Abstract
Each TCR Vβ gene is regulated by an individual Vβ promoter, which becomes active prior to V(D) J recombination and drives germline transcription. It has been shown that Vβ gene locus activation and recombination are dependent on the Vβ promoter. However, transcription factors that regulate Vβ germline transcription remain largely undefined. A major challenge in studying Vβ gene germline transcription is the quantitative assessment of relatively low-level transcripts in T-cell progenitors. Here we used the established Vβ8.2CD2 knock-in mouse model to assess functions of E-protein transcription factors in Vβ8.2 germline transcription. We show that E proteins are required for the activation but not the maintenance of the Vβ8.2 germline transcription during thymocyte development. The activation of Vβ8.2 germline transcription depends more on the E proteins encoded by the E2A gene than by the HEB gene. We further show that IL-7 receptor (IL-7R)-mediated signals are essential for Vβ8.2 germline transcription. We provide evidence that IL-7R expression is only partially controlled by E2A, suggesting a role for E2A in driving Vβ8.2 germline transcription independent of IL-7R activation.
Keywords: E2A, Germline transcription, IL-7 receptor, TCR
Introduction
The TCR-β locus contains multiple V, D, J and C gene segments that span approximately 700 kb on chromosome 6. The two tandem D-J-Cβ clusters are located at the 30′ end of the locus. Most of the Vβ genes are located at the 50′ end of the locus and are separated from the D-J-Cβ clusters by a 250 kb region containing inactive trypsinogen genes. Prior to V(D)J recombination, germline transcription is initiated from each of the V and D gene segments. The germline Dβ promoter (PDβ1) is required for Dβ1 gene transcription and recombination in the context of Eβ [1], which is located downstream of D-J-Cβ clusters. Vβ promoters initiate germline transcription of the individual Vβ segments. Unlike Dβ, Vβ germline transcription seems to be independent of Eβ [2, 3] but absolutely dependent on Vβ promoters. Disruption of a Vβ promoter results in diminished germline transcription and diminished rearrangement of the transcriptionally linked Vβ gene [4]. Although studies suggested that V(D)J recombination can occur independently of germline transcription in transgenic miniloci [5–8], genetic ablation of cis-regulatory elements by gene targeting has revealed a strong link between germline transcription and V(D)J recombination at the endogenous loci [9]. A recent study has shown that blocking transcription elongation through the endogenous TCR-α locus suppresses chromatin remodeling and Va to Ja rearrangement [10], demonstrating that transcription is required for V(D)J recombination of the TCR-α locus. Thus, it is likely that germline transcription plays a similar role in the rearrangement of the TCR-β locus.
During αβ T lineage development, the most immature CD4CD8 double negative (DN) thymocytes differentiate into CD4CD8 double positive (DP) thymocytes before maturing to CD4 or CD8 single positive (SP) thymocytes. TCR-β rearrangement occurs at the DN stage of αβ T-cell development in two steps: first Dβ to Jβ and then Vβ to DJβ. DN thymocytes can be further divided into four populations based on CD44 and CD25 expression [11]. The expression of c-kit can further define proT cells because c-kit is expressed at high levels on some DN1 thymocytes as well as DN2 thymocytes and is downregulated upon thymocytes differentiating into DN3 stage [12]. The most primitive progenitors reside in the CD44+ CD25− (DN1) subset, which still possess differentiation potential to other lymphoid lineages [13, 14]. T-lineage commitment is accompanied with the commencement of TCR-β rearrangement. Low-level Dβ to Jβ rearrangement can be detected at the CD44+CD25+ (DN2) stage [15–17] while V to DJβ rearrangement is not initiated until the CD44−CD25+ (DN3) stage [18]. We previously showed that TCR Vβ gene germline transcription initiates at the DN1 stage [19]. These observations indicate that TCR-β germline transcription is activated prior to rearrangement and is among the first steps during αβ T-cell lineage commitment and differentiation. However, little is known about the transcription factors or signaling pathways that activate germline transcription prior to TCR-β gene rearrangement.
E2A and HEB are basic helix-loop-helix transcription factors that are important for T-cell development [20]. Mice lacking a functional E2A gene exhibit an accumulation of DN1 thymocytes, reduced number of DN2&3 cells and an accelerated transition from the DP to SP stage of development [21]. HEB KO mice display a developmental arrest at the CD8 immature SP stage, a transitional stage between the DN and DP stages of thymocyte development [22]. E2A/HEB heterodimers are the major functional player during T-cell development. A point mutation in the HEB basic region that abolishes HEB DNA-binding activity disrupts the function of both E2A and HEB, resulting in a block in T-cell development at the DN stage. This is the most severe and complete block in T-cell development among the established individual E-protein gene mutations [23]. E2A and HEB have been shown to induce TCRγ and TCRδ gene rearrangement and germline transcription when ectopically expressed in a nonlymphoid cell line [24]. E2A has also been reported to inhibit germline transcription and rearrangement of fetal Vγ and Vδ genes while it activates postnatal Vγ and Vδ genes in adult mice [25]. However, the role of E2A and HEB in regulating Vβ gene germline transcription during thymocyte development remains largely undefined.
We previously generated a TCR Vβ8.2CD2 knock-in allele in which a tail-less human CD2 cDNA driven by an internal ribosome entry site was placed 1.5 kbp upstream of the putative poly(A) site and 104 bp downstream of Vβ8.2 recombination signal sequence. Therefore, the hCD2 marker will be expressed exclusively from germline transcription and will be deleted after recombination involving Vβ8.2 or any upstream Vβ gene. This knock-in mouse model allowed us to monitor germline transcription of TCR Vβ8.2 gene at a single cell level during normal thymocyte development [19]. Here, we used this model to evaluate E-protein function in Vβ germline transcription and identified E proteins as essential regulators for Vβ8.2 germline transcription at distinct stages of thymocyte development.
