Summary
Gαq plays a major role in platelet signal transduction but little is known regarding its transcriptional regulation. We have reported that Gαq is upregulated during phorbol 12-myristate 13-acetate (PMA)-induced megakaryocytic transformation of human erythroleukemia (HEL) cells and regulated by EGR-1, an early growth transcription factor. These studies focused on the initial 238 bp of the 5′ upstream region of Gαq gene. In the present studies we characterize a minimal region −1042/−1037 bp from ATG in the 5′ upstream of the Gαq promoter that is associated with PMA responsiveness. In luciferase reporter gene studies in HEL cells, Gαq 5′ upstream promoter sequence −1042/−1 showed a ∼4-fold increased activity in PMA-treated compared to untreated cells. Deletion of 6-nt −1042/−1037 eliminated the difference. Gel-shift studies on Gαq probe (−1042/−1012 bp) revealed binding of EGR-1 with PMA-treated but not untreated nuclear extracts, and this was dependent on the sequence −1042/−1037. Silencing of endogenous EGR-1 inhibited Gαq induction by PMA. MEK/ERK inhibitor U0126 blocked PMA effect on promoter activity of −1042/−1 construct. Conclusions: EGR-1 binding to sequence −1042/−1037 bp in Gαq promoter mediates the induction of Gαq gene by PMA via the MEK/ERK signaling pathway. These studies provide the first evidence of a PMA-responsive element in Gαq promoter , and new insights into regulation of Gαq gene by EGR-1.
Keywords: EGR-1, Gαq, MAP Kinase, Megakaryocytic differentiation, PMA responsive sequence
Introduction
G proteins are heterotrimeric proteins that play a major role in signal transduction from the surface receptors to effector molecules upon platelet activation and regulate downstream responses (1, 2). They mediate interactions between agonist receptors and intracellular enzymes, such as adenylyl cyclase, phospholipase C and phospholipase A2. During signaling the α subunit dissociates from βγ, associated with replacement of GDP by GTP to produce α-GTP, which then activates the effector molecules. On platelet activation by G-protein coupled receptor agonists, such as thrombin, ADP and thromboxane A2, Gαq activates PLC-β2, the most abundant of the platelet PLC-β isozymes (3), leading to formation of inositoltriphosphate and diacylglycerol. Platelets deficient in Gαq have impaired responses to above agonists (4, 5), including in Ca2+ mobilization, aggregation and secretion. In the Gαq deficient patient described by us (5, 6), platelet Gαq protein and mRNA were decreased but the Gαq coding sequence was normal, suggesting a defect in Gαq transcriptional regulation. However, relatively little is known about the mechanisms regulating Gαq gene expression in megakaryocytes/platelets.
We have previously demonstrated (7) that Gαq is upregulated in human erythroleukemia (HEL) cells undergoing megakaryocytic differentiation induced by phorbol 12-myristate 13-acetate (PMA) and that EGR-1, an early growth response transcription factor, regulates Gαq gene. These findings are important because of the major role of Gαq in platelet activation (1, 2) and of EGR-1 as a master-switch coordinating genes in diverse activities, including cell proliferation, differentiation and apoptosis, and in vascular responses to injury and atherogenesis (8-12).
Phorbol esters modulate protein kinase C (PKC) signaling pathways and diverse cellular responses, such as gene transcription, cellular growth and differentiation, and apoptosis in many cells. PMA can substitute for diacylglycerol, the endogenous PKC activator, and has been widely used to study regulation of cell growth and differentiation by growth factors, hormones and cytokines (13-15). It has been extensively used in studying megakaryocyte biology. PMA induces megakaryocytic phenotype in HEL, K562 and other leukemic cell lines (16-18) and is a potent activator of immediate early response genes (18-20). PMA induces EGR-1 expression in HEL and K562 cells, and this is associated with upregulation of megakaryocyte specific CD41a (18).
PMA-responsive elements (PRE) have been described in the promoters of several genes in different cells (21-24), but not Gαq. We have demonstrated an upregulation of Gαq in HEL cells treated with PMA to induce megakaryocytic transformation (7). Our previous studies characterized the proximal 230 bp 5′ regulatory region of Gαq (7). We now provide new evidence that a minimal region −1042/−1037 (from ATG) in Gαq 5′ upstream region is associated with PMA-responsiveness. More importantly, we provide the first evidence that EGR-1 binds to this element in regulating the PMA effect, and that this is mediated via the MAP kinase signaling pathway.
Materials and Methods
Materials and Molecular Techniques
All chemicals were purchased from Fisher Scientific (Pittsburgh, PA) or Sigma Chemical (St. Louis, MO); restriction endonucleases and protease inhibitors were from Roche (Indianapolis, IN). MAP kinase inhibitor U0126 and Dual Luciferase Assay system were from Promega (Madison, MI). Anti-EGR-1 polyclonal antibody was purchased from Active motif (Carlsbad, CA). PCR primers and oligonucleotides were synthesized by Integrated DNA Technologies, IDT (Coralville, IA).
Cell line and Cell culture
The HEL cell line purchased from ATCC (American Type Cell Culture, Rockville, MD) was cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and antibiotics (pencillin, streptomycin and fungizone, 1% each) (Mediatech, Inc. Virginia, VA). During induction, cells were grown in the presence of 10 nM PMA, a concentration shown to induce megakaryocytic transformation of HEL cells (16, 17, 25).
Plasmid construction
Constructs used for transient transfections were generated using pGL3-Basic (Promega) with a promoterless luciferase reporter gene. The full length promoter region (−1116/−1) and its 5′ deletions with a common 3′ end were generated by PCR using gene specific primers synthesized along with restriction sites Xho I at 5′-end and Hind III at 3′ -end shown in Table 1. Amplified products were cloned into appropriate sites of the pGL3-Basic. The promoter plasmid sequences were verified by sequencing on the ABI Prism 377 (Applied Biosystems, Foster City, CA). All sequences shown in this publication relate to human Gαq or EGR-1.
Table 1.
