Abstract
G- and R-bands of metaphase chromosomes are characterized by profound differences in gene density, CG content, replication timing, and chromatin compaction. The preferential localization of gene-dense, transcriptionally active, and early replicating chromatin in the nuclear interior and of gene-poor, later replicating chromatin at the nuclear envelope has been demonstrated to be evolutionary-conserved in various cell types. Yet, the impact of different local chromatin features on the radial nuclear arrangement of chromatin is still not well understood. In particular, it is not known whether radial chromatin positioning is preferentially shaped by local gene density per se or by other related parameters such as replication timing or transcriptional activity. The interdependence of these distinct chromatin features on the linear deoxyribonucleic acid (DNA) sequence precludes a simple dissection of these parameters with respect to their importance for the reorganization of the linear DNA organization into the distinct radial chromatin arrangements observed in the nuclear space. To analyze this problem, we generated probe sets of pooled bacterial artificial chromosome (BAC) clones from HSA 11, 12, 18, and 19 representing R/G-band-assigned chromatin, segments with different gene density and gene loci with different expression levels. Using multicolor 3D flourescent in situ hybridization (FISH) and 3D image analysis, we determined their localization in the nucleus and their positions within or outside the corresponding chromosome territory (CT). For each BAC data on local gene density within 2- and 10-Mb windows, as well as GC (guanine and cytosine) content, replication timing and expression levels were determined. A correlation analysis of these parameters with nuclear positioning revealed regional gene density as the decisive parameter determining the radial positioning of chromatin in the nucleus in contrast to band assignment, replication timing, and transcriptional activity. We demonstrate a polarized distribution of gene-dense vs gene-poor chromatin within CTs with respect to the nuclear border. Whereas we confirm previous reports that a particular gene-dense and transcriptionally highly active region of about 2 Mb on 11p15.5 often loops out from the territory surface, gene-dense and highly expressed sequences were not generally found preferentially at the CT surface as previously suggested.
Introduction
The enrichment of gene-dense and early replicating chromatin in the nuclear interior and of gene-poor and later replicating chromatin at the nuclear envelope provides an impressive example of higher order organization of chromatin. Such patterns have been found evolutionarily conserved over several hundred millions of years (Alexandrova et al. 2003; Federico et al. 2006; Habermann et al. 2001; Neusser et al. 2007; Postberg et al. 2005; Tanabe et al. 2002) and illustrate that the radial arrangement of chromatin in the interphase nucleus represents a basic principle of nuclear architecture (for review, see Foster and Bridger 2005; Kosak and Groudine 2004; Misteli 2004; Pederson 2004; Zink 2006).
Metaphase chromosomes in contrast show a structured linear organization with segments of high and low gene content and of different replication timing. Some chromosomes carry in their most gene-dense segments clustered regions of increased gene expression (RIDGEs), which are highly expressed in a wide range of cell types (Caron et al. 2001; Versteeg et al. 2003). Segmental differences of chromatin along a chromosome are reflected by consistent chromosome specific banding patterns such as G-dark (G-) and G-light (R-) bands. Compared to G-bands, R-bands have been described as more gene-dense (containing most of the ubiquitously expressed genes), enriched in GC (guanine and cytosine) content and short interspersed element (SINE) sequences (e.g. Alu) and earlier replicating (for review, see Craig and Bickmore 1993; Furey and Haussler 2003; Woodfine et al. 2004). For a subset of R-bands a more open chromatin fiber conformation was described (Gilbert et al. 2004). The underlying mechanisms of the staining patterns yielding G-bands and R-bands are not fully disclosed, but reflect differences in deoxyribonucleic acid (DNA) base composition, chromatin folding, and compactness (Craig and Bickmore 1993; Furey and Haussler 2003; Holmquist et al. 1982; Saitoh and Laemmli 1994). There is still a lack of comprehensive data on how the distinct segments of metaphase chromosomes are folded into variably shaped chromosome territories (CTs). Furthermore, the extent to which chromatin folding and gene positioning within the nuclear and/or CT space are causally related to a given pattern of gene expression has remained a matter of controversial discussions (Bartova and Kozubek 2006; Kosak and Groudine 2004; Lanctot et al. 2007; Parada et al. 2004).