Results
E2A is required for the activation of TCR Vβ8.2 germline transcription
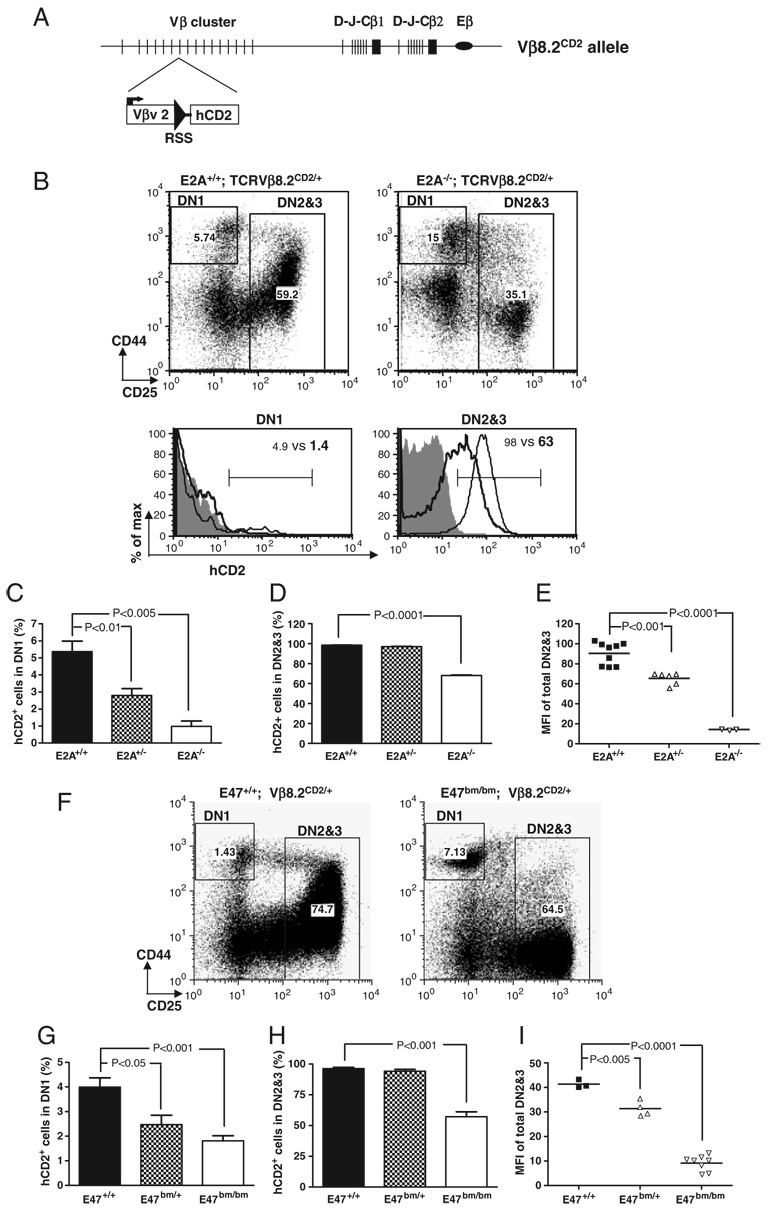
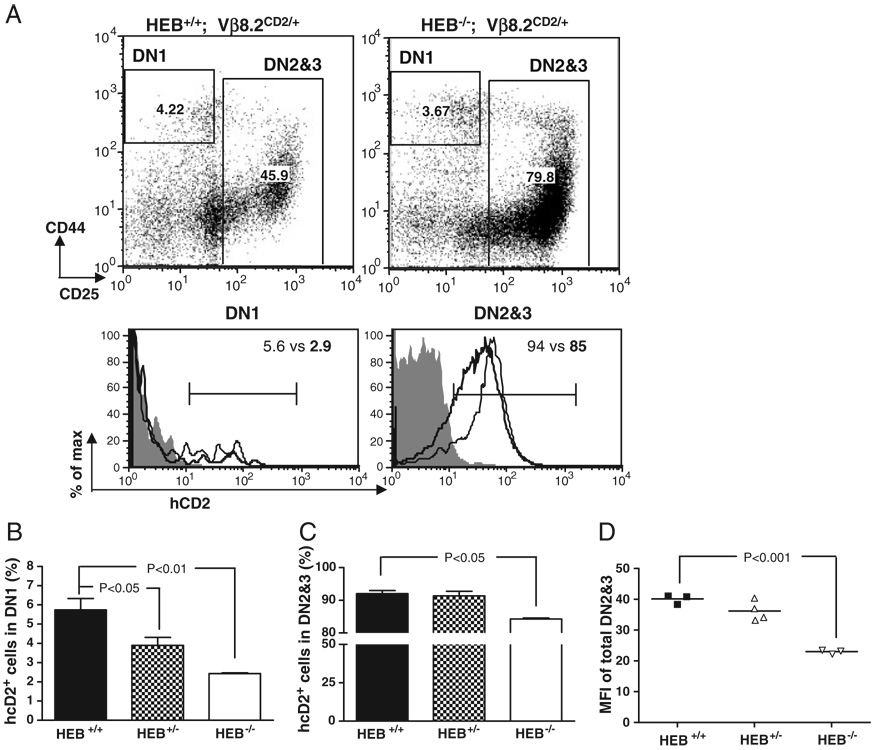
We have previously shown that the germline transcription of Vβ8.2 can first be detected in a small fraction of DN1 thymocytes, whereas all DN2 and DN3 thymocytes undergo active Vβ8.2 germline transcription on both chromosomes [19]. To investigate whether E2A is required for activating Vβ8.2 germline transcription at the DN stage of development, we introduced the Vβ8.2CD2 knock-in allele (Fig. 1A) onto an E2A-deficient background and examined hCD2 expression in DN1 and DN2&3 thymocytes. hCD2 expression from this knock-in allele indicates germline transcription of the endogenous Vβ8.2 gene. Since E2A−/− neonates exhibit low survival rate, E17.5 fetal thymocytes were analyzed for hCD2 expression. We found that the percentage of hCD2+ thymocytes in the DN1 subset was significantly reduced in E2A−/− mice when compared with WT littermates (Fig. 1B and C). hCD2 expression in total DN2&3 thymocytes was also dramatically downregulated in the absence of E2A (Fig. 1B and D). The mean fluorescence intensity (MFI) of hCD2 staining within total DN2&3 thymocyte population from E2A−/− mice was eight to ten fold lower than that from WT littermates. The reduced level of hCD2 expression was also reflected by lower percentage of total DN2&3 thymocytes under the defined hCD2+ gate (Fig. 1B and D). hCD2 expression was also reduced in E2A+/− fetal thymocytes (Fig. 1E), indicating that E2A controls Vβ8.2 germline transcription in a dosage-dependent manner.
Figure 1. Effects of E2A mutations on Vβ8.2 gene germline transcription in DN thymocytes.
(A) Schematic representation of the Vβ8.2CD2 knock-in allele. Vβ8.2 is in the middle of the Vβ cluster. Relative positions of DJCβ1 and DJCβ2 clusters and Eβ (oval) are also shown. An internal ribosome entry site-driven tail-less human CD2 cDNA (hCD2) was inserted immediately downstream of Vβ8.2 recombination signal sequence (solid triangle). (B–E) hCD2 expression in DN thymocytes from E17.5 E2A−/−, E2A+/−, and E2A+/+ embryos. (B) CD44 and CD25 expression of DN thymocytes are displayed in dot plots after being pre-gated on CD3−CD4−CD8−B220−Mac1−Gr1−7AAD− thymocytes. The gating of DN1 and DN2&3 thymocytes is indicated in the dot plots and is used in histogram analysis below the dot plots. hCD2 expression in each DN subset is presented by overlay histograms of Vβ8.2+/+ (shaded area), E2A+/+ Vβ8.2CD2/+ (thin line) and E2A−/− Vβ8.2CD2/+ (thick line). Numbers in the dot plots and histograms represent the percentage of gated population. (C) Bar graph representation of the percentage of hCD2+ cells in DN1. Results are from five independent experiments. (D) Bar graph representation of percentage of hCD2+ cells in DN2&3 thymocytes. Results are from five independent experiments. (E) Scatter plot shows the MFI of hCD2 staining in DN2&3 fetal thymocytes. Each dot represents an individual animal of indicated genotype. p-Value is given when a significant difference is found in the two-tailed t-test. (F–I) hCD2 expression in DN thymocytes from E47bm/bm, E47 bm/+ and E47+/+ mice at 4–6 wk of age. Data are analyzed as in (B–E).
In order to examine whether E2A is also required to activate Vβ8.2 germline transcription in adult thymocytes, the Vβ8.2CD2 knock-in allele was introduced onto the E47bm/bm background. The E47bm allele contains a point mutation in the DNA-binding domain, which disrupts the DNA-binding activity of E47. Accordingly, E47bm/bm mice exhibit lymphocyte development defects that closely resemble what is seen in E2A KO mice. However, E47bm/bm mice exhibit higher survival rates than the E2A−/− mice, permitting the analysis of the postnatal thymocytes. Similar to E2A−/− fetal thymocytes, the percentages of hCD2+ thymocytes in the DN1 and DN2&3 subsets of E47bm/bm mice decreased significantly in comparison with WT littermates (Fig. 1F–I). Therefore, the activation of Vβ8.2 germline transcription requires E2A in adult thymocytes as well. Overall, these results suggest that E2A controls Vβ germline transcription in both fetal and postnatal thymopoiesis.