Oligonucleotide sequences used to generate various Gαq promoter constructs.
| Constructs | Primers (5′-3′) | Positiona | Size (bp) |
|---|---|---|---|
| 1116 bp-Luc | ccac ctcgag AGGCCGCCATATTCCTGTCT caga aagctt CTTCCAAAGTGCCT |
−1116/−1097 −15/+9b |
1116 |
| 1092 bp-Luc | aaa ctcgag CGGCCGGACGGCAGCCGCAGGTC | −1092/−1070 | 1092 |
| 1067 bp-Luc | aaa ctcgag CTGCGGCCCTGGCACGCAACGGCGG | −1067/−1043 | 1067 |
| 1056 bp-Luc | aaa ctcgag GGCACGCAACGGCGGCCGCC | −1056/−1037 | 1056 |
| 1042 bp-Luc | aaa ctcgag CCGCCGGCTCGGGCTGCGCGTCGGC | −1042/−1018 | 1042 |
| 1036 bp-Luc | aaa ctcgag GGCTCGGGCTGCGCGTCGGCCG | −1036/−1012 | 1036 |
| 1011 bp-Luc | aaa ctcgag GGTCTGGCCCCGACTTCG | −1011/−994 | 1011 |
| 941 bp-Luc | aaa ctcgag GCAGTAGGCGTCCGCAG | −941/−925 | 941 |
| 851 bp-Luc | aaa ctcgag GGCCGCGAGCCCAGGAAAGC | −851/832 | 851 |
| 731 bp-Luc | cac ctcgag CCCGGATCTGTGCTCCAGTT | −731/−712 | 731 |
| 238 bp-Luc | cac ctcgag CCGCCTCTCTTCTCCCGTCG | −238/−219 | 238 |
position of the forward primer,
The reverse primer used identical for all the constructs.
The numbering shown is from the ATG site.
Appropriate restriction sites (underlined) were introduced at the 5′-end to facilitate cloning.
Constructs 415 bp-Luc was generated by Sac I enzyme digestion of 1042 bp-Luc construct.
Transfections and reporter assays
Transient transfection assays were conducted using Genfect reagent (Molecular Research Laboratories). HEL cells (2 × 106 cells/ml) were co-transfected with promoter-reporter plasmid (5 μg) and a control vector pRL-TK (0.1 μg) containing Renilla luciferase gene (Promega) at a ratio of 50:1. Promoterless reporter plasmid was used as a negative control. After 3-4 hours at 37°C in 5% CO2, cells were transferred to medium supplemented with PMA (10 nM). Co-transfections with antisense oligonucleotides to EGR-1 (5′-GTCTCCACCAGCACCTTCTC-3′) or an unrelated gene (5′-CCTGGGCGCAGTCAATGTGG-3′) were similarly carried out by appropriately adjusting the amount of DNA/oligonucleotide and Genfect transfection reagent. Cells were lysed 24 hours posttransfection and activity was determined using the Dual-Luciferase Assay system. Promoter activity was calculated by dividing the luciferase activity of the experimental constructs by the internal renilla luciferase activity relative to that of the promoterless vector pGL3-Basic. Transfection studies were performed in triplicate and repeated at least three times.
RNA Extraction and Reverse Transcription (RT)-PCR analysis
Total RNA was extracted from HEL cells using the Trizol reagent (Invitrogen, Carlsbad, CA). For RT-PCR analysis, 5 μg of RNA was reverse transcribed using SuperScript II RT cDNA synthesis kit (Invitrogen). The cDNA sysnthesis was primed by an oligo-(dT) at 65°C for 30 min and reverse transcribed at 50°C for 50 min. PCR analysis was done with the following sets of primers for Gαq (forward 5′-ATGACTCTGGAGTCCATCATGG-3′ and reverse 5′-GGGGTATTCGATCCCTGTGG-3′), for EGR-1 (forward 5′- TTCCCTGAGCCACAAAGCCAG -3′and reverse 5′-GGCTGAAGTTGCGCATGCAG - 3′). GAPDH mRNA was amplified as a control. PCR products were resolved on 1% agarose gel with eithidium bromide staining.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear proteins from HEL cells grown in untreated and PMA (10 nM)-treated conditions for 24 h were prepared by the method of Dignam et al (26) and protein was estimated using the Bio-Rad protein assay reagent. Two double stranded oligonucleotide probes, −1042/−1012 and −1036/−1012, were end-labeled with T4 polynucleotide kinase (Promega). A probe with consensus EGR-1 binding sequence (5-CGCCCT CGCCCCCGC GCCGGG-3′) (12) was labeled similarly. EMSA and immunodepletion (ID) analyses were performed as described (7). Reactions were initiated by adding nuclear protein (5 μg) to oligo probes on ice for 30 min before electrophoresis on a native 8% polyacrylamide gel. For competition assays, 50-100-fold excess nonradioactive oligos were added 30 min prior to the addition of radioactive probe and protein complexes. EGR-1 antibody was pre-incubated with nuclear proteins on ice for 30 min before incubation with radioactive probe to perform super-shift analyses. For EGR-1 depletion, nuclear protein was incubated with anti-EGR-1 antibody for 14 h and protein A sepharose beads for 2 h at 4°C. The supernatant was used in the EMSA reaction. Gels were dried and visualized by autoradiography.
MEK inhibition studies
HEL cells were transfected with promoter-reporter constructs and incubated with MEK specific inhibitor U0126 (10 and 20 μM) for 30 min (27). The medium was replaced with fresh medium with or without PMA (10 nM). After 24 h, cells were lysed and reporter gene activity was measured.
Bioinformatics
Potential transcription factor binding sites in Gαq promoter were analyzed using ‘TFSEARCH’ (http://www.cbrc.jp/research/db/TFSEARCH.html).
