Abstract
Normal brain tissue from 28 individuals and 50 glioma samples were analyzed by real-time Quantitative Methylation-Specific PCR (QMSP). Data from this analysis were compared with results obtained on the same samples by MSP. QMSP analysis demonstrated a statistically significant difference in both methylation level (P = .000009 Mann Whitney Test) and frequencies (P = .0000007, Z-test) in tumour samples as compared with normal brain tissues. Although QMSP and MSP showed similar sensitivity, the specificity of QMSP analysis was significantly higher (93%; CI95%: 84%–100%) as compared with MSP (64%; 95%CI: 46%–82%). Our results suggest that QMSP analysis may represent a powerful tool to identify glioma patients that will benefit from alkylating agents chemotherapy.
1. Introduction
Gliomas are the most common form of primary intracranial malignancy. High-grade gliomas like glioblastoma multiforme (GBM, WHO grade IV) are the most frequent lesions encountered with a median survival rate of less than 1 year [1]. Radiotherapy alone or in association with chemotherapeutical agent is the current option for the treatment of gliomas. Recent clinical trials have demonstrated that the association of radiotherapy to treatment with procarbazine, lomustine, and vincristine can improve progress-free survival and treatment with temozolomide even improves overall survival [2–4]. These results have provided some guide to establish optimal treatment choice for gliomas, but their findings have also highlighted the heterogeneity in response to the treatment in the same patient subgroup.
The epigenetic silencing of the O6-methylguanine-DNA-methyltransferase (MGMT) gene by promoter hypermethylation is emerging as a clinically relevant predictor of response to treatment in glioma patients. MGMT promoter hypermethylation can be detected in approximately half of gliomas and it is associated with longer overall survival in patients who receive alkylating chemotherapy in association with radiotherapy [5–7]. Alkylating agents induce cell death by forming cross links between adjacent DNA strands through the alkylation of the O6 position of guanine. Transcriptional active MGMT rapidly removes the alkyl adducts preventing the formation of cross links thereby causing resistance to alkylating drugs [8]. MGMT promoter hypermethylation with consequent loss of MGMT protein expression reduces the DNA repair activity of glioma cells overcoming resistance to alkylating agents [5].
To translate this finding into clinic there is the need for molecular diagnostic tools that can be reliably applied on a large scale of clinical samples. Several methods for assessing promoter methylation status have been reported [9–16], but the most widely used is Methylation-Specific PCR (MSP) analysis after bisulphite treatment [17]. This method however is not quantitative and bears a significant risk of false positive and false negative results [12, 18, 19]. Real-time Quantitative Methylation-Specific PCR (QMSP) has been used to detect promoter hypermethylation of several genes, including MGMT, in primary tumours and bodily fluid from cancer patients [20–30]. We validated a QMSP approach for determining methylation status of the MGMT promoter in 50 gliomas (28 formalin-fixed paraffin-embedded samples and 22 snap-frozen specimens) from 46 patients. Since promoter hypermethylation is often detected also in normal tissues [31, 32], we determined the specificity of the assay in normal brain tissues obtained from autopsies. The performance of QMSP analysis was compared to conventional Methylation-Specific PCR (MSP).
2. Materials and Methods
2.1. Patients and Samples
A total of 50 tumour samples from 46 patients were obtained from the Department of Pathology and stored in accordance with institutional policies. Of those tumours, 28 were formalin-fixed paraffin-embedded (FFPE) samples and 22 snap-frozen specimens. All patients were treated by surgery at the Department of Neurosurgery of the IRCCS “Casa Sollievo della Sofferenza” San Giovanni Rotondo (FG) between 2006 and 2008. After surgery, patients received chemotherapy with the alkylating agent temozolomide at a dose of 75 mg per square meter of body surface area daily during standard fractionated radiotherapy (60 Gy) for 6-7 weeks. The median age of this cohort was 58 years (Interquartile Range, IQR 47–63 years) and median follow up time was 11 months (IQR 4.75–18 months) Pathological diagnosis included 35 (76%) glioblastoma multiforme (20 males and 15 females), 9 (20%) astrocytomas (4 males and 5 females), and 2 (4%) oligodendrogliomas (2 males). Paired primary and recurrent tumour specimens were available for three patients (G3, G8, and G24). As control 28 snap-frozen histologically confirmed normal brain tissues obtained from autopsies were analyzed (Median age 64 years; IQR 53–75 years).
