Introduction
The SANT (Swi3, Ada2, N-Cor, and TFIIIB) module was first described as a putative DNA-binding domain with strong similarity to the helix-turn-helix DNA binding domain of Myb-related proteins.1–3 The X-ray structure of the C-terminal one third portion of the ATPase ISWI of Drospohila melanogaster, containing both SANT and SLIDE (SANT-Like ISWI Domain), confirmed the overall helix-turn-helix structural architecture of SANT as well as SLIDE.4 However, the DNA-contacting residues in Myb are not conserved in SANT and the structurally corresponding residues in the ISWI SANT domain are acidic, and therefore incompatible with DNA interaction. Recent studies suggested that SANT domains might be a histone-tail-binding module,5 including the DNA binding SANT domain of c-Myb.6 Here we present the X-ray structure of Xenopus laevis ISWI SANT domain, derived from limited proteolysis of a C-terminal fragment of ISWI protein.
Materials and Methods
Identification of the SANT-Containing Domain by Proteolysis
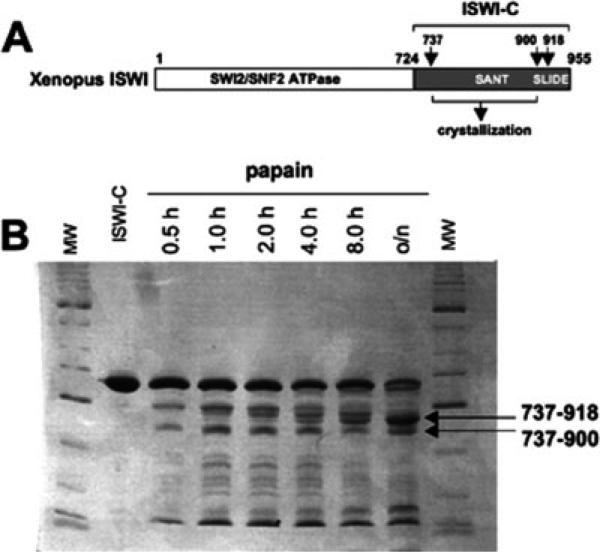
A fragment of Xenopus ISWI corresponding to amino acids 724 through 955 [Fig. 1(A)] was produced in bacteria and purified under denaturing conditions as previously described.7 The purified recombinant protein was renatured by dialysis as follows. First, the protein was dialyzed 1 h versus 1M urea, 2M NaCl, 20 mM Tris HCl, pH 7.9, 1 mM EDTA, 10 mM β-mercaptoethanol. Next, dialysis buffer was changed to 0.5M urea, 2M NaCl, 20 mM Tris HCl, pH 7.9, 1 mM EDTA, 10 mM β-mercaptoethanol, and dialysis was continued for 3 h. Dialysis was performed overnight with buffer containing no urea. Finally, sodium chloride concentration was reduced by dialysis versus 20 mM Tris HCl pH 7.9, 200 mM NaCl, 1 mM EDTA.
Fig. 1.
(A) Schematic representation of Xenopus ISWI. Arrows and residue numbers label proteolytic cleavage sites. (B) A recombinant fragment of Xenopus ISWI-C (amino acids 724−955) was subjected to partial proteolysis using papain protease for various times. The products were resolved by SDS-PAGE.
Renatured Xenopus ISWI fragment (1.25 mg/mL) and papain (2 μg/mL) were incubated on ice for various times in 10 mM Tris HCl, pH 8.0, 10% glycerol, 50 mM NaCl [Fig. 1(B)]. The major fragment resulting from this proteolysis experiment was analyzed by Edman sequencing. Two potential amino termini were assigned corresponding to valine 737 (actual Edman sequence—VSEPKV) and glutamic acid 739 (actual Edman sequence—EPKVPK). Mass spectrometry analysis assigned two potential carboxyl termini corresponding to isoleucine 900 and to leucine 918. In the study described here, we focused our structural analysis on the smaller fragment. The fragment includes the entirety of the classic defined ∼50-residue SANT motif.
Crystallography
Residues 737−900 of Xenopus ISWI protein were fused to the sequence LEHHHHHH at the C-terminus and expressed in a pET expression vector with an N-terminal methionine. Bacterial cultures carrying the expression plasmid were induced with IPTG at 22°C for 16 h. The protein was purified with a 5-mL Ni2+ chelating column (GE-Healthcare), a 6-mL UnoS column (Bio-RAD) and a Superdex S75 column (GE-Healthcare). The protein was concentrated to 60 mg/mL in 20 mM Hepes, 1 mM EDTA, 150 mM NaCl and 5% glycerol. Crystals were obtained by the hanging drop method with 0.1M Hepes pH 6.8, 42.5% PEG 3350, and 0.2M MgCl2 as the mother liquor. The presence of MgCl2 is essential for crystallization.