Pooled BAC clones comprising DNA sequences with defined characteristics provide an excellent tool for 3D flourescent in situ hybridization (FISH) experiments to study links between chromatin organization at the sub-CT level and the global spatial organization of chromatin in the interphase nucleus. Relevant data bases provide detailed information on these distinct chromatin features for any BAC clone and its immediate environment. Comprehensively defined clones provide a source for the generation of BAC pools that can be flexibly composed according to the addressed chromatin features. A BAC clone located within a gene-dense region of a G-dark band and containing a transcribed gene in a given cell type could contribute to a BAC pool selected either for gene-dense segments, for G-band assignment, or for highly expressed genes. This approach allows the topological analysis of DNA within different genomic contexts and opens a way to define chromatin determinants for the global nuclear architecture and for the folding behavior on the subchromosomal level.
In this paper we established BAC pools from human chromosomes (HSA) 11, 12, 18, and 19 with respect to chromosome band assignment, regional gene density, and transcriptional activity. These chromosomes were chosen because of their different properties: HSA 19 has the highest gene density in the human genome (25 genes/Mb), is enriched in Alu sequences, contains only weakly stained G-bands and displays an overall high expression level and early replication timing (Caron et al. 2001; Grimwood et al. 2004). Genes are fairly evenly distributed along the chromosome except for the heteromorphic region 19p12 to 19q12 with <10 genes/Mb that flanks the immediate pericentromeric region over approximately 6 Mb. The exact gene density within this region is not yet available as the repetitive sequence blocks impede complete sequencing (see http://www.ensembl.org/Homo_sapiens). By contrast, HSA 18 is an autosome with an overall low gene density (mean gene content 5.3 genes/Mb) and low expression level. It is proportioned in approximately equal parts of R-bands and G-bands, most of them with high staining intensity (Francke 1994; Nusbaum et al. 2005). HSA 11 and 12 represent chromosomes of intermediate gene density (10 and 13 genes/Mb, respectively) and are comparable to HSA 18 with regard to proportion and staining appearance of R- and G-bands. In contrast to HSA 18, however, the regional gene densities along HSA 11 and 12 differ remarkably within size windows of several megabases. The terminal band 11p15.5 contains a segment of outstanding high gene density (~30 genes/Mb) and ubiquitously increased gene expression (Caron et al. 2001). An overview is provided in Table 1 and more detailed information on gene density along the chromosomes (http://www.ncbi.nlm.nih.gov/genome) in the supplemental Fig. S1.
Table 1.
Overview on common features of chromosomes 12, 18, and 19
| Entire chromosome |
R-bands | G-bands |
|||||
|---|---|---|---|---|---|---|---|
| (G total) | G1 | G2 | G3 | ||||
| Chromosome 12 | |||||||
| Total length (percentage) |
127 Mb (100%) |
56.9 Mb (45%) |
69.3 Mb (55%) |
7.2 Mb (6%) |
33.8 Mb (26%) |
28.3 Mb (23%) |
|
| Mean content of genes /Mb | 10.6/Mb | 12.5/Mb | 7.6/Mb | 21.2/Mb | 7.0/Mb | 4.6/Mb | |
| Mean replication timing | 1.54 | 1.59 | 1.49 | 1.67 | 1.50 | 1.43 | |
| Chromosome 18 | |||||||
| Total length (percentage) |
74.3 Mb (100%) |
37.5 Mb (50.5%) |
36.8 Mb (49.5%) |
6.8 Mb (9.1%) |
17.1 Mb (23%) |
12.9 Mb (17.4%) |
|
| Mean content of genes /Mb | 5.4/Mb | 6.5/Mb | 4.0/Mb | 5.4/Mb | 4.4/Mb | 3.6/Mb | |
| Mean replication timing | 1.47 | 1.49 | 1.39 | 1.44 | 1.42 | 1.32 | |
| Chromosome 19 | Hetero-morphic region 19p12–q12 |
||||||
| Total length (percentage) |
60 Mb (100%) |
23.5 Mb (39%) |
22.5 Mb (37.5%) |
22.5 Mb (37.5%) |
– | – | 14 Mb (23.5%) |
| Mean content of genes /Mb | 27.5/Mb | 30.4/Mb | 38.5/Mb | 35/Mb | – | – | ?/Mb (Incomplete data) |
| Mean replication timing | 1.59 | 1.64 | 1.61 | 1.61 | – | – | 1.37 |
Band composition, gene density, and replication timing of chromosomes 12, 18, and 19. The total length (Mb) of a chromosome does not include pericentromeric regions. Values for the replication timing range from 1 (latest replication) to 2 (earliest replication; compare Woodfine et al. 2004). Classification of G-bands (G1–G3) according to different staining intensity (modified from Francke 1994).
