immunohistochemical analysis confirmed synthesis of collagen types II and X at the protein level, which were lost in the presence of IL-1β but recovered by transduction with Ad.srIκB (Figure 4B).
ALP activity does not parallel chondrogenic marker trends
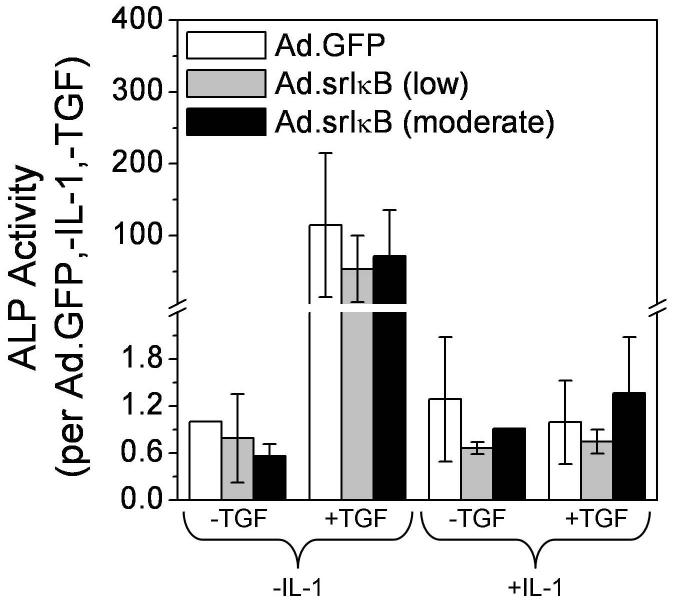
Increased ALP activity is an indicator of osteoblastic differentiation of MSCs as well as hypertrophic differentiation of growth plate chondrocytes. ALP activity in aggregate lysates was increased ∼100-fold by TGF-β1 stimulation, but returned to near-control levels with IL-1β co-stimulation (Figure 5). Surprisingly, the delivery of srIκB did not reverse fully the effects of IL-1β, in contrast to expression of chondrogenic markers (Figure 4A, Table 1). This suggests that suppression of NF-κB activity could reduce only certain aspects of hypertrophic differentiation within cartilage generated by differentiating chondroprogenitors.
Figure 5.
DISCUSSION
For reasons of injury or disease, IL-1 and TNF-α are likely to be present in joints where cartilage is undergoing attempted repair or regeneration. It has been known for a long time that IL-1 and TNF-α increase the breakdown of the extracellular matrix of articular cartilage, while inhibiting its synthesis (5, 6). As shown by the present data, these cytokines also inhibit the chondrogenic differentiation of human MSCs. These circumstances combine to present enormous biological challenges to the regeneration of cartilage in human joints and may explain why existing methods fail to provide a reliably successful, long-term clinical outcome.
Our data implicate NF-κB in the mechanism through which IL-1β and TNF-α inhibit chondrogenesis. This is consistent with the ability of NF-κB to block the expression of sox9 (8), a transcription factor that is essential for chondrogenesis, and to down-regulate expression of TGF-β receptor II (27). There is also evidence that NF-κB inhibits the phosphorylation of stimulatory Smad 3/4 in response to TGF-β (28) and enhances the activity of the inhibitory Smad 7 (27, 29).
These observations suggest possible strategies for improving the clinical outcome of cartilage repair procedures. The most immediate are to inhibit the activities of IL-1 and TNF-α within the joint space, or to block the actions of NF-κB within chondrocytes and cells undergoing chondrogenesis. These two general approaches are not mutually incompatible.
Various antagonists of IL-1 and TNF-α are already in clinical use for the treatment of rheumatoid arthritis and other inflammatory diseases (30). They include the IL-1 receptor antagonist (IL-1Ra; anakinra) (31), antibodies directed against TNF-α (infliximab, adalimubab), and bivalent, soluble TNF receptor-IgG (etanercept) (32). Although these are delivered systemically for the treatment of rheumatic diseases, they could be injected intraarticularly into joints undergoing cartilage repair to avoid the side effects and costs associated with their systemic application. However, the intra-articular dwell time of these recombinant proteins is unlikely to be sufficient for the purposes of cartilage regeneration. Gene transfer, either to the synovium (33) or, when using ex vivo strategies, directly to repair chondrocytes (34) or chondroprogenitors (25), offers one technology for obviating this limitation. The feasibility and safety of transferring IL-1Ra cDNA to the synovial lining of human joints has already been demonstrated (35).