2.2. DNA Extraction and Bisulphite Conversion
Sections, 5-μm-thick, were cut from paraffin blocks or OCT embedded frozen specimens to ensure that tumour samples contained at least 70% cancer cells and to confirm that tissues obtained from autopsies were histological normal. Tumour specimens were then carefully dissected, under a microscope from six to ten 12-μm-thick sections to enrich for areas that contained tumour cells. FFPE samples were subsequently placed in xylene to remove the paraffin as described previously [33]. Tumour and normal tissues were digested with SDS/proteinase K for 24 hours at 48°C, and DNA was extracted with phenol/chloroform [34]. DNA concentration was quantified using the Nanodrop spectrophotometer.
DNA extracted from normal brain or tumour samples was subjected to bisulphite treatment and DNA purification using the Epitect Bisulfite kit (Qiagen Sci, Md, USA) according to manufacturer instruction. Bisulphite-modified DNA from the same treatment was used as template for fluorescence-based real-time quantitative Methylation-Specific PCR (QMSP) and conventional Methylation-Specific PCR (MSP).
2.3. Quantitative Methylation-Specific PCR (QMSP)
Real-time PCR was performed for MGMT using the following primer/probe set [22]: forward 5′-CGAATATACTAAAACAACCCGCG-3′, reverse 5′-GTATTTTTTCGGGAGCGAGGC-3′; probe 5′-FAMAATCCTCGCGATACGCACCGTTTACGTAMRA-3′, yielding a 122 bp amplicon. As reference gene a primer/probe set specific for the unmethylated promoter region of the ACTB gene was used [22]: forward 5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′; reverse 5′-AACCAATAAAACCTACTCCTCCCTTAA-3′; probe 5′-FAMACCACCACCCAACACACAATAACAAACACATA-MRA-3′, yielding an amplicon size of 133 bp Leukocyte DNA from a healthy blood donor was methylated in vitro with excess SssI methyltransferase (New England Biolabs Inc., Beverly, Mass, USA) to generate completely methylated DNA. Serial dilutions (90-0.009 ng) of this DNA were used to construct calibration curves for MGMT and ACTB genes.
Amplification reactions were carried out in triplicate in a volume of 20 μL that contained 50 ng bisulphite-modified DNA, 600 nM forward and reverse primers, 200 nM probe, 0.6 U of Platinum Taq polymerase (Invitrogen, Inc., Rockville, Md, USA), 200 μM each of dATP, dCTP, dGTP, dTTP, and 2 μL of PCR buffer [35]. PCR conditions were as follows: one step at 95°C for 3 minutes, 50 cycles at 95°C for 15 seconds, and 60°C to 62°C for 1 minute. PCR reactions were performed in 96-well plates in a 7900 Sequence detector (Applied Biosystems, Carlsband, Calif, USA) and were analyzed by SDS 2.1.1 software (Applied Biosystems, Carlsband, Calif, USA). Each plate included calibration curves for the ACTB and MGMT genes, patient DNA samples, positive (CpGenome Universal Methylated DNA, Serologicals Corp., Norcross, Ga, USA) and negative (Universal Unmethylated DNA, Serologicals Corp., Norcross, Ga, USA) controls, and multiple water blanks. The relative level of methylated DNA was determined as a ratio of MGMT to ACTB and then multiplied by 1000 for easier tabulation (average value of triplicates of gene of interest/average value of triplicates of ACTB × 1000). For each sample QMSP analysis was repeated on three separate plates and median values were considered for statistical analyses.
2.4. Methylation-Specific PCR (MSP) Analysis
Conventional MSP was carried out as described previously [36]. A PCR reaction for the ACTB gene promoter region not containing CpG was also performed as control of the bisulphite conversion. Forward and reverse primers for the bisulphite-converted methylated sequence of MGMT and ACTB were the same nonfluorogenic oligonucleotides used for QMSP. PCR conditions were as follows: 35 cycles at 95°C for 1 minute, 64°C and 72°C for 1 minute. For each PCR reaction CpGenome Universal Methylated DNA (Serologicals Corp., Norcross, Ga, USA) was used as positive control, and Universal Unmethylated DNA (Serologicals Corp., Norcross, Ga, USA) was used as negative control. PCR products were run on a 3% Agarose gel stained with ethidium bromide and visualized at UV light.