Results
PMA effect on Gαq promoter activity
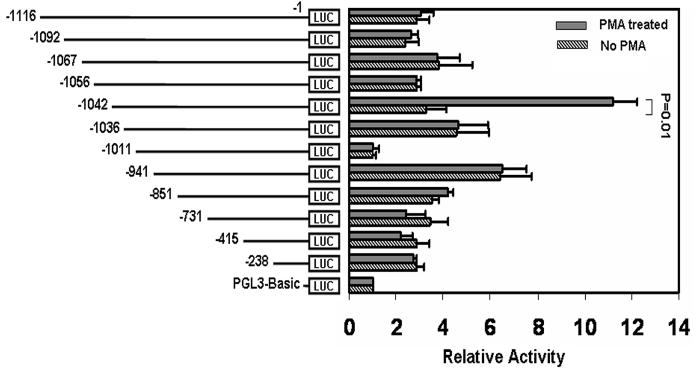
The full-length construct −1116/−1 exhibited equal promoter activity in PMA-treated and untreated HEL cells (Fig. 1). Similar equal activity was obtained following serial 5′ deletions from −1116 to −1056. A 14-nt deletion from −1056 to −1042 showed markedly enhanced activity in PMA-treated but not in untreated cells. The construct −1042/−1 showed a 3-4 fold increase in activity in PMA-treated compared to untreated cells. Further deletion of 6-nt (−1042/−1037) caused marked reduction in reporter activity and the construct −1037/−1 had equal activity in treated and untreated cells. These findings suggested that the sequence −1042/−1037 bp conferred PMA-responsiveness. Deletion of 25-nt between −1036 and −1012 showed a marked loss of activity in both PMA treated and untreated cells suggesting the presence of positive regulator(s) for the gene activity. Deletion of 99-nts between −1011 and −942 showed enhanced activity that was comparable in untreated and treated cells suggesting the presence of negative regulator(s) in this region. Further deletions down to −238 nts showed similar activity under both the conditions. Together, these data revealed that the region −1042/−1012 bp was important in the positive regulation of the Gαq gene; and, more importantly, that the 6 nucleotides −1042/−1037 constituted a PMA responsive element (PRE) (Fig. 1). We studied this region in greater detail.
Figure 1. Mapping of PMA-responsive sequence in Gαq promoter.
Promoter (luciferase) activity of sequential deletions of Gαq 5′ upstream region in PMA (10 nM)- treated and untreated HEL cells. Shown mean±SE values of three experiments in triplicate. p=0.01 relative to the construct −1042/−1-Luc.
EMSA on the region (−1042/−1012)
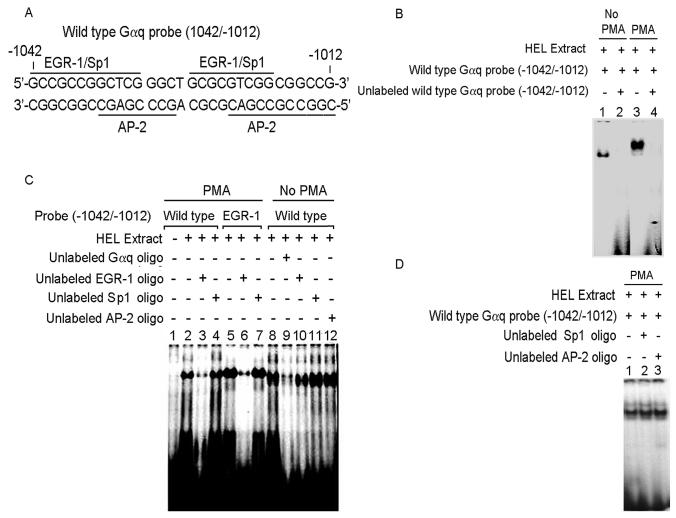
Analyses using TFSEARCH displayed consensus sites for EGR-1 at −1042/−1031 and −1023/−1015 overlapping with consensus sites for Sp1 and AP-2 (Fig. 2A). Gel-shift studies using nuclear extracts from PMA-treated and untreated HEL cells (Fig. 2B) showed protein binding to the probe with both extracts (Lanes 1 and 3) that was competed with excess unlabeled oligo (Lanes 2 and 4). The protein-oligo complex migrated faster with untreated nuclear extracts compared to PMA-treated cells suggesting that the protein binding was different. This was further assessed by competition studies with EGR-1, Sp1 and AP-2 consensus oligos (Fig. 2C). With PMA-treated extracts the protein-binding (Lane 2) was competed with EGR-1 consensus oligo (Lane 3) but not by Sp1 consensus oligo (Lane 4). In control studies, protein binding was observed when PMA-treated extracts were reacted with a labeled EGR-1 consensus oligo (Lane 5) and this was markedly reduced by competition with unlabeled EGR-1 oligo (Lane 6) but not by Sp1 oligo (Lane 7). With untreated extracts, protein binding (Lane 8) was competed by unlabeled identical oligo (Lane 9) but not by oligos with EGR-1, Sp1 or AP-2 consensus sites. Figure 2D shows that the AP-2 oligo did not alter the protein binding observed with PMA-treated extracts. Overall, these experiments suggested that the putative protein bound to the region −1042/−1012 with PMA-treated extracts was EGR-1, and that the unknown protein bound with untreated extracts was displaced by EGR-1 in PMA treated extracts.
Figure 2. Electrophoretic mobility shift assay (EMSA) with Gαq oligonucleotide (−1042/−1012) and nuclear proteins from untreated and PMA-treated HEL cells.
(A) Gαq oligonucleotide sequence showing two predicted EGR-1 sites overlapping with Sp1 and AP-2 sites. (B) EMSA with Gαq oligonucleotide and nuclear extracts from untreated (Lanes 1-2) and PMA-treated (Lanes 3-4) HEL cells. Lanes 1 and 3: protein complex formed between the Gαq oligonucleotide and HEL nuclear extracts. Lanes 2 and 4: competition with excess unlabeled Gαq oligonucleotide indictating the specificity of binding. (C) EMSA with Gαq oligonucleotide with PMA-treated (Lanes 1-7) and untreated (Lanes 8-12) nuclear extracts and competition with EGR-1, Sp1 and AP-2 consensus oligos. Lane 1: labeled oligonucleotide probe alone. Lane 2: binding with PMA-treated nuclear extract. Lane 3: competition with excess unlabeled EGR-1 oligo. Lane 4: no competition with excess unlabeled Sp1 oligo. Lane 5: protein complex formed between labeled EGR-1 oligo and PMA-treated extract. Lane 6: competition with excess unlabeled EGR-1 oligo. Lane 7: no competion with Sp1 oligo. Lane 8: binding with untreated nuclear extract. Lane 9: competition with excess unlabeled oligonucleotide. Lanes 10-12: no competition with EGR-1, Sp1 and AP-2 consensus oligos respectively. (D) Lane 1: binding between Gαq oligonucleotide and PMA-treated nuclear extract. Lanes 2 and 3: no competition with Sp1 and AP-2 consensus oligos.