E2A activates Vβ8.2 germline transcription in a T-cell intrinsic manner
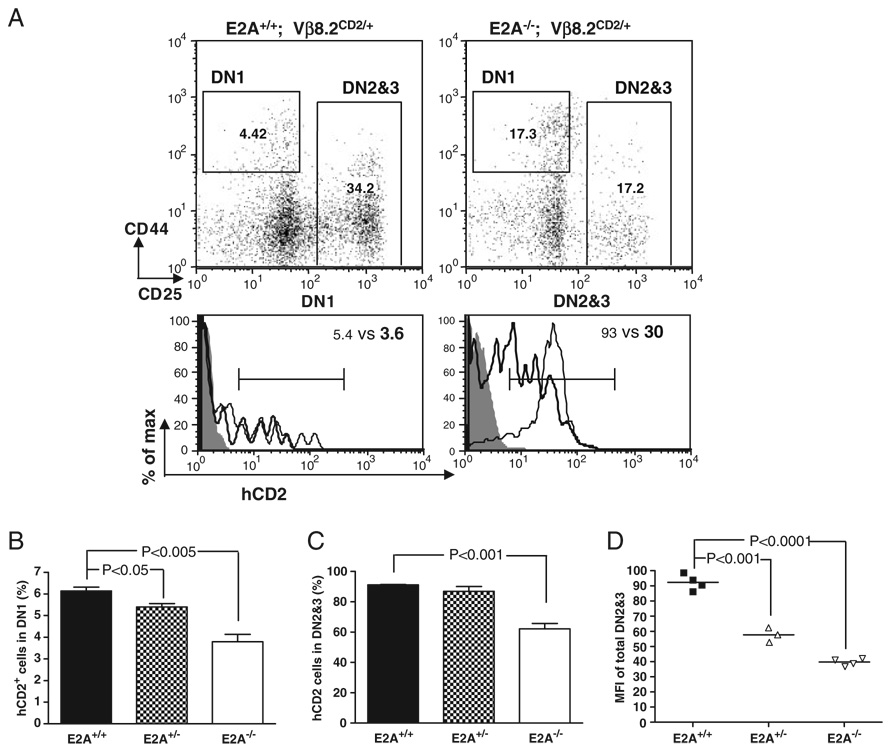
Since E2A expression has been detected in thymic stromal cells [26], it remains a question as to whether E2A plays a T-cell intrinsic or extrinsic role in activating Vβ8.2 germline transcription. To address this question, we performed adoptive transfer experiments with E17.5 fetal liver cells from E2A+/+ Vβ8.2CD2, E2A+/− Vβ8.2CD2 or E2A−/− Vβ8.2CD2 embryos. Similar to what has been observed in E2A−/− mice, the percentages of hCD2+ cells in E2A−/− donor-derived DN1 and DN2&3 thymocytes were significantly reduced compared with their E2A+/+ donor-derived counterparts (Fig. 2). In addition, the expression of hCD2 in E2A−/− donor-derived DN2&3 thymocytes was also significantly lower than in E2A+/+ donor-derived DN2&3 thymocytes. The reduction of hCD2 expression in E2A+/− donor-derived DN2&3 thymocytes was less dramatic than E2A−/− donor-derived DN2&3 thymocytes but was still statistically significant (Fig. 2D). These results demonstrate a T-cell intrinsic role for E2A in activating Vβ8.2 germline transcription.
Figure 2. Adoptive transfer assay for T-cell intrinsic function of E2A in Vβ8.2 gene germline transcription.
(A) hCD2 expression in donor-derived DN thymocytes. The gating of DN1 and DN2&3 thymocytes is indicated in the dot plots. Numbers represent the percentage of gated population. Cells are pre-gated on CD45.1−CD3−CD4−CD8−B220−Mac1−Gr1−7AAD− thymocytes. hCD2 expression in each DN subset is presented by overlay histograms of Vβ8.2+/+ (shaded area), E2A+/+ Vβ8.2CD2/+ (thin line) and E2A−/− Vβ8.2CD2/+ (thick line). Results are representative of at least three separate experiments. (B) Percentage of hCD2+ cells in DN1 subsets from E2A+/+, E2A+/− and E2A−/− fetal liver-derived thymocytes. Results are from five independent experiments. (C) Percentage of hCD2+ cells in DN2&3 fraction of donor-derived cells. Results are from five independent experiments. (D) Scattered dot plot shows the MFI of hCD2 staining in total DN2&3 thymocytes derived from E2A+/+, E2A+/− and E2A−/− donors. p-value is indicated when significant difference is found. Number of animals used for each genotype: n = 4 for E2A+/+, n = 3 for E2A+/−, n = 4 for E2A−/−.
Vβ8.2 germline transcription in DP and SP thymocytes is independent of E2A
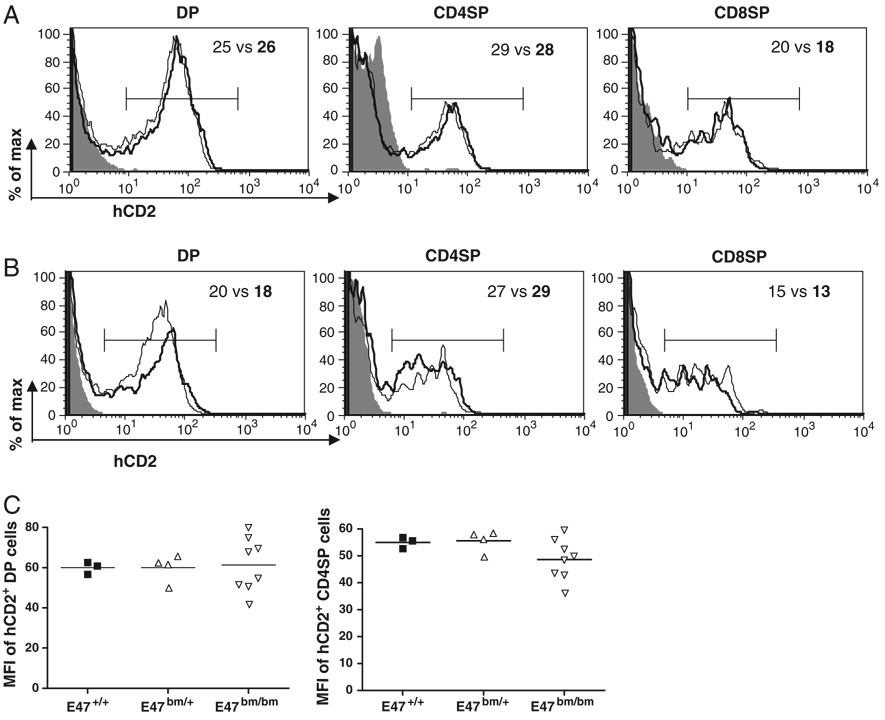
Our earlier study showed that hCD2 was expressed in a fraction of DP, SP thymocytes and peripheral T cells [19]. In contrast to DN thymocytes, germline transcription in DP, SP thymocytes and peripheral T cells seems to be dependent on the TCR-β enhancer (Eβ) [27]. We evaluated hCD2 expression in DP and SP thymocytes of E2A-sufficient or E2A-deficient background. In contrast to the defect of Vβ8.2 germline transcription in E2A-deficient DN thymocytes, the DP, CD4 SP and CD8 SP subsets derived from E2A−/− donor cells displayed comparable hCD2 expression to their counterparts derived from WT donor cells (Fig. 3A). This result was further confirmed in E47bm/bm DP and SP thymocytes (Fig. 3B). To further examine the abundance of germline transcripts at the DP and SP stages, the MFI of hCD2 staining was also analyzed. We found that the level of hCD2 expression in hCD2+ DP and hCD2+ CD4+ SP thymocytes of E47bm/bm mice was not altered (Fig. 3C). We conclude that Vβ8.2 germline transcription in DP and SP thymocytes is independent of E2A.
Figure 3. E2A is dispensable for Vβ8.2 germline transcription in DP and SP thymocytes.
(A) hCD2 expression in DP, CD4+SP and CD8+SP thymocytes derived from Vβ8.2+/+ (shaded area) E2A+/+ Vβ8.CD2/+ (thin line), E2A−/− Vβ8.2CD2/+ (thick line) fetal thymus. Numbers in the plots are percentages of hCD2+ cells (E2A+/+ versus. E2A−/−) defined by the brackets. Results are representative of at least three separate experiments. (B) hCD2 expression in DP, CD4+SP and CD8+SP thymocytes derived from Vβ8.2+/+ (shaded area), E47+/+ Vβ8.2CD2/+ (thin line), E47bm/bmVβ8.2CD2/+ (thick line) mice at 6–8 wk of age. (C) Scattered dot plots show the MFI of hCD2 staining (as defined in B) in hCD2+ DP or hCD2+ CD4SP thymocytes from E47+/+, E47bm/+, E47bm/bm adult mice. Each dot represents one individual animal of indicated genotype.