2.5. Statistical Analyses
Multiple comparisons among normal brain tissues, formalin-fixed paraffin-embedded samples (FFPE) and snap-frozen tumour specimens, in terms of MGMT methylation levels, were performed through the nonparametric Kruskall-Wallis and Dunn tests. Results from these analyses allowed also to the application of the Mann-Whitney test. Nonparametric analysis was implemented because results of Kolmogorov-Smirnov and Shapiro tests rejected the Gaussian distribution assumption. The optimal cutoff in terms of sensitivity and specificity for detecting MGMT methylation by Quantitative Methylation-Specific PCR (QMSP) was determined by drawing the ROC curve using methylation levels in normal brain tissues and tumour samples. The area under the ROC curve, computed numerically and tested for statistically significance, was assumed as a measure of goodness of the test. The ROC curves, built separately for the FFPE and snap-frozen tumour group, were compared by testing the difference between the two areas under the ROC curves using the Gaussian approximation. Promoter methylation frequencies determined, in each group, by MSP and QMSP were compared using the Z-test for proportions' difference. To measure the degree of concordance of the results between QMSP and MSP the κ statistic and its 95% confidence interval (95%CI) were calculated. κ values were interpreted as follows: κ = 0.00 no agreement; κ = 0.00–0.20 slight agreement; κ = 0.21–0.40 fair agreement; κ = 0.41–0.60 moderate agreement; κ = 0.61–0.80 substantial agreement; κ = 0.81–1.00 almost perfect agreement [37]. The McNemar test was used to test for the null hypothesis that there was no concordance between the two methods. All probabilities were 2-tailed, and P values <.05 were considered statistically significant. Statistical calculations were performed with the R statistical software package.
3. Results
3.1. Real-Time Quantitative Methylation-Specific PCR (QMSP) is a Sensitive and Specific Method to Determine MGMT Methylation Status in Gliomas
DNA from 50 brain tumours (28 FFPE samples and 22 snap-frozen specimens) and 28 normal brain tissues obtained from autopsies were analyzed by QMSP assay to determine MGMT promoter methylation status. For each sample the analysis was repeated in three separate plates and the median value of the MGMT/ACTB ratio of the three plates was used for statistical analyses. The median MGMT/ACTB ratios for the negative and positive controls used in each PCR reaction (a total of 20 determinations) were as follows: unmethylated DNA median MGMT/ACTB ratio 0 (IQR 0-0) and methylated DNA median MGMT/ACTB ratio 710.64 (IQR 562.80–933.00) (Figure 1).
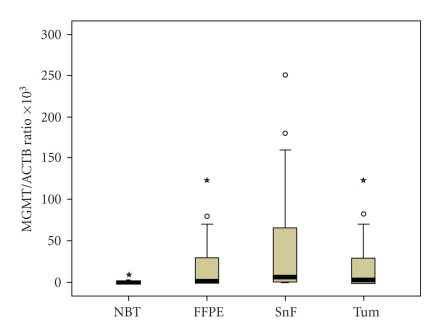
Figure 1.
Box plot for the MGMT/ACTB ratios determined by Quantitative Methylatiom Specific PCR in normal brain tissues and tumour samples. NBT: Normal brain tissues (n = 28); FFPE: Formalin fixed paraffin-embedded samples (n = 28); SnF: Snap-frozen specimens (n = 22); Tum: Total tumour samples (n = 50). The boxes mark the interquartile range, (interval between the 25th and 75th percentile). The lines inside the boxes denote median values. The whiskers represent the interval between the 10th and 90th percentiles. *indicates the extreme cases with more than three boxes length upper or down from the interquartile range.
The median values and Interquartile ranges (IQRs) of MGMT/ACBT ratios for the three type of sample groups were as follows: 0 (IQR 0–0.019) for the normal brain tissue group, 1.16 (IQR 0–34.27) for FFPE, and 5.98 (IQR 0–70.46) for snap-frozen specimens (P < .0007 Kruskall Wallis) (Figure 1). The Dunn test demonstrated statistically significant differences between healthy controls and both FFPE (P < .01) and snap-frozen (P < .01) specimens groups, but no statistically significant difference was found when FFPE and snap-frozen samples groups were compared. The statistically significant difference with the normal brain tissue group was maintained when FFPE and fresh-frozen specimens were considered as one group with a median MGMT/ACTB ratio of 2.48 (IQR 0–50.39) (P < .000009 Mann-Whitney Test).