EGR-1 binding to the oligo −1042/−1012
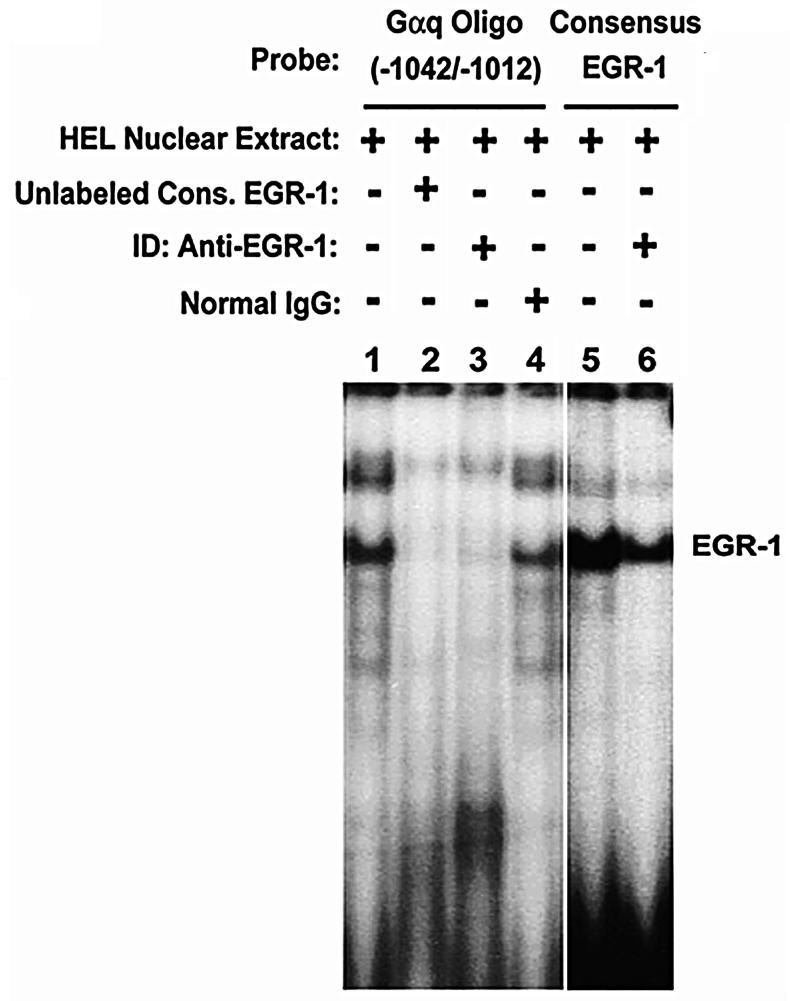
Immunodepletion experiments were performed using anti-EGR-1 polyclonal antibody and PMA-treated extracts (Fig. 3). Protein binding observed with the probe (Lane 1) was competed by excess unlabeled consensus EGR-1 oligo (Lane 2) and abolished by immunodepletion with anti-EGR-1 antibody (Lane 3) but not normal IgG (Lane 4). In control studies, protein binding to labeled EGR-1 consensus oligo (Lane 5) was inhibited by EGR-1 immunodepletion (Lane 6). These findings confirm that EGR-1 binds to the region −1042/−1012 in PMA treated condition. Of note, while two bands were observed in binding studies with the Gαq oligo (lane 1), only one prominent band was noted with consensus EGR-1 oligo (Lane 5). We believe that the loss of or a fainter upper band with the EGR-1 consensus oligo is likely due to the differences in the two probes: the former (Gαq −1042/−1012) is longer (31 bp) than EGR-1 oligo (21) and has a different sequence (except with respect to the EGR-1 consensus motif). It is likely that the additional band with the Gαq probe reflects oligomerization of EGR1 or combination of EGR-1with another protein (9). We have observed this also in earlier studies with a Gαq probe from a different location (7).
Figure 3. EGR-1 binding to Gαq oligonucleotide (−1042/−1012).
EMSA performed with Gαq oligonucleotide (Lanes 1-4) or EGR-1 consensus oligo (Lanes 5-6). Lane 1: protein complex formed between labeled Gαq oligonucleotide and PMA-treated HEL nuclear extract. Lane 2: competition with excess unlabeled EGR-1 consensus oligo. Lane 3: protein binding to Gαq oligonucleotide is inhibited by immunodepletion (ID) of EGR-1. Lane 4: ID with normal IgG. Lane 5: protein complex formed between labeled EGR-1 consensus oligo and PMA-treated HEL extract. Lane 6: reduced protein binding with ID of EGR-1.