E2A upregulation correlates with initial activation of Vβ8.2 germline transcription
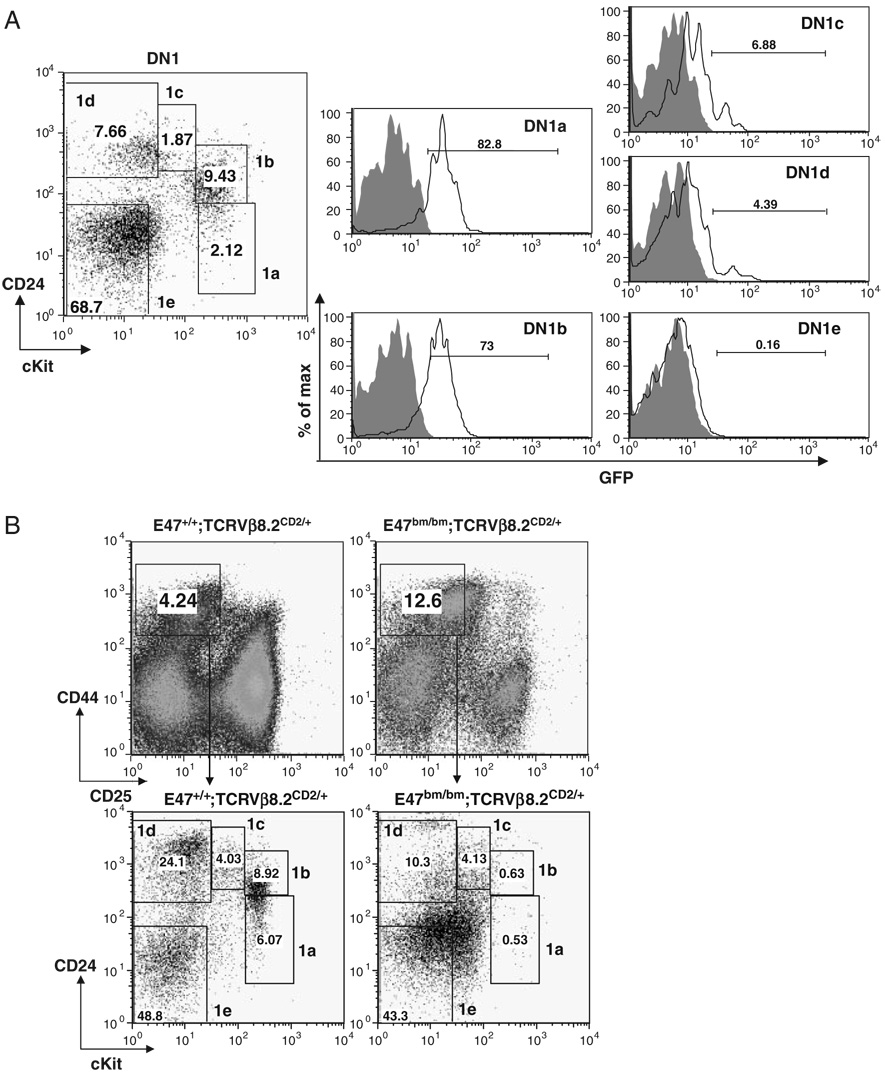
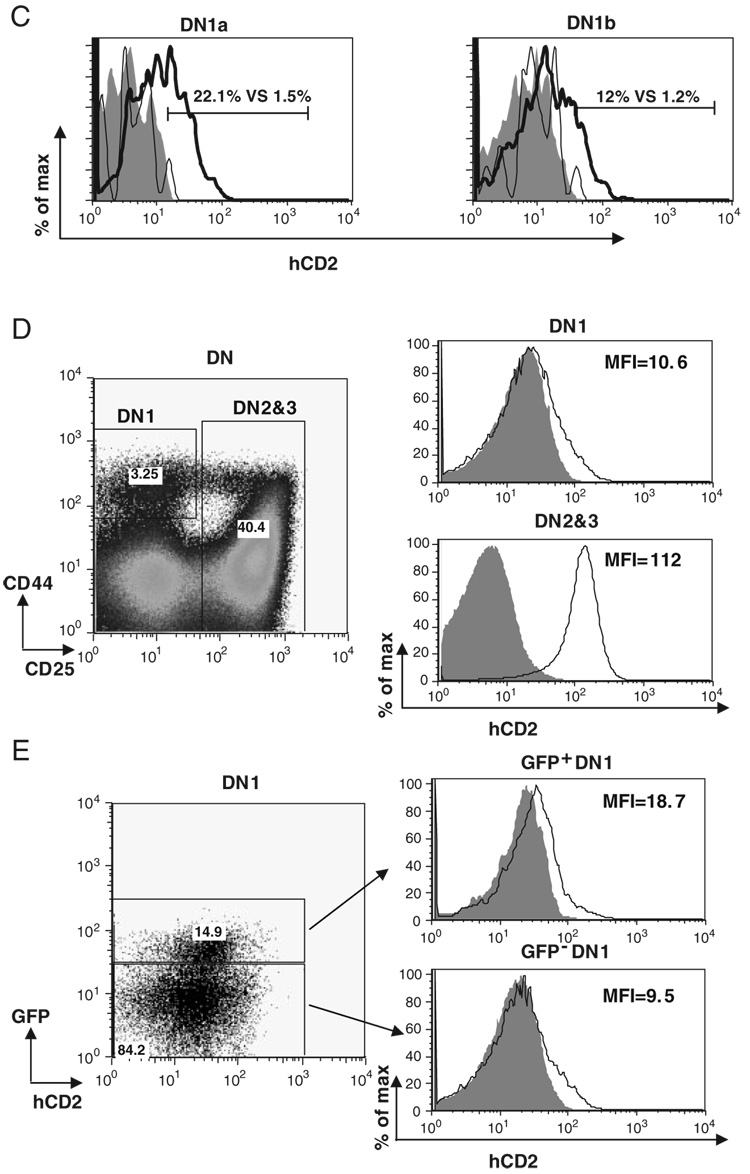
The most immature thymocytes are CD4++CD25− DN1 thymocytes. DN1 cells have been shown to be a heterogeneous population that contains DN1a–DN1e five different subsets [14]. Among those subsets, DN1a and DN1b cells were characterized as canonical T-cell progenitors. Vβ8.2 germline transcription has been shown to be activated in both DN1a and DN1b populations at low levels [19]. However, it remains to be addressed whether E2A is also expressed in these early T-cell progenitors and whether E2A expression in DN1 thymocytes correlates with the activation of Vβ8.2 germline transcription. To answer these questions, we first examined E2A expression in subsets of DN1 thymocytes. For this purpose, E2AGFP knock-in allele, which contains the GFP coding sequence inserted in-frame at the carboxyl end of the E2A-coding sequence, was utilized [26]. The E2A–GFP fusion proteins are functionally equivalent to the WT E2A proteins and E2A expression can be determined in individual cells by the fluorescence intensity of GFP [28]. We found that E2A is predominantly expressed in DN1a and DN1b subsets while other DN1 subsets have very little, if any, E2A expression (Fig. 4A). Consistent with this expression pattern, the distribution of DN1 subsets from E47bm/bm mice is significantly altered in comparison with the WT control. Most strikingly, DN1a and DN1b subsets are dramatically reduced in E47bm/bm DN1 thymocytes, suggesting that E2A expression may play an important role in producing or maintaining these early T cell progenitors (Fig. 4B). We further evaluated Vβ germline transcription using the hCD2 marker among the DN1a and DN1b cells. The percentage of hCD2+ cells was reduced approximately ten fold in comparison with the WT controls (Fig. 4C). This result is consistent with a defect in Vβ germline transcription in T-cell progenitors upon E2A deletion. Next, we examined E2A expression together with hCD2 expression in DN1 thymocytes prepared from E2AGFP/GFP Vβ8.2CD2/CD2 mice. The level of hCD2 expression in DN1 thymocytes is much lower than that in DN2&3 thymocytes (Fig. 4D). However, E2A expressing DN1 cells display a higher level of hCD2 expression than DN1 cells that do not express E2A, suggesting that the activation of Vβ8.2 germline transcription correlates with E2A expression in the earliest stage of thymocyte development (Fig. 4E).