The MGMT/ACTB ratios in the normal brain tissues and in tumour samples were used to draw an ROC curve (Figure 2(a)), the AUC value of the curve was 0.75 (P < .000001). Based on the ROC curve an optimal cutoff value of 0.35 yielded a 58% (95%CI: 45%–71%) sensitivity and a 93% (95%CI: 84%–100%) specificity. Comparison of ROC curves construed by considering FFPE and snap-frozen specimens as separate groups indicate that the test performs better on frozen specimens (AUC 0.80; P = .0000001) than on FFPE samples (AUC 0.71; P = .001) but this difference was not statistically significant (P = .26) (Figure 2(b)).
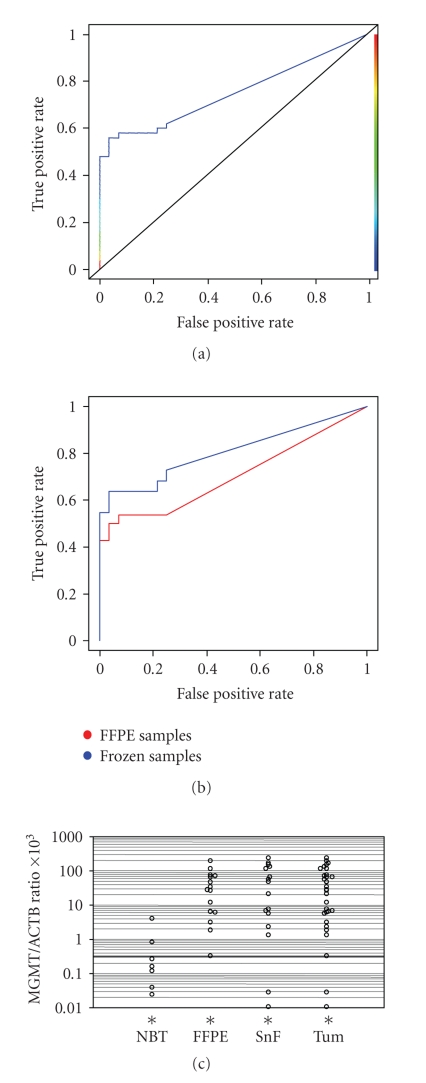
Figure 2.
ROC curve analysis of Quantitative Methylation-specific PCR results. (a) ROC curve for QMSP assay was designed on the basis of MGMT/ACTB ratios in determined in normal brain tissues (n = 28) and total tumour samples (n = 50). The area under the curve (AUC) is 0.74 (P = .000001). (b) Comparison of ROC curves were designed on the basis of MGMT/ACTB ratios determined in normal brain tissues (n = 28) and FFPE (n = 28) or snap-frozen specimens (n = 22). The area under the curve (AUC) is 0.70 (P = .001) for FFPE samples and 0.79 (P = .0000001) for snap-frozen specimens. (c) Scatter plot in logarithm scale of MGMT/ACTB ratios in normal brain tissue (NBT), tumour FFPE samples, and snap-frozen (SnF) specimens. *indicates samples with an MGMT/ACTB ratio of 0 which cannot be plotted on a logarithm scale (n = 21 for NBT, n = 13 for FFPE and n = 6 for SnF). The horizontal line indicates the optimal cutoff value of 0.35.
As shown in Figure 2(c) the majority of tumour samples showed methylation levels well above the cutoff value of 0.35. When compared with the normal brain tissue group (Table 1), methylation frequencies determined by QMSP were significantly different for FFPE (P = .000009, Z-test), snap-frozen specimens (P = .000001, Z-test) as well as for the two groups combined (P = .0000007, Z-test), whereas no statistically significant difference was demonstrated between the FFPE and the snap-frozen groups. In the three patients for which metachrone lesions were available, G3 showed methylation in the primary tumour and in the two recurrences, G8 did not show methylation in either the primary tumour or recurrence, whereas for G24 methylation was detected in the recurrence but not in the primary glioblastoma (Table 2). Methylation levels above the cutoff value of 0.35 were detected in 24 glioblastoma multiforme samples (48% of the tumours analyzed), 3 astrocytomas (6% of the tumour analyzed), and 2 oligodendrogliomas (4% of the tumour analyzed).
Table 1.