Crystals were protected by paraffin oil before flash freezing in a cold nitrogen stream. Selenomethionine incorporated crystals did not yield sufficient phasing power as the protein contains only one SeMet at position 884, which is located in the flexible C-terminal helix αG (see below). Molecular replacement was performed using residues 734−845 of the structure of Drosophila melanogaster ISWI (PDB code 1OFC) as the search model. Two solutions were found using CNS8 with Patterson-correlation refinement, treating residues 734−799 and 800−845 of the search model as separate rigid bodies. The structure was refined using CNS (Table I).
TABLE I.
Statistics of X-ray Data Reduction and Refinement
| Resolution range (Å) (highest resolution shell) | 30.0−2.0 (2.07−2.0) |
| Beamline | IMCA-CAT at the APS |
| Crystal-to-detector distance | 200 mm |
| Wavelength | 0.9789 Å |
| Exposure | 30 s per 0.5° rotation |
| Space group | P21 |
| Unit cell | a = 34.61 Å |
| b = 109.50 Å | |
| c = 46.07 Å | |
| β = 94.62° | |
| Measured reflections | 52,726 |
| Unique reflections | 21,300 |
| Redundancy | ∼2.4 |
| <I/s> | 20.4 |
| Completeness (%) | 93.8 (59.7) (note: 96% complete for 30−2.07 Å |
| Rlinear | 0.043 (0.226) |
| R-factora | 0.212 (0.263) |
| R-freeb | 0.274 (0.362) |
| Nonhydrogen atoms | |
| Protein | 1248 |
| Hetrogen | 2 (Mg2+) |
| Water | 154 |
| r.m.s. deviation from ideality | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.2 |
| Dihedral (°) | 19.6 |
| Improper (°) | 0.9 |
| Estimated coordinate error | |
| From Luzzati plot (Å) | 0.24 |
| From σ (Å) | 0.14 |
Rlinear =Σ (I – 〈I〉/Σ〈I〉, where I is the measured intensity of each reflection and 〈I〉 is the intensity averaged from multiple observations of symmetry related reflections.
R-factor = Σ(Fo – Fc)/Σ Fc, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively, with 90% of the data included.
R-free = same as R-factor for 5% of randomly selected data that is not used in structure refinement.
Results and Discussion
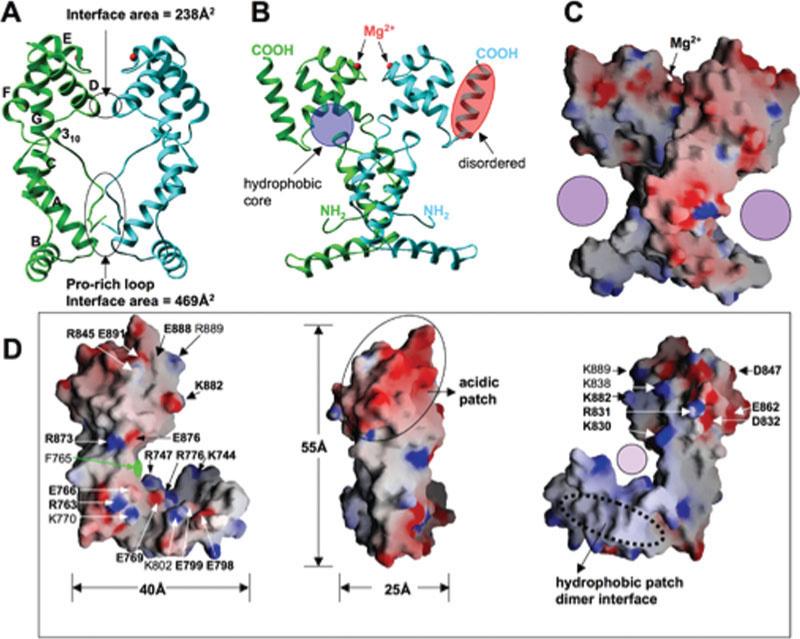
The SANT domain of Xenopus ISWI (residues 737−900 identified by limited proteolysis) was crystallized with two molecules in the crystallographic asymmetric unit [Figs. 2(A–C)]. The overall monomeric structure consists of seven helices (αA to αG) and an N-terminal 3-residue 310 helix, forming a L-shape single domain with dimensions of 55 × 40 × 25 Å3 [Fig. 2(D)]. The N-terminal part of the molecule is largely responsible for the L-appearance: the N-terminal loop with six-prolines forms a nearly 90° bend at the double proline P748-P749. Helix αA and its following loop lie nearly perpendicular to helices αB and αC. The C-terminal part of the molecule is a tightly packed 3-helix bundle (αD, αE, and αF), which corresponds to the classically defined SANT motif spanning some 50 amino acids.10
Fig. 2.