Using an in vitro model of equine cartilage degeneration, Haupt et al. (36) demonstrated the protective effect upon articular cartilage of delivering IL-1Ra cDNA to synovial cells. IL-1Ra has a molecular weight of 20-25 kDa, depending on the degree of glycosylation, and is small enough to diffuse freely into cartilage, or sites of cartilage regeneration, when synthesized by synovium (37). Antibody-based drugs, such as infliximab, etanercept and adalimubab are probably too large to do so. Other, more general anti-inflammatory agents, such as autologous conditioned serum (38), might be more useful in this context, although high doses of steroids (39) and NSAIDs (40) are probably counter-indicated because they inhibit the synthesis of the cartilaginous matrix .
NF-κB provides an alternative target. Because it is induced by a number of different inflammatory agents, blocking this transcription factor may provide more comprehensive protection for regenerating cartilage within the joint than targeting individual cytokines. The intra-cellular location of NF-κB restricts the types of inhibitors that can be used for this purpose. Gene delivery is one possibility when using ex vivo strategies, and the adenoviral construct used in the present paper confirms the effectiveness of this approach. Alternatively, it is possible to use peptide antagonists of NF-κB containing peptide transduction domains (41), or oligonucleotide decoys (42). A clinical trial on the use of NF-κB decoys, injected into rheumatoid joints, is presently underway. Interestingly, the neutraceutical glucosamine that is commonly taken to treat osteoarthritis, shows activity against NF-κB (43) and enhances matrix production by chondrocytes (44). Because of the importance of NF-κB in the host immune response, it is likely that any suppression of this transcription factor to aid cartilage repair will need to be highly localized.
The literature contains two papers reporting in vitro data consistent with our conclusions. In the first of these studies (36), IL-1Ra was shown to enhance the ability of IGF-1 to regenerate the matrix of equine articular cartilage. The second study, recently published by McNulty et al. (45), shows that IL-1 and TNF-α impair the repair of meniscus using a simulated autologous plug approach and that this can be reversed with IL-1Ra or neutralizing antibody against TNF-α. In vivo studies of the effects of such agents on cartilage repair in suitable animal models seem warranted.
Our data may also be relevant to the observation that articular cartilage has very limited intrinsic ability to repair spontaneously. Increasing data suggest that chondroprogenitor cells exist at the surface of the cartilage (46) and, possibly, also within the deeper layers (47). The local presence of inhibitors such as IL-1 could prevent these cells from differentiating into chondrocytes following partial thickness injury or during disease-induced erosion.
ACKNOWLEDGEMENTS
We are very grateful to Paul Robbins and Gary Gibson for supplying Ad.srIκB and antibody to type X collagen, respectively; and to Mark Vrahas and Mitchell Harris for recovering the intramedullary reamings. This work was done as part of NW’s dissertation which is in preparation at the medical school of Ludwig-Maximilians University of Munich, Germany.
This work was supported in part by the AO Research Fund (Project number 04-B86) of the AO Foundation (Davos, Switzerland). RMP was supported by a NIH postdoctoral fellowship (F32 EB005566) from the NIBIB.