Identification of critical sequence in −1042/−1012 oligo for EGR-1 binding
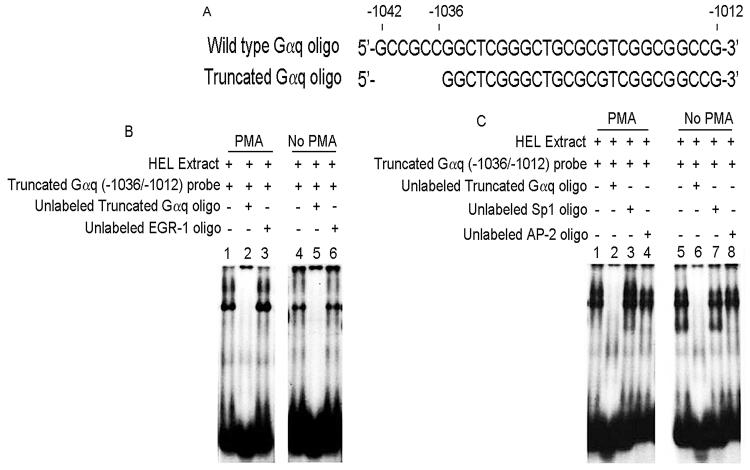
Our promoter studies (Fig. 1) showed loss of PMA-effect on truncation of −1042/−1037. Therefore, a truncated oligo (−1036/−1012) was designed (Fig. 4A) by eliminating the PRE (−1042/−1037). Protein binding was observed (Fig. 4B) with PMA-treated and untreated extracts (Lanes 1 and 4). This was competed by excess unlabeled identical oligo under both conditions (Lanes 2 and 5), but not by consensus EGR-1 oligo (Lanes 3 and 6) indicating that EGR-1 was not involved. This contrasts the findings with the −1042/−1012 probe (Fig. 3) where the binding with PMA-extracts was eliminated by EGR-1 oligo, and is due to EGR binding −1 to the region −1042/−1037. Further, protein binding to this oligo was not competed by Sp1 or AP2 consensus oligos in studies with either PMA-treated or untreated extracts (Fig. 4C). Together, these studies show that EGR-1 binding is lost with the elimination of PRE (−1042/−1037 bp) indicating that these 6 nts are critical for EGR-1 binding.
Figure 4. Effect of truncation of PMA responsive element (PRE) −1042/−1037 bp from Gαq oligonucleotide in EMSA.
(A) Nucleotide sequence of wild type and truncated Gαq oligonucleotides. (B) EMSA using Gαq truncated oligonucleotide and nuclear extracts from PMA-treated (lanes 1-3) and untreated (lanes 4-6) HEL cells. Lanes 1 and 4: binding with the Gαq truncated oligo probe. Lanes 2 and 5: competition with excess unlabeled oligo indicating the specificity. Lane 3 and 6: no competition shown with unlabeled EGR-1 oligonucleotide. (C) EMSA showing competition with excess unlabeled Gαq truncated oligo (lanes 2 and 6), but not with Sp1 and AP-2 oligos.
Downregulation of PMA-induced Gαq Expression by EGR-1 anti-sense oligonucleotide
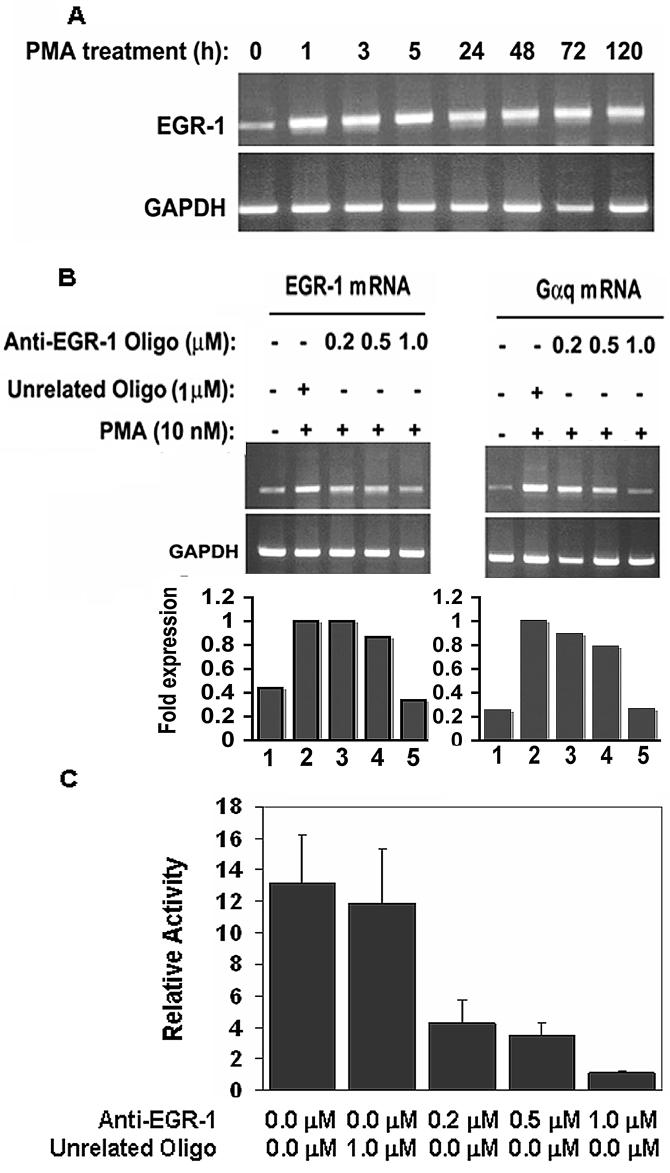
EGR-1 mRNA levels were measured in PMA-treated HEL cells at different time points (0-120 h) (Fig. 5A). At 0 h (no PMA added) EGR-1 mRNA expression was relatively low; it increased with PMA by 1 h and was consistently present thereafter. We have previously shown that Gαq mRNA levels are also induced by PMA and peak at 5 h (7). To establish EGR-1 involvement in Gαq activation, HEL cells were transfected with either control unrelated antisense oligo (1.0 μM) or anti-EGR-1 oligo (0.2 to 1.0 μM) and, subsequently treated with PMA. Gαq mRNA expression increased with PMA (Fig. 5B) and was inhibited by increasing anti-EGR-1 oligo (0.2 to 1.0 μM) associated with the decreased EGR-1 mRNA (Fig. 5B). Effect of EGR-1 oligo on Gαq transcription was further investigated through promoter-reporter studies (Fig. 5C). The reporter construct −1042/−1-Luc was cotransfected with anti-EGR-1 oligo (0.2 to 1.0 μM). Promoter expression was markedly decreased with increasing anti-EGR-1 oligo, but not by a control oligo. These studies indicate that PMA-induction of Gαq promoter is mediated by EGR-1. We have previously shown (7) that silencing of EGR-1 decreases Gαq protein in HEL cells.
Figure 5. Expression of EGR-1 mRNA and GAPDH mRNA in HEL cells in response to PMA (10 nM).