MGMT methylation frequencies in normal brain tissues and glioma samples.
| MGMT* status | Normal brain tissues n = 28 | FFPE samples#n = 28 | Frozen samples n = 22 | Total tumour samples n = 50 | ||||
|---|---|---|---|---|---|---|---|---|
| QMSP | MSP | QMSP | MSP | QMSP | MSP | QMSP | MSP | |
| neg | 26 (92.9%) | 18 (64.3%) | 13 (46.4%) | 15 (53.6%) | 8 (36.4%) | 7 (31.8%) | 21 (42.0%) | 22 (44.0%) |
| pos | 2 (7.1%) | 10 (35.7%) | 15 (53.6%) | 13 (46.4%) | 14 (63.6%) | 15 (68.2%) | 29 (58.0%) | 28 (56.0%) |
*neg: Methylation levels for QMSP are below the cutoff point of 0.35, no band is seen in gel electrophoresis for MSP; pos: Methylation levels for QMSP exceed the cutoff point of 0.35, a band is seen in gel electrophoresis for MSP.
#FFPE: Formalin-fixed paraffin-embedded.
Table 2.
MGMT promoter methylation status in primary tumour and recurrences by quantitative methylation-specific PCR (QMSP) and Methylation-Specific PCR (MSP).
| Patient | Hystological* type | Months§ | Sample# | QMSP¶ | QMSP¶ | MSP¶ | |
|---|---|---|---|---|---|---|---|
| Median | IQR | ||||||
| G3 | GB | 0 | FFPE | 28.41 | 13.28–47.82 | pos | pos |
| GB | 28 | FFPE | 12.8 | 11.07–16.00 | pos | pos | |
| GB | 29 | FFPE | 209.37 | 101.70–226.68 | pos | pos | |
| G8 | GB | 0 | FFPE | 0 | 0-0 | neg | neg |
| GB | 10 | FFPE | 0 | 0-0 | neg | neg | |
| G24 | GB | 0 | FFPE | 0 | 0-0 | neg | neg |
| GB | 9 | SF | 5.98 | 4.37–7.60 | pos | pos | |
*GB: Glioblastoma.
§number of months from first surgery.
#FFPE: Formalin fixed paraffin-embedded sample; SF: Snap-frozen pecimen.
¶IQR: Interquartile range; pos: Methylation levels for QMSP exceed the cutoff point of 0.35, a band is seen in gel electrophoresis for MSP; neg: Methylation levels for QMSP are below the cutoff point of 0.35, no band is seen in gel electrophoresis for MSP.
3.2. Determination of MGMT Status by Methylation-Specific PCR (MSP)
Methylation frequencies determined by MSP are shown in Table 1. The overall sensitivity and specificity of the assay were, respectively, 56% (95%CI: 42%–70%) and 64% (95%CI: 46%–82%). A statistically significant difference in methylation frequencies was found between the normal brain tissue group and the two tumour groups combined (P = .04 Z-test). However, when the tumour groups were considered separately, MSP analysis was able to demonstrate statistical significant differences with the normal brain tissue group only for the fresh frozen specimen group (P = .01 Z-test) but not for the FFPE specimen group (P = .21 Z-test) (Table 1).
3.3. QMSP is More Specific Than MSP in Determining MGMT Methylation Status in Gliomas
When promoter methylation frequencies determined by MSP and QMSP were compared, no differences were demonstrated for tumour groups but a statistically significant difference was found in the normal brain tissue group (P = .005 Proportions' Difference Z-Test) (Table 1). The measurement of the degree of agreement between the two technique demonstrated a good concordance in tumour samples considered as a separate groups or combined in one group, whereas the agreement was poor in the normal brain sample group (Table 3).
Table 3.
Comparison of MGMT methylation status as determined by Quantitative Methylation-Specific PCR (QMSP) and. Methylation-Specific PCR (MSP).
| NBT* n = 28 | FFPE* n = 28 | SF* n = 22 | Tumours* n = 50 | Total samples* n = 78 | |||||||
| MSP + | MSP- | MSP + | MSP- | MSP + | MSP- | MSP + | MSP- | MSP + | MSP- | ||
| QMSP | <0.35 | 8 | 18 | 0 | 13 | 3 | 5 | 3 | 18 | 11 | 36 |
| >0.35 | 2 | 0 | 13 | 2 | 12 | 2 | 25 | 4 | 27 | 4 | |
| κ# | 0.243 | 0.860 | 0.495 | 0.715 | 0.613 | ||||||
| 95%CI | 0.084–0.40 | 0.73–0.99 | 0.286–0.704 | 0.59–0.84 | 0.505–0.712 | ||||||
| P | .049 | P < .0001 | P = .00195 | <.0001 | <.0001 | ||||||
*NBT: Normal brain tissue; FFPE: Formalin fixed paraffin embedded specimens, SF: Snap-frozen specimen; MSP +: A band is seen in gel electrophoresis; -: No band is seen in gel electrophoresis.