Figure 4. E2A expression in DN1 thymocytes correlates the activation of Vβ8.2 germline transcription.
(A) E2A expression in DN1 thymocytes. Gating of subsets of DN1 thymocytes is indicated in the dot plot. GFP expression in each subset of DN1 fromWT (shaded area) and E2AGFP/GFP (black line) is presented by overlay histograms. Percentage of GFP+ cells is shown in each histogram. (B) Effects of E2A disruption on the distribution of DN1 subsets. Thymocytes from E47+/+ Vβ8.2CD2/+ and E47bm/bm Vβ8.2CD2/+ mice were used in the analysis. Gating of total DN1 thymocytes and subsets of DN1 thymocytes is indicated in the upper and lower dot plot, respectively. Numbers within each gate represent the percentages of each subset in the dot plot. (C) hCD2 expression in DN1a and DN1b cells on E47bm/bm background. hCD2 expression in DN1a and DN1b subsets defined in (B) is shown in overlayed histograms. Numbers (E2AWT vs. E47 mutant) within the histograms represent percentages of hCD2+ cells in total DN1a or DN1b thymocytes from E2A WT (thick line) or E47 bm/bm (thin line) mice. Vβ8.2+/+ mice (shaded area) are used as the negative control for hCD2 expression. (D) Total DN1 thymocytes and total DN2&3 thymocytes from Vβ8.2CD2/CD2 E2AGFP/GFP mice are gated in the dot plot of CD44 and CD25 (left) and analyzed for hCD2 expression in the histogram. Samples from Vβ8.2+/+ E2AGFP/GFP mice (shade) and Vβ8.2CD2/CD2 E2AGFP/GFP mice (line) are shown in each overlay histogram. MFI of hCD2 staining is indicated in each plot. (E) DN1 thymocytes from Vβ8.2CD2/CD2 E2AGFP/GFP mice shown in (D) are further separated into GFP+ and GFP− fractions before the analysis of hCD2 expression. Samples from Vβ8.2+/+ E2AGFP/GFP and Vβ8.2CD2/CD2 E2AGFP/GFP mice are shown as shaded area and solid line, respectively, in the overlay histogram.
A comparison of E2A and HEB in regulating Vβ8.2 germline transcription
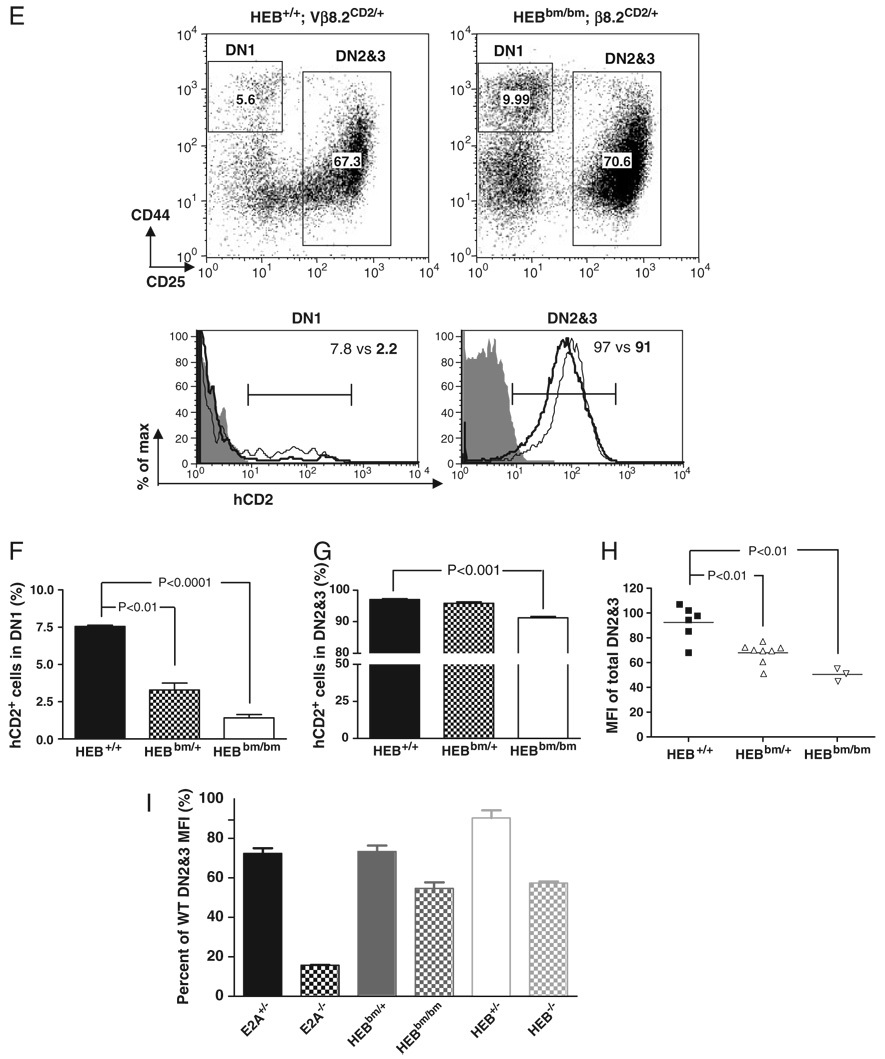
Because HEB is required for thymocyte development as a dimerization partner for E2A, we next investigated the role of HEB in activating Vβ8.2 germline transcription. For this purpose, the Vβ8.2CD2 knock-in allele was introduced onto an HEB-deficient background. Due to the poor survival rate of HEB−/− mice after birth, newborn mice were analyzed. Similar to what was observed in E2A−/− fetal thymocytes, the percentage of hCD2+thymocytes in the DN1 subset also showed a significant decrease in the absence of HEB (Fig. 5A and B). A reduction of hCD2+ cells in HEB−/− DN2&3 thymocytes was also observed although the phenotype was much milder than was observed for E2A−/− DN2&3 thymocytes. More than 80% of HEB−/− DN2&3 cells expressed hCD2+ (Fig. 5C) while only 50–60% of E2A-deficient DN2&3 thymocytes did so. The level of hCD2 expression on HEB−/− DN2&3 thymocytes was reduced by less than 50% compared with HEB+/+ DN2&3 thymocytes (Fig. 5D). Furthermore, unlike E2A−/− DN2&3 thymocytes, which revealed a dosage effect on hCD2 expression, HEB+/− DN2&3 thymocytes expressed hCD2 at levels that were similar to WT DN2&3 thymocytes (Fig. 5C and D).
Figure 5. Effect of HEB mutations on Vβ8.2 gene germline transcription in DN thymocytes.
(A)–(D) hCD2 expression in DN thymocytes from newborn HEB+/+, HEB+/− and HEB−/− mice. Data are presented in the same format as in Fig. 1(A)–(D). Samples in overlay histograms are Vβ8.2+/+ (shaded area), HEB+/+ Vβ8.2CD2/+ (thin line) HEB−/− Vβ8.2CD2/+ (thick line). (E)–(H) hCD2 expression in DN fetal thymocytes of HEB+/+ and HEBbm/bm mice. Data are presented in the same format as in Fig. 1(A)–(D). Samples in overlay histograms are Vβ8.2+/+ (shaded area), HEB+/+ Vβ8.2CD2/+ (thin line), HEBbm/bm Vβ8.2CD2/+ (thick line). (I) A side-by-side comparison of the effect of different E-protein gene mutations on hCD2 expression in DN2&3 thymocytes. The relative MFI of hCD2 staining is represented as the ratio of mutant MFI to WT MFI. Results presented in (A)–(I) are representative of at least three independent experiments.