(A) RT-PCR amplification of EGR-1 mRNA. GAPDH was amplified as an internal control. (B) Effect of antisense oligonucleotide to EGR-1 and unrelated oligonucleotide on EGR-1 and Gαq mRNA in PMA-treated HEL cells. The bars represent densitometric analysis. Gαq levels were normalized with GAPDH levels and presented as fold expression relative to levels in the presence of unrelated antisense oligo alone. (C) Effect of anti-EGR-1 oligonucleotide and unrelated antisense oligonucleotide on reporter promoter activity (construct −1042/−1) in HEL cells. Reporter activity was decreased by EGR-1 oligo in a dose dependent manner.
MEK pathway mediates PMA-induced Gαq promoter activity
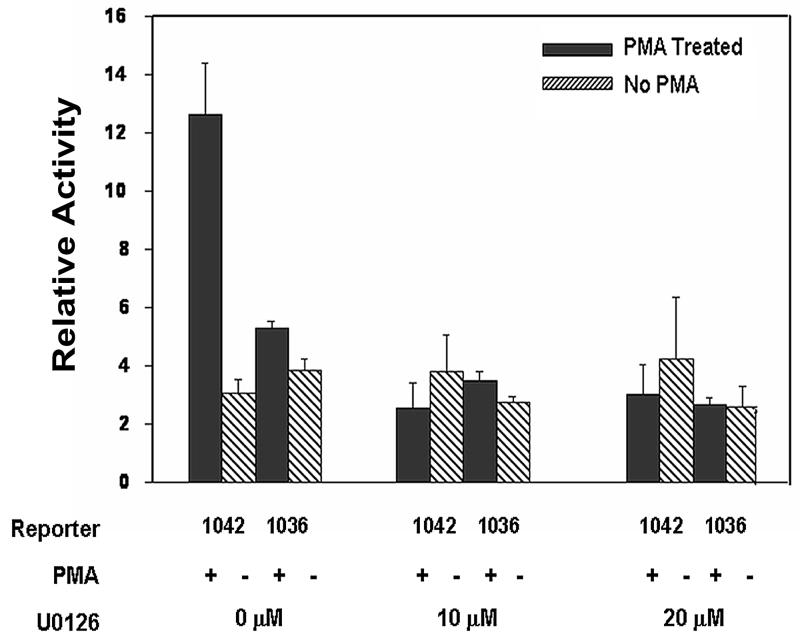
PMA activates downstream signaling pathways, including the MAP kinase pathways (28, 29). To determine the effect of MAP kinase activation on Gαq transcription, PMA-responsive reporter activity was studied in the presence of U0126, a specific inhibitor for MEK1/MEK2, a MAP kinase kinase (MEKK) for extracellular signal-regulated protein kinase (ERK). The promoter reporter construct (−1042/−1) was transiently transfected into PMA-treated HEL cells in the presence and absence of MEK inhibitor U0126 (10 and 20 μM). To determine the specific role of PRE (−1042/−1037), studies were also performed using the truncated construct (−1036/−1). In the absence of the inhibitor, the wild type reporter plasmid (−1042/−1) showed ∼4-fold increased activity in PMA-treated cells compared to untreated cells (Fig. 6). The increased PMA-induced activity was abolished by U0126 indicating the effect of MEK pathway. There was no effect of U0126 on the activity in untreated HEL cells. In studies with the truncated plasmid (−1036/−1) there was no difference in activity in the presence or absence of PMA, and U0126 had no effect in either scenario. Together, these results suggest that MEK pathway is involved in PMA-activation of Gαq promoter and this is mediated via the PRE.
Figure 6.
Effect of MEK inhibitor U0126 on reporter promoter activity of Gαq wild type construct (−1042/−1) and truncated construct (−1036/−1) in PMA-treated and untreated HEL cells. Shown mean±SE (n=3-4).
Discussion
In the present study we characterize a hitherto undescribed PMA-responsive element (−1042/−1037) in Gαq 5′ upstream region and demonstrate that it is regulated by EGR-1. In addition, we provide new evidence that the PMA effect is MAPK-dependent. In reporter studies the construct −1042/−1 showed 3-4 fold increased activity in PMA-treated HEL cells compared to untreated cells and this increase was lost on truncation at −1036 (Fig. 1) suggesting a PMA-responsive element in the region. Gel shift assays revealed binding of EGR-1 to −1042/−1037 with nuclear extracts from PMA-treated cells but not untreated cells. Silencing of EGR-1 downregulated Gαq mRNA (Fig. 5B) as well as Gαq promoter activity (Fig. 5C). Together, these studies provide evidence for a PRE in Gαq promoter (−1042/−1036) and its regulation by EGR-1. PREs have been described in the promoter regions of several genes (21-24) but not of Gαq.
Treatment by phorbol ester is an extracellular stimulus known to activate MAPK signaling pathway, and MAP kinases regulate multiple cellular responses and genes (30-33). MEK/ERK signaling is required for induction of megakaryocytic differentiation of K562 cells by PMA (34, 35). EGR-1 is activated by the ERK1/2 pathway in different cell types (30, 36). In our studies in PMA-treated cells, MEK inhibitor U0126 inhibited promoter activity of the wildtype Gαq construct but had no effect on construct lacking the PRE (Fig. 6). These findings suggest that PMA-induced upregulation of Gαq is mediated via induction of EGR-1 and dependent on MEK.
Gαq gene activation by EGR-1 assumes importance because of the prominent role of EGR-1 in cell cycle progression, migration and proliferation of vascular cells, and atherosclerosis. Egr-1 regulates cyclin D1, a critical component in cell cycle progression (37). Egr-1 and its targeted genes are highly expressed in atherosclerosis (10) and induces the expression of several genes, including PDGF-B, during vascular injury (9, 38). Blocking Egr-1 attenuates arterial neointimal formation following angioplasty (39). Khachigian et al (38) have shown that, in response to inducible stimulus, EGR-1 binds to a cryptic element overlapping a Sp1 site in the PDGF-B promoter and displaces Sp1 to upregulate gene expression. This displacement has been proposed as a common mechanism of inducible gene expression, observed with other PMA-stimulated genes as well, such as PDGF-A, tissue factor, human transforming growth factor-β1, urokinase-type plasminogen activator and 5-lipoxygenase (38, 40, 41). Our studies revealed that EGR-1 regulation of Gαq similarly involves interplay of at least two proteins that differ between untreated and PMA-treated conditions in their binding. The protein bound to the promoter sequence −1042/−1012 in untreated condition is distinct from that in PMA-treated cells (EGR-1) (Fig. 2) and is displaced by EGR-1 in PMA-stimulated state suggesting applicability of this model to Gαq as well. Further studies are required to establish the identity of the protein displaced; our studies suggest that Sp1 may not be the protein (Fig. 2).