Structure of Xenopus SANT. (A) A dimer of SANT domain formed in the crystallographic asymmetric unit. The dimer interface is formed between the N-terminal Pro-rich loop—hydrophobic interactions involving proline residues (P743, P746, P748, and P749) and helix αA (Tyr775 and Tyr781) - and the helix αD (two salt bridges between invariant Arg803 of one protomer and invariant Asp804 of another protomer). The Stokes radius of the domain was consistent with a solution dimer: the 21-kDa protein eluted from S75 gel filtration column with apparent molecular weight of 41 kDa. The total area of dimer interface of 707 Å2 is within the limits, though at the lower end, of the 32 homodimers examined.9 (B) A 90° rotated view from panel A. A Mg2+ ion, required for crystallization, is bound at the loop region between helices αE and αF. Three main chain carbonyl oxygen atoms of Ala852, Val855, and Lys858 and two water molecules (w) coordinate the metal atom. (C) GRASP surface charge distribution is displayed with blue for positive, red for negative, and white for neutral. Filled circle indicates the opening of the concave L-surface. (D) Three views of the monomer structure of Xenopus SANT domain.
The N- and C-terminal halves of each protomer are packed through a hydrophobic core [Fig. 2(B)], mediated by many aromatic residues from the N-terminal 310 helix (F757, F759, and F760), helices αD (W828, F833, and F836) and αF (Y866, F870, and W871). The importance of these intramolecular interactions is supported by the observation that most residues involved are invariant or conserved among the ISWI SANT sequences examined (Supplemental Figure S1). We suggest that the two halves behave as a single, folded structural module, explaining its resistance to proteolysis [Fig. 1(B)].
Except for the last helix αG, the two protomers in the asymmetric unit are very similar: least squares superposition gave a root-mean-square-deviation of 0.55 Å for 104 pairs of Cα atoms (residues 739−878). While helix αG is ordered in molecule A [green in Fig. 2(B)], which loosely packed against the 3-helix bundle, the corresponding helix is unstructured in molecule B (light blue) with residual, disconnected densities. This flexibility might be due to the missing C-terminal SLIDE domain (see below) or an indication of flexibility.
Structural Comparison of Xenopus ISWI Structure with that of Drosophila
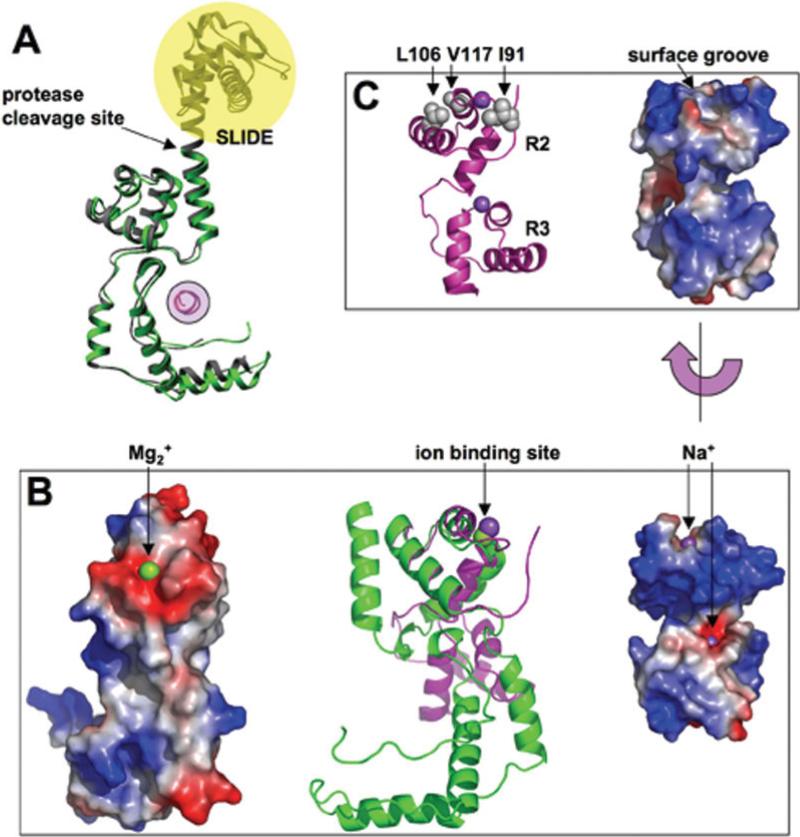
The structured portion of the Drosophila protein contains an additional N-terminal helix [red in Fig. 3(A)] and a C-terminal SLIDE domain.4 The red helix reposes on the concave L-surface. The residues flanking the red helix are disordered in the Drosophila structure. The interaction between the red helix and the rest of the protein is primarily mediated via a nonconserved residue corresponding to F765 of Xenopus ISWI, which complements a small area on the concave L-surface [Fig. 2(D), left panel]. The concave L-surface is highly polarized, containing many invariant or conserved charged residues including solvent exposed R747, R873, and E876 [Fig. 2(D), left panel], suggesting that they are involved in a conserved function and these invariant residues in the Drosophila ISWI do not contact the red-flagged helix.