#κ: Measure of the degree of concordance between MSP and QMSP (see Materials and Methods), 95%CI, Confidence interval.
In the normal brain tissue group, 8 out of the 10 cases positive by MSP displayed MGMT/ACTB ratios below the 0.35 cutoff value by QMSP (Figure 3). In FFPE samples QMSP as compared with MSP detected two additional positive (G17 median 3.35, IQR 2.81–3.89 and G22 median 0.35, IQR 0.24–0.46) (Figure 3). In snap-frozen specimens, three cases were positive at MSP but not at QMSP (G34 median 0.01, IQR 0–0.04; G35 median 0.03, IQR 0–0.18; G42 median 0, IQR 0-0), whereas two cases were negative at MSP and positive at QMSP (G39 median 1.39, IQR 1.30–1.50 and G45 median 2.48, IQR 2.03–2.93) (Figure 3). The analysis of methylation status in the three patients for which primary tumour and recurrences were available showed MSP and QMSP analyses concordant results (Table 2, Figure 3).
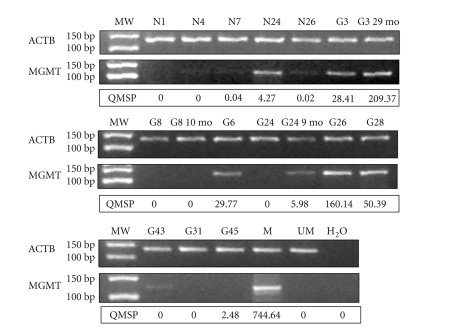
Figure 3.
Comparison of QMSP analysis MSP . Representative results of Methylation-Specific PCR analysis and comparison with Quantitative Methylation-Specific PCR. As control of the bisulphite conversion the upper panels show PCR reactions specific for the ACTB gene promoter region not containing CpG; the middle panels show PCR reactions specific for the methylated sequence of the MGMT promoter; lower panels show results of QMSP. MW: Molecular weight marker. M, positive control (CpGenome Universal Methylated DNA); UM: Negative control (Universal Unmethylated DNA); N: Normal brain tissue; G: Glioma sample. Normal brain tissues N4, N7, N26 and snap-frozen tumour sample G43 show faint bands in the gel electrophoresis but MGMT/ACTB ratios below the cutoff value of 0.35 by QMSP, snap-frozen sample G45 show no methylated band by MSP but an MGMT/ACTB ratio above the cutoff value of 0.35 by QPCR.
4. Discussion
Determination of MGMT promoter methylation status is likely to become a pivotal tool in the management of chemotherapy in glioma patients [7]. Thus there is the need for sensitive and specific diagnostic tools that can be easily implemented into the clinical setting. In this study we applied real-time Quantitative Methylation-Specific PCR (QMSP) analysis to determine MGMT methylation status in a series of paraffin-embedded and fresh frozen specimens from glioma patients. As compared with the real-time-based MSP approach recently described by Vlassenbroeck et al. [30] the specificity of our assay was evaluated by determining methylation status in 28 normal brain tissues obtained from autopsies. Very low levels of methylation were detected in normal brain whereas MGMT/ACTB ratios were significantly higher in tumours. The ROC curve construed by using the MGMT/ACTB ratios in normal brain tissues and tumour samples allowed the definition of an optimal cutoff value of 0.35, which yielded a 93% (95%CI: 84%–100%) specificity and a 58% (95%CI: 45%–71%) sensitivity. As expected the comparison of ROC curves indicate that the test performs better on snap-frozen specimens than on FFPE samples, however this difference was not statistically significant suggesting that QMSP could be a reliable method to determine methylation status in the clinical setting. Although a 58% sensitivity may seem too low for clinical testing it has to be considered that MGMT analysis is a predictive and not a diagnostic tool. MGMT methylation is reported in approximately one half of the glioblastoma samples thus our results are consistent with published data [5, 7] The analysis of our normal and tumour samples by conventional MSP with the same primer set used for QMSP showed a similar sensitivity (56%; 95%CI: 42%–70%) but a much lower specificity (64%; 95%CI: 46%–82%).