We have previously shown two functional EGR-1 binding sites at −202/−187 and −164/−150 bp in PMA-treated cells (7). We now show EGR-1 binding to −1042/−1037 with gene induction. Constructs (−203/−1) with mutations of the proximal EGR-1 sites show loss of PMA responsiveness (Jalagadugula and Rao, unpublished results). Thus, EGR-1 binds at multiple sites in regulating Gαq promoter, as also observed in promoters of other genes, such as tissue factor, 5-lipoxygenase and gonadotropin-releasing hormone (GnRH) (41-43), induced by EGR-1. The number and relative positions of EGR-1 consensus sites are essential determinants of EGR-1-induced gene transcription (9).
In summary, our studies delineate a hitherto undescribed PMA-responsive element (−1042/−1037) in Gαq promoter during megakaryocytic transformation of HEL cells. This element is regulated by EGR-1, an important transcription factor with diverse effects on multiple cellular responses, and through a mechanism involving the MAP kinase pathway. These findings enhance understanding of the regulation of Gαq in megakaryocytes and platelets, about which little is currently known.
Acknowledgements
The authors gratefully acknowledge the assistance of Denise Tierney in manuscript preparation.
This work was supported by grant NIH R01 HL56724 (AKR)
References
- 1.Brass LF, Manning DR, Cichowski K, et al. Signaling through G proteins in platelets: to the integrins and beyond. Thrombosis and haemostasis. 1997;78:581–9. [PubMed] [Google Scholar]
- 2.Offermanns S. Activation of platelet function through G protein-coupled receptors. Circ Res. 2006;99:1293–304. doi: 10.1161/01.RES.0000251742.71301.16. [DOI] [PubMed] [Google Scholar]
- 3.Lee SB, Rao AK, Lee K-H, et al. Decreased expression of phospholipase C-β2 isozyme in human platelets with impaired function. Blood. 1996;88:1684–91. [PubMed] [Google Scholar]
- 4.Offermanns S, Toombs CF, Hu YH, et al. Defective platelet activation in Gαq-deficient mice. Nature. 1997;389:183–6. doi: 10.1038/38284. [DOI] [PubMed] [Google Scholar]
- 5.Gabbeta J, Yang X, Kowalska MA, et al. Platelet signal transduction defect with Gα subunit dysfunction and diminished Gαq in a patient with abnormal platelet responses. Proc Natl Acad Sci U S A. 1997;94:8750–5. doi: 10.1073/pnas.94.16.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabbeta J, Vaidyula VR, Dhanasekaran DN, et al. Human platelet Gαq deficiency is associated with decreased Gαq gene expression in platelets but not neutrophils. Thrombosis and haemostasis. 2002;87:129–33. [PubMed] [Google Scholar]
- 7.Jalagadugula G, Dhanasekaran DN, Kim S, et al. Early growth response transcription factor EGR-1 regulates Galphaq gene in megakaryocytic cells. J Thromb Haemost. 2006;4:2678–86. doi: 10.1111/j.1538-7836.2006.02229.x. [DOI] [PubMed] [Google Scholar]
- 8.Yan SF, Pinsky DJ, Mackman N, et al. Egr-1: is it always immediate and early? J Clin Invest. 2000;105:553–4. doi: 10.1172/JCI9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman ES, Collins T. Pathways of Egr-1-mediated gene transcription in vascular biology. Am J Pathol. 1999;154:665–70. doi: 10.1016/S0002-9440(10)65312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCaffrey TA, Fu C, Du B, et al. High-level expression of Egr-1 and Egr-1-inducible genes in mouse and human atherosclerosis. J Clin Invest. 2000;105:653–62. doi: 10.1172/JCI8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu M, Zhu X, Zhang J, et al. Egr-1 target genes in human endothelial cells identified by microarray analysis. Gene. 2003;315:33–41. doi: 10.1016/s0378-1119(03)00730-3. [DOI] [PubMed] [Google Scholar]
- 12.Cao XM, Koski RA, Gashler A, et al. Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Mol Cell Biol. 1990;10:1931–9. doi: 10.1128/mcb.10.5.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. Faseb J. 1995;9:484–96. [PubMed] [Google Scholar]
- 14.Jaken S. Protein kinase C isozymes and substrates. Curr Opin Cell Biol. 1996;8:168–73. doi: 10.1016/s0955-0674(96)80062-7. [DOI] [PubMed] [Google Scholar]
- 15.Parekh DB, Ziegler W, Parker PJ. Multiple pathways control protein kinase C phosphorylation. Embo J. 2000;19:496–503. doi: 10.1093/emboj/19.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long MW, Heffner CH, Williams JL, et al. Regulation of megakaryocyte phenotype in human erythroleukemia cells. J Clin Invest. 1990;85:1072–84. doi: 10.1172/JCI114538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong Y, Martin JF, Vainchenker W, et al. Inhibition of protein kinase C suppresses megakaryocytic differentiation and stimulates erythroid differentiation in HEL cells. Blood. 1996;87:123–31. [PubMed] [Google Scholar]
- 18.Cheng T, Wang Y, Dai W. Transcription factor egr-1 is involved in phorbol 12-myristate 13-acetate-induced megakaryocytic differentiation of K562 cells. J Biol Chem. 1994;269:30848–53. [PubMed] [Google Scholar]