Fig. 3.
Structural comparisons. (A) Xenopus and Drosophila SANT domains are colored in green and grey, respectively. The additional N-terminal helix in the Drosophila protein is in red, and the C-terminal SLIDE domain is shaded in yellow. The proteolytic cleavage site in the Xenopus protein is indicated. (B) Xenopus SANT domain and cMyb R2R3 are colored in green and magenta, respectively (middle panel). Structural alignment of helices αD, αE, and αF of Xenopus SANT and that of R2 of c-Myb R2R3 (PDB 1GV2) derived the superimposition. The left panel shows the acidic surface patch of Xenopus SANT and the right panel shows the corresponding region of basic DNA binding surface of R2 of c-Myb R2R3. (C) X-ray structure of c-Myb R2R311 with the locations of three hydrophobic residues whose mutation to polar or charged side chains affecting the ability of v-Myb interaction with histone (left panel). A surface groove on the opposite side of the DNA binding might be the binding groove for histone H3 peptide.
In the Drosophila ISWI structure,4 the structurally similar SANT and SLIDE domains are connected by a continuous long helix [Fig. 3(A)]. In our Xenopus structure, the proteolysis-derived fragment was cleaved right in the middle of the helix and the resulting shorter helix αG is ordered in one protomer and disordered in the other [Fig. 2(B)]. This observation indicates that the long αG could become unstructured to allow the individual, structurally similar domains to significantly move with respect to each other, reminiscent of the textbook example of the conformational changes in calmodulin-target interactions.12–15 Calmodulin has two domains, each consisting of a pair of EF-hand motifs joined by a long flexible helix. Upon binding to the C-terminal α-helix of a calmodulin-dependent protein kinase, the long helix linking the two EF-hands unwinds and folds back onto itself, allowing the two halves of calmodulin to clamp down around the target helix.
Structural Comparison with c-Myb
The c-Myb transcriptional factor, a proto-oncogene, has three consecutive N-terminal SANT-type repeats (R1, R2, R3), each repeat containing a 3-helix bundle. The second and third helices of the R2 and R3 form a canonical helix-turn-helix DNA binding motif with the third helix being a recognition helix that is closely packed in the DNA major groove.2 We compared our Xenopus SANT helices (αD, αE, and αF) with that of the X-ray structure of c-Myb R2R3 (PDB 1GV2).11 The three helices of R2 or R3 are superimposed with that of Xenopus within 1.0 Å of root-mean-square deviation for approximately 40 pairs of Cα atoms [Fig. 3(B), middle panel]. While the DNA binding surface in R2 is enriched with basic residues [Fig. 3(B), right panel], the corresponding convex surface on the Xenopus SANT domain is enriched in acidic residues [Fig. 3(B), left panel] including invariant D832, D847, and E862 [Fig. 2(D), middle and right panels], the primary reason that the SANT domain is incompatible with DNA contacts.4
The retroviral oncoprotein v-Myb of avian myeloblastosis virus is an abducted and mutated version of c-Myb, the cellular predecessor. Recently, Mo et al. showed that, in addition to the recognition of cis-regulatory Myb-binding sites on DNA, the cellular c-Myb R2R3, but not its leukemogenic v-Myb cousin, interacts with the N-terminal tail of histone H3.6 The three mutations—affecting the ability of v-Myb to interact with histone—are located in the R2 repeat, replacing hydrophobic side chains I91, L106, and V117 by N, H, and D, respectively. Structural mapping showed that these three critical amino acid substitutions are located on the opposite side of Myb that faces away from the DNA [Fig. 3(C)]. A surface groove in c-Myb is evident that could be the binding groove for histone H3 peptide.