REFERENCES
- 1.Simon TM, Jackson DW. Articular cartilage: injury pathways and treatment options. Sports Med Arthrosc. 2006;14(3):146–54. doi: 10.1097/00132585-200609000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Steinert AF, Ghivizzani SC, Rethwilm A, Tuan RS, Evans CH, Noth U. Major biological obstacles for persistent cell-based regeneration of articular cartilage. Arthritis Res Ther. 2007;9(3):213. doi: 10.1186/ar2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djouad F, Mrugala D, Noel D, Jorgensen C. Engineered mesenchymal stem cells for cartilage repair. Regen Med. 2006;1(4):529–37. doi: 10.2217/17460751.1.4.529. [DOI] [PubMed] [Google Scholar]
- 4.Pountos I, Corscadden D, Emery P, Giannoudis PV. Mesenchymal stem cell tissue engineering: techniques for isolation, expansion and application. Injury. 2007;38(Suppl 4):S23–33. doi: 10.1016/s0020-1383(08)70006-8. [DOI] [PubMed] [Google Scholar]
- 5.Saklatvala J. Tumour necrosis factor alpha stimulates resorption and inhibits synthesis of proteoglycan in cartilage. Nature. 1986;322(6079):547–9. doi: 10.1038/322547a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyler JA. Articular cartilage cultured with catabolin (pig interleukin 1) synthesizes a decreased number of normal proteoglycan molecules. Biochem J. 1985;227(3):869–78. doi: 10.1042/bj2270869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murakami S, Lefebvre V, de Crombrugghe B. Potent inhibition of the master chondrogenic factor Sox9 gene by interleukin-1 and tumor necrosis factor-alpha. J Biol Chem. 2000;275(5):3687–92. doi: 10.1074/jbc.275.5.3687. [DOI] [PubMed] [Google Scholar]
- 8.Sitcheran R, Cogswell PC, Baldwin AS., Jr. NF-kappaB mediates inhibition of mesenchymal cell differentiation through a posttranscriptional gene silencing mechanism. Genes Dev. 2003;17(19):2368–73. doi: 10.1101/gad.1114503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majumdar MK, Wang E, Morris EA. BMP-2 and BMP-9 promotes chondrogenic differentiation of human multipotential mesenchymal cells and overcomes the inhibitory effect of IL-1. J Cell Physiol. 2001;189(3):275–84. doi: 10.1002/jcp.10025. [DOI] [PubMed] [Google Scholar]
- 10.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps ML. Construction of adenovirus vectors through Cre-lox recombination. J Virol. 1997;71(3):1842–9. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouze JN, Stoddart MJ, Gouze E, Palmer GD, Ghivizzani SC, Grodzinsky AJ, et al. In vitro gene transfer to chondrocytes and synovial fibroblasts by adenoviral vectors. Methods Mol Med. 2004;100:147–64. doi: 10.1385/1-59259-810-2:147. [DOI] [PubMed] [Google Scholar]
- 12.Porter RM, Liu F, Pilapil C, Betz O, Harris MB, Vrahas MS, Evans CH. Osteogenic Potential of Reamer Irrigator Aspirator (RIA) Aspirate Collected from Patients Undergoing Hip Arthroplasty. J Orthop Res. 2008 doi: 10.1002/jor.20715. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solchaga LA, Penick K, Porter JD, Goldberg VM, Caplan AI, Welter JF. FGF-2 enhances the mitotic and chondrogenic potentials of human adult bone marrow-derived mesenchymal stem cells. J Cell Physiol. 2005;203(2):398–409. doi: 10.1002/jcp.20238. [DOI] [PubMed] [Google Scholar]
- 14.Palmer GD, Gouze E, Gouze JN, Betz OB, Evans CH, Ghivizzani SC. Gene transfer to articular chondrocytes with recombinant adenovirus. Methods Mol Biol. 2003;215:235–46. doi: 10.1385/1-59259-345-3:235. [DOI] [PubMed] [Google Scholar]
- 15.Palmer GD, Stoddart MJ, Gouze E, Gouze JN, Ghivizzani SC, Porter RM, et al. A simple, lanthanide-based method to enhance the transduction efficiency of adenovirus vectors. Gene Ther. 2008;15(5):357–63. doi: 10.1038/sj.gt.3303092. [DOI] [PubMed] [Google Scholar]
- 16.Yoo JU, Barthel TS, Nishimura K, Solchaga L, Caplan AI, Goldberg VM, et al. The chondrogenic potential of human bone-marrow-derived mesenchymal progenitor cells. J Bone Joint Surg Am. 1998;80(12):1745–57. doi: 10.2106/00004623-199812000-00004. [DOI] [PubMed] [Google Scholar]