While on tumour tissues QMSP and MSP showed a good concordance, results on normal brain tissues indicate a poor agreement. A faint shadow band was observed on gel electrophoresis of MSP analysis in 8 normal brain tissues. In all these instances methylation levels determined by QMSP were negative or well below the cutoff value, suggesting that conventional MSP bears a higher risk of false positive results as compared with QMSP. This leads us to speculate that the three tumour samples showing faint bands on gel electrophoresis by MSP but MGMT/ACTB ratios below the cutoff value are likely to represent false positive results that would wrongly classify the patients as responder to alkylating agents chemotherapy. Four glioma samples were negative by MSP but displayed MGMT/ACTB ratios above (G17, G39, and G45) or equal (G22) to the cutoff value. These data suggest that on larger sample cohorts QMSP analysis could be also more sensitive than MSP in the determination of MGMT methylation status.
5. Conclusions
QMSP combines the advantages of MSP (high sensitivity, applicability to any CpGs) [12] and real-time PCR (rapidity, small quantity of starting DNA, large dynamic range) [26]. Moreover our results indicate that QMSP is more specific than MSP in the determination of MGMT status in gliomas. The robustness of QMSP analysis was confirmed on FFPE samples which makes it possible to investigate MGMT promoter methylation of archival tissues in a cost efficient and accurate way. As compared to other methods for aberrant promoter methylation analysis, QMSP is definitely less labour-intensive and time-consuming than bisulphite sequencing and COBRA analysis and does not require costly dedicated equipments as for Pyrosequencing and MALDI-TOF [15, 16, 38, 39]. In addition QMSP analysis is suitable for large-scale applications and may catalyze the widespread use of MGMT analysis into clinical practice.
Acknowledgments
This work was supported by a grant from the Italian Ministry of Health (RC2009) and from a grant from the Italian Ministry of Health (Progetto Integrato Oncologia, PIO 2006).
Nonstandard Abbreviations
- QMSP:
Quantitative methylation-specific PCR
- MSP:
Methylation-specific PCR
- FFPE:
Formal-fixed paraffin-embedded
- ROC:
Receiver operating characteristic
- AUC:
Area under curve
References
- 1.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathologica. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England Journal of Medicine. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.van den Bent MJ, Carpentier AF, Brandes AA, et al. Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. Journal of Clinical Oncology. 2006;24(18):2715–2722. doi: 10.1200/JCO.2005.04.6078. [DOI] [PubMed] [Google Scholar]
- 4.Cairncross G, Berkey B, Shaw E, et al. Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group trial 9402. Journal of Clinical Oncology. 2006;24(18):2707–2714. doi: 10.1200/JCO.2005.04.3414. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. The New England Journal of Medicine. 2000;343(19):1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 6.Hegi ME, Diserens A-C, Godard S, et al. Clinical trial substantiates the predictive value of O6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clinical Cancer Research. 2004;10(6):1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 7.Hegi ME, Diserens A-C, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. The New England Journal of Medicine. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 8.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nature Reviews Cancer. 2004;4(4):296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 9.Singer-Sam J, LeBon JM, Tanguay RL, Riggs AD. A quantitative HpaII-PCR assay to measure methylation of DNA from a small number of cells. Nucleic Acids Research. 1990;18(3):p. 687. doi: 10.1093/nar/18.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang TH-M, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Human Molecular Genetics. 1999;8(3):459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 12.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.An Q, Liu Y, Gao Y, et al. Detection of p16 hypermethylation in circulating plasma DNA of non-small cell lung cancer patients. Cancer Letters. 2002;188(1-2):109–114. doi: 10.1016/s0304-3835(02)00496-2. [DOI] [PubMed] [Google Scholar]
- 14.Ogino S, Odze RD, Kawasaki T, et al. Correlation of pathologic features with CpG island methylator phenotype (CIMP) by quantitative DNA methylation analysis in colorectal carcinoma. American Journal of Surgical Pathology. 2006;30(9):1175–1183. doi: 10.1097/01.pas.0000213266.84725.d0. [DOI] [PubMed] [Google Scholar]