One interesting finding of the comparison is that the position of Mg2+ ion in the SANT domain is equivalent to that of a sodium ion (Na+) in the c-MybR2or R3 structure [Fig. 3(B), middle panel], interacting mainly through main chain atoms. While its importance is currently unclear, other than the essence for the crystallization, we suggest that the ion binding site may be a docking site for the histone H3 N-terminus. The main chain carbonyl oxygen atoms could form a hydrogen bond “cage” that recognizes the amino () terminus of H3, as observed in PHD finger and H3 peptide interactions.16,17
Conclusions
We have described the crystallographic structure of the conserved region of the Xenopus ISWI SANT domain. The majority of the conserved residues are involved in structural stability, formation of a concave L-surface and a convex acidic surface that might provide a binding surface for highly basic histone tails.
Supplementary Material
Acknowledgments
We acknowledge the help of Dr. Lisa J. Keefe in use of the IMCA-CAT beamline (supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Center for Advanced Radiation Sources at the University of Chicago) at the Advanced Photon Source (supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38).
Grant sponsor: National Institutes of Health (NIH); Grant numbers: GM68680, GM49245, and HD01238; Grant sponsor: Intramural Research Program of NIEHS, NIH.
Footnotes
The Supplementary Material referred to in this article can be found at http://www.interscience.wiley.com/jpages/0887-3585/suppmat/
Coordinates have been deposited in the Protein Data Bank (accession code 2NOG).
REFERENCES
- 1.Tahirov TH, Sato K, Ichikawa-Iwata E, Sasaki M, Inoue-Bungo T, Shiina M, Kimura K, Takata S, Fujikawa A, Morii H, Kumasaka T, Yamamoto M, Ishii S, Ogata K. Mechanism of c-Myb-C/EBP beta cooperation from separated sites on a promoter. Cell. 2002;108:57–70. doi: 10.1016/s0092-8674(01)00636-5. [DOI] [PubMed] [Google Scholar]
- 2.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 3.Ogata K, Morikawa S, Nakamura H, Hojo H, Yoshimura S, Zhang R, Aimoto S, Ametani Y, Hirata Z, Sarai A, Ishii S, Nishimura Y. Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb. Nat Struct Biol. 1995;2:309–320. doi: 10.1038/nsb0495-309. [DOI] [PubMed] [Google Scholar]
- 4.Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Muller CW. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12:449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 5.Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 6.Mo X, Kowenz-Leutz E, Laumonnier Y, Xu H, Leutz A. Histone H3 tail positioning and acetylation by the c-Myb but not the v-Myb DNA-binding SANT domain. Genes Dev. 2005;19:2447–2457. doi: 10.1101/gad.355405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guschin D, Geiman TM, Kikyo N, Tremethick DJ, Wolffe AP, Wade PA. Multiple ISWI ATPase complexes from Xenopus laevis. Functional conservation of an ACF/CHRAC homolog. J Biol Chem. 2000;275:35248–35255. doi: 10.1074/jbc.M006041200. [DOI] [PubMed] [Google Scholar]
- 8.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D: Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 9.Jones S, Thornton JM. Principles of protein–protein interactions. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aasland R, Stewart AF, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- 11.Tahirov TH, Morii H, Uedaira H, Sasaki M, Sarai A, Adachi S, Park SY, Kamiya N, Ogata K. Crystal Structure of C-Myb DNA-binding domain: specific Na+ binding and correlation with NMR structure. (PDB 1GV2). In Press. DOI 10.2210/pdb1gv2/pdb.
- 12.Osawa M, Tokumitsu H, Swindells MB, Kurihara H, Orita M, Shibanuma T, Furuya T, Ikura M. A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase. Nat Struct Biol. 1999;6:819–824. doi: 10.1038/12271. [DOI] [PubMed] [Google Scholar]
- 13.Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 14.Meador WE, Means AR, Quiocho FA. Modulation of calmodulin plasticity in molecular recognition on the basis of X-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 15.Yamauchi E, Nakatsu T, Matsubara M, Kato H, Taniguchi H. Crystal structure of a MARCKS peptide containing the calmodulin-binding domain in complex with Ca2+-calmodulin. Nat Struct Biol. 2003;10:226–231. doi: 10.1038/nsb900. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.