- 17.Penick KJ, Solchaga LA, Welter JF. High-throughput aggregate culture system to assess the chondrogenic potential of mesenchymal stem cells. Biotechniques. 2005;39(5):687–91. doi: 10.2144/000112009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YJ, Sah RL, Doong JY, Grodzinsky AJ. Fluorometric assay of DNA in cartilage explants using Hoechst 33258. Anal Biochem. 1988;174(1):168–76. doi: 10.1016/0003-2697(88)90532-5. [DOI] [PubMed] [Google Scholar]
- 19.Farndale RW, Buttle DJ, Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883(2):173–7. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Teixeira CC, Hatori M, Leboy PS, Pacifici M, Shapiro IM. A rapid and ultrasensitive method for measurement of DNA, calcium and protein content, and alkaline phosphatase activity of chondrocyte cultures. Calcif Tissue Int. 1995;56(3):252–6. doi: 10.1007/BF00298620. [DOI] [PubMed] [Google Scholar]
- 22.Iimuro Y, Nishiura T, Hellerbrand C, Behrns KE, Schoonhoven R, Grisham JW, et al. NFkappaB prevents apoptosis and liver dysfunction during liver regeneration. J Clin Invest. 1998;101(4):802–11. doi: 10.1172/JCI483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jobin C, Panja A, Hellerbrand C, Iimuro Y, Didonato J, Brenner DA, et al. Inhibition of proinflammatory molecule production by adenovirus-mediated expression of a nuclear factor kappaB super-repressor in human intestinal epithelial cells. J Immunol. 1998;160(1):410–8. [PubMed] [Google Scholar]
- 24.Tamanini A, Rolfini R, Nicolis E, Melotti P, Cabrini G. MAP kinases and NF-kappaB collaborate to induce ICAM-1 gene expression in the early phase of adenovirus infection. Virology. 2003;307(2):228–42. doi: 10.1016/s0042-6822(02)00078-8. [DOI] [PubMed] [Google Scholar]
- 25.Palmer GD, Steinert A, Pascher A, Gouze E, Gouze JN, Betz O, et al. Gene-induced chondrogenesis of primary mesenchymal stem cells in vitro. Mol Ther. 2005;12(2):219–28. doi: 10.1016/j.ymthe.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Pelttari K, Winter A, Steck E, Goetzke K, Hennig T, Ochs BG, et al. Premature induction of hypertrophy during in vitro chondrogenesis of human mesenchymal stem cells correlates with calcification and vascular invasion after ectopic transplantation in SCID mice. Arthritis Rheum. 2006;54(10):3254–66. doi: 10.1002/art.22136. [DOI] [PubMed] [Google Scholar]
- 27.Bauge C, Legendre F, Leclercq S, Elissalde JM, Pujol JP, Galera P, et al. Interleukin-1beta impairment of transforming growth factor beta1 signaling by down-regulation of transforming growth factor beta receptor type II and up-regulation of Smad7 in human articular chondrocytes. Arthritis Rheum. 2007;56(9):3020–32. doi: 10.1002/art.22840. [DOI] [PubMed] [Google Scholar]
- 28.Roman-Blas JA, Stokes DG, Jimenez SA. Modulation of TGF-beta signaling by proinflammatory cytokines in articular chondrocytes. Osteoarthritis Cartilage. 2007;15(12):1367–77. doi: 10.1016/j.joca.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauge C, Attia J, Leclercq S, Pujol JP, Galera P, Boumediene K. Interleukin1beta up-regulation of Smad7 via NF-kappaB activation in human chondrocytes. Arthritis Rheum. 2008;58(1):221–6. doi: 10.1002/art.23154. [DOI] [PubMed] [Google Scholar]
- 30.Siddiqui MA. The efficacy and tolerability of newer biologics in rheumatoid arthritis: best current evidence. Curr Opin Rheumatol. 2007;19(3):308–13. doi: 10.1097/01.bor.0000265447.48722.04. [DOI] [PubMed] [Google Scholar]
- 31.Gabay C. IL-1 inhibitors: novel agents in the treatment of rheumatoid arthritis. Expert Opin Investig Drugs. 2000;9(1):113–27. doi: 10.1517/13543784.9.1.113. [DOI] [PubMed] [Google Scholar]
- 32.Toussirot E, Wendling D. The use of TNF-alpha blocking agents in rheumatoid arthritis: an update. Expert Opin Pharmacother. 2007;8(13):2089–107. doi: 10.1517/14656566.8.13.2089. [DOI] [PubMed] [Google Scholar]