- 15.Colella S, Shen L, Baggerly KA, Issa J-PJ, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. BioTechniques. 2003;35(1):146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 16.Mikeska T, Bock C, EI-Maarri O, et al. Optimization of quantitative MGMT promoter methylation analysis using Pyrosequencing and combined bisulfite restriction analysis. Journal of Molecular Diagnostics. 2007;9(3):368–381. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cankovic M, Mikkelsen T, Rosenblum ML, Zarbo RJ. A simplified laboratory validated assay for MGMT promoter hypermethylation analysis of glioma specimens from formalin-fixed paraffin-embedded tissue. Laboratory Investigation. 2007;87(4):392–397. doi: 10.1038/labinvest.3700520. [DOI] [PubMed] [Google Scholar]
- 18.Derks S, Lentjes MH, Hellebrekers DM, de Bruïne AP, Herman JG, van Engeland M. Methylation-specific PCR unraveled. Cellular Oncology. 2004;26(5-6):291–299. doi: 10.1155/2004/370301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Research . 1999;59(4):793–797. [PubMed] [Google Scholar]
- 20.Harden SV, Sanderson H, Goodman SN, Partin AAW, Walsh PC, Epstein JI, Sidransky D. Quantitative GSTP1 methylation and the detection of prostate adenocarcinoma in sextant biopsies. Journal of the National Cancer Institute. 2003;95(21):1634–1637. doi: 10.1093/jnci/djg082. [DOI] [PubMed] [Google Scholar]
- 21.Reesink-Peters N, Wisman GBA, Jeronimo C, et al. Detecting cervical cancer by quantitative promoter hypermethylation assay on cervical scrapings: a feasibility study. Molecular Cancer Research. 2004;2(5):289–295. [PubMed] [Google Scholar]
- 22.Hoque MO, Begum S, Topaloglu O, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. Journal of the National Cancer Institute. 2006;98(14):996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 23.Schmiemann V, Böcking A, Kazimirek M, et al. Methylation assay for the diagnosis of lung cancer on bronchial aspirates: a cohort study. Clinical Cancer Research. 2005;11(21):7728–7734. doi: 10.1158/1078-0432.CCR-05-0999. [DOI] [PubMed] [Google Scholar]
- 24.Kahn SL, Ronnett BM, Gravitt PE, Gustafson KS. Quantitative methylation-specific PCR for the detection of aberrant DNA methylation in liquid-based Pap tests. Cancer. 2008;114(1):57–64. doi: 10.1002/cncr.23258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fackler MJ, McVeigh M, Mehrotra J, et al. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Research. 2004;64(13):4442–4452. doi: 10.1158/0008-5472.CAN-03-3341. [DOI] [PubMed] [Google Scholar]
- 26.Lo YMD, Wong IHN, Zhang J, Tein MSC, Ng MHL, Hjelm NM. Quantitative analysis of aberrant p16 methylation using real-time quantitative methylation-specific polymerase chain reaction. Cancer Research. 1999;59(16):3899–3903. [PubMed] [Google Scholar]
- 27.Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Research. 2006;66(6):3338–3344. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 28.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Research. 2000;28(8, article E32):1–8. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeronimo C, Usadel H, Henrique R, et al. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. Journal of the National Cancer Institute. 2001;93(22):1747–1752. doi: 10.1093/jnci/93.22.1747. [DOI] [PubMed] [Google Scholar]
- 30.Vlassenbroeck I, Califice S, Diserens A-C, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. Journal of Molecular Diagnostics. 2008;10(4):332–337. doi: 10.2353/jmoldx.2008.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoque MO, Kim MS, Ostrow KL, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Research. 2008;68(8):2661–2670. doi: 10.1158/0008-5472.CAN-07-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu M, Yao J, Cai L, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nature Genetics. 2005;37(8):899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 33.Parrella P, Xiao Y, Fliss M, et al. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Research. 2001;61(20):7623–7626. [PubMed] [Google Scholar]
- 34.Parrella P, Sidransky D, Merbs SL. Allelotype of posterior uveal melanoma: implications for a bifurcated tumor progression pathway. Cancer Research. 1999;59(13):3032–3037. [PubMed] [Google Scholar]
- 35.van der Riet P, Karp D, Farmer E, et al. Progression of basal cell carcinoma through loss of chromosome 9q and inactivation of a single p53 allele. Cancer Research. 1994;54(1):25–27. [PubMed] [Google Scholar]
- 36.Parrella P, Poeta ML, Gallo AP, et al. Nonrandom distribution of aberrant promoter methylation of cancer-related genes in sporadic breast tumors. Clinical Cancer Research. 2004;10(16):5349–5354. doi: 10.1158/1078-0432.CCR-04-0555. [DOI] [PubMed] [Google Scholar]
- 37.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. [PubMed] [Google Scholar]
- 38.Fraga MF, Esteller M. DNA methylation: a profile of methods and applications. BioTechniques. 2002;33(3):632–649. doi: 10.2144/02333rv01. [DOI] [PubMed] [Google Scholar]
- 39.Ehrich M, Nelson MR, Stanssens P, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]