- 19.Simmons DL, Levy DB, Yannoni Y, et al. Identification of a phorbol ester-repressible vsrc-inducible gene. Proc Natl Acad Sci U S A. 1989;86:1178–82. doi: 10.1073/pnas.86.4.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irving SG, June CH, Zipfel PF, et al. Mitogen-induced genes are subject to multiple pathways of regulation in the initial stages of T-cell activation. Mol Cell Biol. 1989;9:1034–40. doi: 10.1128/mcb.9.3.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angel P, Imagawa M, Chiu R, et al. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–39. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 22.Keen JC, Cianferoni A, Florio G, et al. Characterization of a novel PMA-inducible pathway of interleukin-13 gene expression in T cells. Immunology. 2006;117:29–37. doi: 10.1111/j.1365-2567.2005.02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malakooti J, Sandoval R, Memark VC, et al. Zinc finger transcription factor Egr-1 is involved in stimulation of NHE2 gene expression by phorbol 12-myristate 13-acetate. Am J Physiol Gastrointest Liver Physiol. 2005;289:G653–63. doi: 10.1152/ajpgi.00010.2005. [DOI] [PubMed] [Google Scholar]
- 24.D'Angelo DD, Oliver BG, Davis MG, et al. Novel role for Sp1 in phorbol ester enhancement of human platelet thromboxane receptor gene expression. J Biol Chem. 1996;271:19696–704. doi: 10.1074/jbc.271.33.19696. [DOI] [PubMed] [Google Scholar]
- 25.Cupit LD, Schmidt VA, Gnatenko DV, et al. Expression of protease activated receptor 3 (PAR3) is upregulated by induction of megakaryocyte phenotype in human erythroleukemia (HEL) cells. Exp Hematol. 2004;32:991–9. doi: 10.1016/j.exphem.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Kawai Y, Hanson RW, et al. Sodium butyrate induces transcription from the G alpha(i2) gene promoter through multiple Sp1 sites in the promoter and by activating the MEK-ERK signal transduction pathway. J Biol Chem. 2001;276:25742–52. doi: 10.1074/jbc.M102821200. [DOI] [PubMed] [Google Scholar]
- 28.Ueda Y, Hirai S, Osada S, et al. Protein kinase C activates the MEK-ERK pathway in a manner independent of Ras and dependent on Raf. J Biol Chem. 1996;271:23512–9. doi: 10.1074/jbc.271.38.23512. [DOI] [PubMed] [Google Scholar]
- 29.Schonwasser DC, Marais RM, Marshall CJ, et al. Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol Cell Biol. 1998;18:790–8. doi: 10.1128/mcb.18.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eyries M, Agrapart M, Alonso A, et al. Phorbol ester induction of angiotensin-converting enzyme transcription is mediated by Egr-1 and AP-1 in human endothelial cells via ERK1/2 pathway. Circ Res. 2002;91:899–906. doi: 10.1161/01.res.0000042703.39845.b4. [DOI] [PubMed] [Google Scholar]
- 31.Blenis J. Signal transduction via the MAP kinases: proceed at your own RSK. Proc Natl Acad Sci U S A. 1993;90:5889–92. doi: 10.1073/pnas.90.13.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson G, Robinson F, Beers Gibson T, et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–83. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 33.Schramek H. MAP kinases: from intracellular signals to physiology and disease. News Physiol Sci. 2002;17:62–7. doi: 10.1152/nips.01365.2001. [DOI] [PubMed] [Google Scholar]
- 34.Racke FK, Lewandowska K, Goueli S, et al. Sustained activation of the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway is required for megakaryocytic differentiation of K562 cells. J Biol Chem. 1997;272:23366–70. doi: 10.1074/jbc.272.37.23366. [DOI] [PubMed] [Google Scholar]
- 35.Kim KW, Kim SH, Lee EY, et al. Extracellular signal-regulated kinase/90-KDA ribosomal S6 kinase/nuclear factor-kappa B pathway mediates phorbol 12-myristate 13-acetate-induced megakaryocytic differentiation of K562 cells. J Biol Chem. 2001;276:13186–91. doi: 10.1074/jbc.M008092200. [DOI] [PubMed] [Google Scholar]
- 36.Dieckgraefe BK, Weems DM. Epithelial injury induces egr-1 and fos expression by a pathway involving protein kinase C and ERK. Am J Physiol. 1999;276:G322–30. doi: 10.1152/ajpgi.1999.276.2.G322. [DOI] [PubMed] [Google Scholar]
- 37.Guillemot L, Levy A, Raymondjean M, et al. Angiotensin II-induced transcriptional activation of the cyclin D1 gene is mediated by Egr-1 in CHO-AT(1A) cells. J Biol Chem. 2001;276:39394–403. doi: 10.1074/jbc.M103862200. [DOI] [PubMed] [Google Scholar]
- 38.Khachigian LM, Lindner V, Williams AJ, et al. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–31. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 39.Santiago FS, Lowe HC, Kavurma MM, et al. New DNA enzyme targeting Egr-1 mRNA inhibits vascular smooth muscle proliferation and regrowth after injury. Nat Med. 1999;5:1264–9. doi: 10.1038/15215. [DOI] [PubMed] [Google Scholar]
- 40.Cui MZ, Parry GC, Oeth P, et al. Transcriptional regulation of the tissue factor gene in human epithelial cells is mediated by Sp1 and EGR-1. J Biol Chem. 1996;271:2731–9. doi: 10.1074/jbc.271.5.2731. [DOI] [PubMed] [Google Scholar]
- 41.Silverman ES, Du J, De Sanctis GT, et al. Egr-1 and Sp1 interact functionally with the 5-lipoxygenase promoter and its naturally occurring mutants. Am J Respir Cell Mol Biol. 1998;19:316–23. doi: 10.1165/ajrcmb.19.2.3154. [DOI] [PubMed] [Google Scholar]
- 42.DiVall SA, Radovick S, Wolfe A. Egr-1 binds the GnRH promoter to mediate the increase in gene expression by insulin. Mol Cell Endocrinol. 2007;270:64–72. doi: 10.1016/j.mce.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mechtcheriakova D, Wlachos A, Holzmuller H, et al. Vascular endothelial cell growth factor-induced tissue factor expression in endothelial cells is mediated by EGR-1. Blood. 1999;93:3811–23. [PubMed] [Google Scholar]