To specifically evaluate the role of E2A/HEB heterodimers in activating Vβ8.2 germline transcription, we tested hCD2 expression on the HEBbm/bm background in which the mutant HEB proteins form nonfunctional dimers with E2A [23]. The percentage of hCD2+ cells in the DN1 subset was reduced on the HEBbm/bm background to about the same extent as on E2A-deficiency background (Fig. 5E and F). However, the percentage of hCD2+ cells, as well as the expression level of hCD2, was only slightly decreased in HEBbm/bm DN2&3 thymocytes (Fig. 5G and H). Direct comparison of the MFI of hCD2 staining in total DN2&3 thymocytes of E2A−/−, HEB−/− and HEBbm/bm mice revealed that HEB−/− and HEBbm/bm DN2&3 thymocytes retained much higher hCD2 expression than E2A−/− DN2&3 thymocytes (Fig. 5I). Overall these results indicate that while both E2A and HEB are involved in the activation of Vβ8.2 germline transcription in DN thymocytes, E2A homodimers seem to play a greater role than other E-protein dimers.
IL-7 receptor signaling is required for Vβ8.2 germline transcription
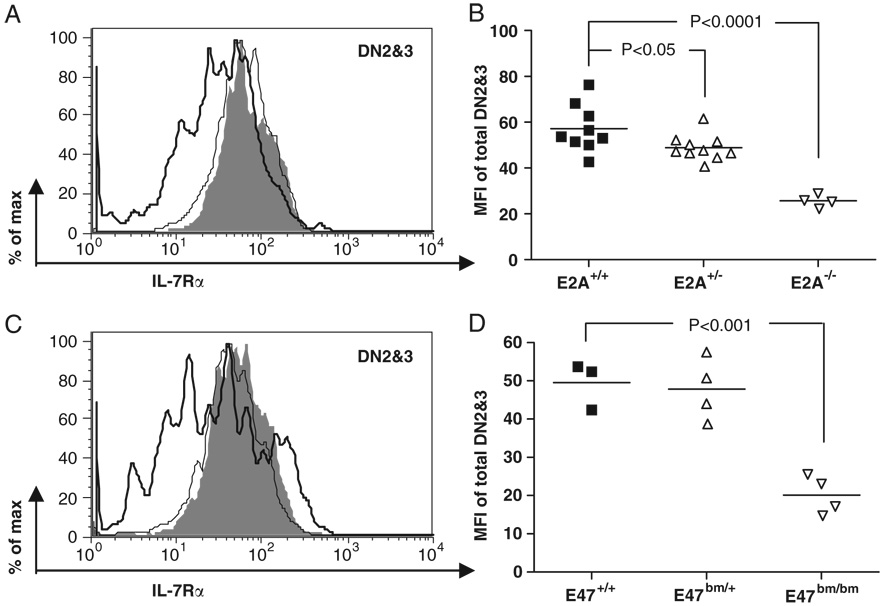
IL-7 receptor (IL-7R) signaling has been implicated to be important for TCR-b gene rearrangement [29, 30]. It has also been suggested that IL-7 receptor α(IL-7Rα) is a downstream target regulated by E2A [31]. In order to further address the relationship between E2A and IL-7R signaling in regulating Vβ8.2 germline transcription, we first examined the level of IL-7Rα expression in E2A-deficient thymocytes by FACS. Levels of IL-7Rα expression are reduced approximately two fold in E2A−/− fetal DN2&3 thymocytes and E47bm/bm adult DN2&3 thymocytes (Fig. 6), suggesting that E2A is involved but not absolutely required for IL-7Rα expression in DN2&3 thymocytes. Next, we investigated the effect of IL-7R signaling on TCR Vβ8.2 germline transcription. For this purpose, the Vβ8.2CD2 knock-in allele was introduced onto an IL-7Rα−/− background. We found that the percentages of hCD2+ thymocytes in the DN1 and DN2&3 subsets were significantly decreased in IL-7Rα−/− mice (Fig. 7A–C). The level of hCD2 expression in IL-7Rα−/− DN2&3 thymocytes was also greatly reduced (Fig. 7D). These results suggest that IL-7 signaling is indispensable for the activation of Vβ8.2 germline transcription. Since common gamma chain (γc) is the essential signal transduction component in the IL-7R complex, we conducted the same analysis in mice. In agreement with what was seen in IL-7Rα−/− mice, the percentages of both hCD2+ cells in DN1 and DN2&3 thymocytes and the level of hCD2 expression in DN2&3 thymocytes were dramatically decreased compared with WT littermates (Fig. 7E–H). Thus, results from both IL-7Rα−/− and mice demonstrated that IL-7 signaling is indispensable for the activation of Vβ8.2 germline transcription at the DN stage of thymocyte development.
Figure 6. Effects of E2A mutations on IL-7Rα expression in DN thymocytes.
(A) A representative overlay histogram shows IL-7Rα expression in DN2&3 thymocytes from E2A+/+ (shaded area), E2A+/− (thin line), E2A−/− (thick line) E17.5 embryos. (B) Scatter plot displays MFI of IL-7Rα expression in DN2&3 thymocytes among multiple litters. (C) A representative overlay histogram shows IL-7Rα expression in DN2&3 thymocytes from E47+/+ (shaded area), E47bm/+ (thin line), E47bm/bm (thick line) adult mice at 4–6wk of age. (D) A scatter dot plot summarizes the MFI of IL-7Rα in DN2&3 cells from multiple litters.
Figure 7. Requirements of IL-7R for Vβ8.2 gene germline transcription.
(A) Representative dot plots of DN thymocytes of 4–6 wk-old IL-7Rα+/+ and IL-7Rα−/− mice. (B) Percentage of hCD2+cells in DN1 fraction shown in (A). (C) Percentage of hCD2+ cells in DN2&3 fraction shown in (A). (D) Scatter plot shows the MFI of hCD2 staining in total DN2&3 thymocytes from IL-7Rα+/+, IL-7Rα+/− and IL-7Rα−/− mice. Each dot represents an individual animal of indicated genotype. (E)–(H) Analysis of hCD2 expression in DN thymocytes of , and mice. Data are presented in the same format as in (A)–(D). hCD2 expression in DN1 and DN2&3 thymocytes isolated from Vβ8.2+/+ (shaded area), Vβ8.2CD2/+ (thin line) or Vβ8.2CD2/+ (thick line) mice is shown in overlay histogram; 6–8 thymi of the same genotype were pulled together for flow cytometry analysis.
Discussion
Our data showed that E proteins are required for the optimal activation of Vβ8.2 germline transcription at the DN1 stage, during which T lineage commitment occurs. Because E proteins, particularly E2A, are required for T lineage commitment [21], it remains a possibility that the reduced number of hCD2+ DN1 cells on E2A or HEB mutant background is due to an impairment of T lineage commitment. In fact, deletion of E2A resulted in a severe reduction of T-cell progenitors defined as the DN1a and DN1b fractions. Analysis of these earliest T-cell progenitors also showed reduced frequency of hCD2 expression. This result is consistent with the idea that E2A is not only important for T lineage commitment but also required for optimal activation of Vβ germline transcription. DN2&3 cells are relatively pure T-cell progenitors and precursors. Our data showed that full activation of Vβ8.2 germline transcription among DN2&3 cells is also dependent on E proteins, especially E2A. Interestingly, the germline transcription of Vβ8.2 becomes independent of E2A as thymocytes differentiate to DP and SP stages. This result correlates with our previous findings that E2A expression is significantly downregulated at the DP stage and is further diminished at SP the stage [26]. It also suggests that Vβ8.2 germline transcription is regulated by different transcription factors in pre versus post-β rearrangement stages.