- 33.Evans CH, Ghivizzani SC, Robbins PD. Gene therapy for arthritis: what next? Arthritis Rheum. 2006;54(6):1714–29. doi: 10.1002/art.21886. [DOI] [PubMed] [Google Scholar]
- 34.Kang R, Marui T, Ghivizzani SC, Nita IM, Georgescu HI, Suh JK, et al. Ex vivo gene transfer to chondrocytes in full-thickness articular cartilage defects: a feasibility study. Osteoarthritis Cartilage. 1997;5(2):139–43. doi: 10.1016/s1063-4584(97)80007-6. [DOI] [PubMed] [Google Scholar]
- 35.Evans CH, Robbins PD, Ghivizzani SC, Wasko MC, Tomaino MM, Kang R, et al. Gene transfer to human joints: progress toward a gene therapy of arthritis. Proc Natl Acad Sci U S A. 2005;102(24):8698–703. doi: 10.1073/pnas.0502854102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haupt JL, Frisbie DD, McIlwraith CW, Robbins PD, Ghivizzani S, Evans CH, et al. Dual transduction of insulin-like growth factor-I and interleukin-1 receptor antagonist protein controls cartilage degradation in an osteoarthritic culture model. J Orthop Res. 2005;23(1):118–26. doi: 10.1016/j.orthres.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 37.Arend WP, Evans CH. Interleukin-1 receptor antagonist. In: Thompson AW, Lotze MT, editors. The Cytokine Handbook. Academic Press; London: 2003. pp. 669–708. [Google Scholar]
- 38.Wehling P, Moser C, Frisbie D, McIlwraith CW, Kawcak CE, Krauspe R, et al. Autologous conditioned serum in the treatment of orthopedic diseases: the orthokine therapy. BioDrugs. 2007;21(5):323–32. doi: 10.2165/00063030-200721050-00004. [DOI] [PubMed] [Google Scholar]
- 39.Celeste C, Ionescu M, Robin Poole A, Laverty S. Repeated intraarticular injections of triamcinolone acetonide alter cartilage matrix metabolism measured by biomarkers in synovial fluid. J Orthop Res. 2005;23(3):602–10. doi: 10.1016/j.orthres.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Brandt KD. Effects of nonsteroidal anti-inflammatory drugs on chondrocyte metabolism in vitro and in vivo. Am J Med. 1987;83(5A):29–34. doi: 10.1016/0002-9343(87)90848-5. [DOI] [PubMed] [Google Scholar]
- 41.Rehman KK, Bertera S, Bottino R, Balamurugan AN, Mai JC, Mi Z, et al. Protection of islets by in situ peptide-mediated transduction of the Ikappa B kinase inhibitor Nemo-binding domain peptide. J Biol Chem. 2003;278(11):9862–8. doi: 10.1074/jbc.M207700200. [DOI] [PubMed] [Google Scholar]
- 42.Miagkov AV, Kovalenko DV, Brown CE, Didsbury JR, Cogswell JP, Stimpson SA, et al. NF-kappaB activation provides the potential link between inflammation and hyperplasia in the arthritic joint. Proc Natl Acad Sci U S A. 1998;95(23):13859–64. doi: 10.1073/pnas.95.23.13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gouze JN, Bianchi A, Becuwe P, Dauca M, Netter P, Magdalou J, et al. Glucosamine modulates IL-1-induced activation of rat chondrocytes at a receptor level, and by inhibiting the NF-kappa B pathway. FEBS Lett. 2002;510(3):166–70. doi: 10.1016/s0014-5793(01)03255-0. [DOI] [PubMed] [Google Scholar]
- 44.Derfoul A, Miyoshi AD, Freeman DE, Tuan RS. Glucosamine promotes chondrogenic phenotype in both chondrocytes and mesenchymal stem cells and inhibits MMP-13 expression and matrix degradation. Osteoarthritis Cartilage. 2007;15(6):646–55. doi: 10.1016/j.joca.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 45.McNulty AL, Moutos FT, Weinberg JB, Guilak F. Enhanced integrative repair of the porcine meniscus in vitro by inhibition of interleukin-1 or tumor necrosis factor alpha. Arthritis Rheum. 2007;56(9):3033–42. doi: 10.1002/art.22839. [DOI] [PubMed] [Google Scholar]
- 46.Dowthwaite GP, Bishop JC, Redman SN, Khan IM, Rooney P, Evans DJ, et al. The surface of articular cartilage contains a progenitor cell population. J Cell Sci. 2004;117(Pt 6):889–97. doi: 10.1242/jcs.00912. [DOI] [PubMed] [Google Scholar]
- 47.Alsalameh S, Amin R, Gemba T, Lotz M. Identification of mesenchymal progenitor cells in normal and osteoarthritic human articular cartilage. Arthritis Rheum. 2004;50(5):1522–32. doi: 10.1002/art.20269. [DOI] [PubMed] [Google Scholar]