A comparison between E2A and HEB KO backgrounds indicated that Vβ8.2 germline transcription in DN2&3 thymocytes depends more on E2A than HEB. It has been shown that most E proteins present in the thymus are in the form of E2A and HEB heterodimers. However, E2A and HEB have overlapping but not identical expression patterns during the early phase of thymocyte development. E2A expression can first be detected in a fraction of DN1 thymocytes. High-level E2A expression is maintained throughout DN2 and DN3 stages of thymocyte development [26, 32]. In contrast, HEB expression was not upregulated until the DN3 stage [33]. Therefore, E2A and HEB heterodimers become the major form of E-protein dimers only starting at the DN3 stage. Homodimers of either E2A or HEB can clearly substitute for the function of heterodimers when one partner is missing in the HEB or E2A KO condition, respectively [23]. It is possible that the differential requirement of E2A versus HEB reflects the timing and level of their expression during thymocyte development. At the DN1 and DN2 stages, hCD2 expression from the Vβ8.2CD2 allele is primarily determined by E2A homodimers simply because E2A gene products contribute most of functional E proteins at this early stage of thymocyte development. This interpretation implies that E-protein dimers play a less important role in the maintenance of Vβ8.2 germline transcription at the DN3 stage. This idea is consistent with the fact that hCD2 expression in HEBbm/bm DN3 cells is only slightly affected even though both E2A and HEB functions are effectively blocked by the dominant negative HEBbm allele at the DN3 stage of thymocyte development [23].
E2A and the IL-7 signal have been shown to work independently in driving B-cell development [34]. We showed that both E2A and IL-7 signaling are required for Vβ8.2 germline transcription. We further showed that IL-7Rα expression is only mildly downregulated in E2A-deficient DN2&3 thymocytes. Therefore, the effect of E2A on activating Vβ8.2 germline transcription cannot be explained by this subtle change in IL-7Rα expression. In addition, E2A-deficient DN1 thymocytes, which contain less hCD2+ cells percentage-wise, express comparable level of IL-7Rα as WT DN1 thymocytes (data not shown). Furthermore, γc is expressed at a normal level in both DN1 and DN2&3 thymocytes in the absence of E2A (Supporting Information Fig. 1), suggesting that there is no further perturbation on the expression of overall IL-7R complex. Finally, the adoptive transfer test showed that the effect of E2A KO on Vβ8.2 germline transcription is intrinsic to the T-cell lineage, indicating that E2A does not play a major role in regulating IL-7 production from the stromal cells. Together, these results suggest that E2A-mediated Vβ8.2 germline transcription is independent of the IL-7 signal. Whether the two pathways converge downstream of the IL-7R is still a subject of further investigation.
Sequence analysis revealed five E-box sites in the putative promoter region of Vβ8.2. We have asked whether E2A can directly activate the Vβ8.2 promoter using a LUC reporter assay. However, our study showed that neither E2A nor these E-box sites seem to play significant roles in activating the Vβ8.2 promoter in either fibroblasts or a pro-T-cell line (data not shown). It is highly possible that these in vitro assays are not ideal to recapitulate the physiological condition of DN1 and DN2 thymocytes. We speculate that E2A alone is not sufficient to directly activate Vβ8.2 germline transcription. A very recent publication from Murre’s group [35] showed that E2A does not have a strong affinity to the Vβ8.2 gene in the ChIP assay of DN thymocytes. Nonetheless, E2A seems to play a global role in regulating Vβ germline transcription and chromatine status. In addition to the E-boxes, potential binding sites for other transcription factors such as T-cell factor-1, E26 transformation-specific gene (Ets) and acute myelogenous leukemia gene-1 (AML1/Runx1) are also revealed by the TFSearch program [36]. We evaluated the mRNA expression of these transcription factors in E2A-deficient DN2&3 thymocytes (Supporting Information Fig. 2). T-cell factor-1 and Ikaros are expressed at comparable levels in E47bm/bm DN2&3 thymocytes as in WT DN2&3 thymocytes. The expression of Runx1, Ets-1 and Ets-2 are significantly reduced in E47bm/bm DN2&3 thymocytes, while the expression of GATA3 is increased. It is possible that the Vβ8.2 promoter is regulated by a complex network of transcription factors and changes in expression of multiple transcription factors together lead to the defect in the activation of Vβ8.2 germline transcription in E2A-deficient DN2&3 thymocytes. Alternatively, the disregulated expression pattern of these transcription factors may simply reflect the impaired thymocyte differentiation in the absence of E2A. Further delineation of these possibilities clearly requires genetic studies of individual candidate genes listed here. The genetic approach involving the Vβ8.2CD2 marker described here provides an effective means for further dissection of the regulatory network leading to Vβ locus activation.
Materials and methods
Mice
E2A−/+, E47bm/+, HEB−/+, HEBbm/+ were maintained on a C57B6 background and genotyped as described [22, 23, 26, 37]. IL-7Rα−/− and γc KO (γc−/−) mice were gifts from Dr. Motonari Kondo. TCR Vβ8.2CD2/+ mice were maintained on a 129/C57B6 mixed background and genotyped as described [19]. TCR Vβ8.2CD2/+ mice were crossed to the above mouse strains and progeny were then intercrossed to obtain mice for analysis. C57BL/6 mice congenic for the CD45.1 allotype marker were originally purchased from Jackson Laboratories. All mice were used in accordance with protocols approved by the Duke University Animal Care and Use Committee.
PCR genotyping
Protocol used for the WT and IL-7Rα KO allele genotyping were from Jackson Laboratory (website: http://jaxmice.jax.org/pub-cgi/protocols/protocols.sh?objtype=protocol&protocol_id=249). Genotyping of the WT and γc KO alleles was 800 and 1080 bp fragment, respectively. PCR was conducted by amplifying toe DNA with primers gamma 15/6 F: 5′-GAG TAC CAC AAT GCA AGG GTC-3′, gamma E8 R 5′-CGT GGC AGA ACC GTT CAC TG-3′ and pMC1-neo R 5′-GTA CGT GCT CGC TCG ATG CG-3′.
Flow cytometry
Thymocytes were isolated from E17.5 embryos or 4–6 wk old mice in PBS supplemented with 5% bovine calf serum and were used immediately for FACS analysis. Cell suspensions were stained with a combination of a FITC-conjugated antibody (anti-mouse CD44 or anti-mouse CD8α monoclonal antibody), a PE-conjugated antibody (anti-human CD2 monoclonal antibody) and an APC-conjugated antibody (anti-mouse CD25 or anti-mouse CD4 monoclonal antibody) plus 7-aminoactinomycin D (7AAD; Molecular Probes) and analyzed on an FACS Caliber (Becton Dickinson). Five-color analysis of DN1 subsets were performed on an FACS-Diva (Becton Dickinson). PE-Cy5-conjugated anti-CD3, CD4, CD8, B220, Gr-1 and Mac-1 antibodies were also used together with 7AAD as the dump channel in the analysis of DN thymocytes. Antibodies were purchased from Caltag and eBioscience. For DN2&3 thymocyte purification, total DN thymocytes were enriched by Dynal beads through CD4/CD8 depletion. Enriched DN thymocytes were then subject to sorting by FACS-Diva.
Irradiation and stem cell reconstitution
C57BL/6 mice congenic for the CD45.1 allotype marker were used at age 6–8 wk as host mice. Mice were irradiated with 900 rads 1 day before stem cell transfusion and were maintained thereafter in sterile bedding with antibiotics added in the drinking water. Donor cells were prepared from freshly isolated fetal liver cells or frozen stocks. About 3 × 105 total fetal liver cells were delivered to the host in 0.2mL of PBS through tail vein injection. For each donor type, three to five recipients were used in the test. Mice were sacrificed 6wk after irradiation for flow cytometry analysis.
Quantitative RT-PCR
Total RNA were extracted using TRI Reagent (Sigma) from purified DN2&3 thymoyctes and reverse transcribed into cDNA using M-MLV reverse transcriptase (Invitrogen). mRNA expression of transcription factors was determined by quantitative PCR analysis using Roche LightCycler and a FastStart DNA master SYBR green I kit (Roche), and the data are normalized to GAPDH expression. Serial dilutions of cDNA extracted from RAG2−/− total thymocytes were used as the source of standard curve for each primer set. All results are confirmed by PCR reactions run in separate batches. The primers used for quantitative PCR are listed and described in Supporting Information Table 1.
Software and statistics
Flow cytometry data management and calculations were performed using Flowjo 6.3. Graphs were generated using GraphPad Prism 4.0c. Values are expressed as mean with SEM. Significant differences between two genotypes were calculated using unpaired two-tailed t-tests with 95% confidence interval.
Acknowledgements
The authors thank Dr. Michael Krangel and Mary Elizabeth Jones for critical reading of the manuscript. The authors are grateful to Dr. Motonari Kondo for providing IL-7Rα−/− mice and mice and to Dr. Weiguo Zhang for sharing LAT−/− mice. The work has been supported by grants from the National Institute of Health to Y. Z.
Abbreviations
- DN
double negative
- DP
double positive
- IL-7Rα
IL-7 receptor α
- 7AAD
7-aminoactinomycin D
- SP
single positive
Footnotes
Conflict of interest: The authors declare no potential commercial or financial conflict of interest.
Supporting Information for this article is available at http://www.wiley-vch.de/contents/jc_2040/2008/38144_s.pdf
References
- 1.Whitehurst CE, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the D beta 1 gene segment at the TCR beta locus by a germline promoter. Immunity. 1999;10:313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- 2.Bouvier G, Watrin F, Naspetti M, Verthuy C, Naquet P, Ferrier P. Deletion of the mouse T-cell receptor beta gene enhancer blocks alphabeta T-cell development. Proc. Natl. Acad. Sci. USA. 1996;93:7877–7881. doi: 10.1073/pnas.93.15.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathieu N, Hempel WM, Spicuglia S, Verthuy C, Ferrier P. Chromatin remodeling by the T cell receptor (TCR)-beta gene enhancer during early T cell development: implications for the control of TCR-beta locus recombination. J. Exp. Med. 2000;192:625–636. doi: 10.1084/jem.192.5.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryu CJ, Haines BB, Lee HR, Kang YH, Draganov DD, Lee M, Whitehurst CE, et al. The T-cell receptor beta variable gene promoter is required for efficient V beta rearrangement but not allelic exclusion. Mol. Cell Biol. 2004;24:7015–7023. doi: 10.1128/MCB.24.16.7015-7023.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sikes ML, Meade A, Tripathi R, Krangel MS, Oltz EM. Regulation of V(D)J recombination: a dominant role for promoter positioning in gene segment accessibility. Proc. Natl. Acad. Sci. USA. 2002;99:12309–12314. doi: 10.1073/pnas.182166699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kallenbach S, Babinet C, Pournin S, Cavelier P, Goodhardt M, Rougeon F. The intronic immunoglobulin kappa gene enhancer acts independently on rearrangement and on transcription. Eur. J. Immunol. 1993;23:1917–1921. doi: 10.1002/eji.1830230828. [DOI] [PubMed] [Google Scholar]
- 7.Coquilleau I, Cavelier P, Rougeon F, Goodhardt M. Comparison of mouse and rabbit Ei kappa enhancers indicates that different elements within the enhancer may mediate activation of transcription and recombination. J. Immunol. 2000;164:795–804. doi: 10.4049/jimmunol.164.2.795. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez JD, Anderson SJ, Loh DY. V(D)J recombination and allelic exclusion of a TCR beta-chain minilocus occurs in the absence of a functional promoter. J. Immunol. 1995;155:1191–1202. [PubMed] [Google Scholar]
- 9.Sleckman BP, Gorman JR, Alt FW. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu. Rev. Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 10.Abarrategui I, Krangel MS. Regulation of T cell receptor-alpha gene recombination by transcription. Nat. Immunol. 2006;7:1109–1115. doi: 10.1038/ni1379. [DOI] [PubMed] [Google Scholar]
- 11.Godfrey DI, Zlotnik A. Control points in early T-cell development. Immunol. Today. 1993;14:547–553. doi: 10.1016/0167-5699(93)90186-O. [DOI] [PubMed] [Google Scholar]
- 12.Godfrey DI, Zlotnik A, Suda T. Phenotypic and functional characterization of c-kit expression during intrathymic T cell development. J. Immunol. 1992;149:2281–2285. [PubMed] [Google Scholar]
- 13.Shortman K, Wu L. Early T lymphocyte progenitors. Annu. Rev. Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29. [DOI] [PubMed] [Google Scholar]
- 14.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Godfrey DI, Kennedy J, Mombaerts P, Tonegawa S, Zlotnik A. Onset of TCR-beta gene rearrangement and role of TCR-beta expression during CD3-CD4-CD8- thymocyte differentiation. J. Immunol. 1994;152:4783–4792. [PubMed] [Google Scholar]
- 16.Capone M, Hockett RD, Jr., Zlotnik A. Kinetics of T cell receptor beta, gamma, and delta rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44(+)CD25(+) Pro-T thymocytes. Proc. Natl. Acad. Sci. USA. 1998;95:12522–12527. doi: 10.1073/pnas.95.21.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak F, Tourigny M, Schatz DG, Petrie HT. Characterization of TCR gene rearrangements during adult murine T cell development. J. Immunol. 1999;162:2575–2580. [PubMed] [Google Scholar]
- 18.Tourigny MR, Mazel S, Burtrum DB, Petrie HT. T cell receptor (TCR)-beta gene recombination: dissociation from cell cycle regulation and developmental progression during T cell ontogeny. J. Exp. Med. 1997;185:1549–1556. doi: 10.1084/jem.185.9.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia J, Kondo M, Zhuang Y. Germline transcription from T-cell receptor Vbeta gene is uncoupled from allelic exclusion. Embo J. 2007;26:2387–2399. doi: 10.1038/sj.emboj.7601671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 21.Bain G, Engel I, Robanus Maandag EC, te Riele HP, Voland JR, Sharp LL, Chun J, et al. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol. Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barndt R, Dai MF, Zhuang Y. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during alpha beta thymopoiesis. J. Immunol. 1999;163:3331–3343. [PubMed] [Google Scholar]
- 23.Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol. Cell Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh JK, Romanow WJ, Murre C. Induction of a diverse T cell receptor gamma/delta repertoire by the helix-loop-helix proteins E2A and HEB in nonlymphoid cells. J. Exp. Med. 2001;193:769–776. doi: 10.1084/jem.193.6.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bain G, Romanow WJ, Albers K, Havran WL, Murre C. Positive and negative regulation of V(D)J recombination by the E2A proteins. J. Exp. Med. 1999;189:289–300. doi: 10.1084/jem.189.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan L, Hanrahan J, Li J, Hale LP, Zhuang Y. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J. Immunol. 2002;168:3923–3932. doi: 10.4049/jimmunol.168.8.3923. [DOI] [PubMed] [Google Scholar]
- 27.Jackson AM, Krangel MS. Allele-specific regulation of TCR beta variable gene segment chromatin structure. J. Immunol. 2005;175:5186–5191. doi: 10.4049/jimmunol.175.8.5186. [DOI] [PubMed] [Google Scholar]
- 28.Zhuang Y, Jackson A, Pan L, Shen K, Dai M. Regulation of E2A gene expression in B-lymphocyte development. Mol. Immunol. 2004;40:1165–1177. doi: 10.1016/j.molimm.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 29.Sleasman JW, Harville TO, White GB, George JF, Barrett DJ, Goodenow MM. Arrested rearrangement of TCR V beta genes in thymocytes from children with X-linked severe combined immunodeficiency disease. J. Immunol. 1994;153:442–448. [PubMed] [Google Scholar]
- 30.Muegge K, Vila MP, Durum SK. Interleukin-7: a cofactor for V(D)J rearrangement of the T cell receptor beta gene. Science. 1993;261:93–95. doi: 10.1126/science.7686307. [DOI] [PubMed] [Google Scholar]
- 31.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J. Exp. Med. 2006;203:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 2001;194:733–745. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 34.Kee BL, Bain G, Murre C. IL-7Ralpha and E47: independent pathways required for development of multipotent lymphoid progenitors. EMBO J. 2002;21:103–113. doi: 10.1093/emboj/21.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. Regulation of T cell receptor beta gene rearrangements and allelic exclusion by the helix-loop-helix protein, e47. Immunity. 2007;27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 36.Chen F, Rowen L, Hood L, Rothenberg EV. Differential transcriptional regulation of individual TCR V beta segments before gene rearrangement. J. Immunol. 2001;166:1771–1780. doi: 10.4049/jimmunol.166.3.1771. [DOI] [PubMed] [Google Scholar]
- 37.Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol. Cell Biol. 1997;17:7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]