Using multicolor 3D-FISH and 3D image analysis, we determined the nuclear positioning of probe sets generated from the respective pooled BAC clones representing either R- or G-band-assigned chromatin, segments with different gene density, or gene loci with different expression levels. Plotting the mean distance to the nuclear center of each sequence pool against its mean gene density, GC content, replication timing, and expression level allowed for a direct weighting of different local chromatin features and revealed regional gene density as the decisive parameter determining the radial nuclear positioning of chromatin. Finally, the positioning of chromatin delineated by the differently assigned BAC pools was assessed with regard to their respective core CTs. Whereas the particular gene-dense and transcriptionally active region of about 2 Mb on 11p15.5 often looped out from the territory surface, gene-dense and/or highly expressed sequences from the other chromosomal regions were not generally found preferentially at the CT surface.
Material and methods
Cell material and slide preparation
Primary human diploid fibroblasts with a normal female karyotype, obtained from early passages from a skin biopsy, were grown in Dulbecco's modified Eagle's medium (DMEM)/10% fetal calf serum (FCS). Human lymphocytes (G0 cells) were obtained by a Ficoll® gradient from the peripheral blood of a healthy donor and directly placed on a polylysine coated slide as previously described in detail in (Cremer et al. 2006b). PHA-stimulated lymphocytes from fresh peripheral blood were grown for 65 h in RPMI/10% FCS before placing them on a polylysine-coated slide. For the preparation of three-dimensionally (3D) preserved nuclei lymphocytes were fixed in 4% paraformaldehyde/0.3×phosphate-buffered saline (PBS), the fibroblasts in 4% paraformaldehyde/1×PBS. Permeabilization steps for all cell cultures included treatment in 0.5% Triton X-100 in PBS, 20% glycerol in PBS, repeated freezing/thawing in liquid nitrogen, incubation in 0.1 M HCl, pepsin treatment (2 mg/ml pepsin in 0.01 N HCl at 37° for 7–10 min) and equilibration in 50% formamide/2×SSC (pH 7.0). For the identification of S-phase, cells were pulse-labeled with BrdU (5 μM final concentration) 1 h before fixation.
Preparation of complex DNA probes and FISH
The painting probes delineating human chromosomes 12, 18, and 19, originally generated from sorted chromosomes were amplified and hapten-labeled by degenerate oligonucleotide primer-polymerase chain reaction (DOP-PCR) using the 6MW primer as described in detail (Cremer et al. 2006b). Human BAC clones for chromosomes 12, 18, and 19 were in part kindly provided by T. Ried, (NCI, Bethesda, MD, USA) and in part purchased from BACPAC Resources Center (Oakland, CA, USA). The identification number of each BAC clone used in the different experiments is listed in the supplementary material. Clones assigned to either G-dark or G-light (R) band domains of HSA 12, 18, and 19 were selected according to the data provided by the Cancer Genome Anatomy project (http://cgap.nci.nih.gov/Chromosomes/BAC_Clone_Map), mapped on an 850-band resolution. As transitions from G to R bands are not exactly defined at the molecular level (Furey and Haussler 2003), only BACs from central regions within a band domain were included.
All BAC clones were verified by testing their correct position on metaphase chromosomes and by the comparison of restriction patterns, mostly SWAI and EcoRV, with in silico patterns obtained by the software pDRAW32 (http://www.acaclone.com). Genomic DNA from each BAC was amplified by a modified DOP-PCR using two different primers DOP-2 and DOP-3 (Fiegler et al. 2003) under conditions described in (Cremer et al. 2006b). Primary PCR products (approximately 100 ng/μl) of up to 27 BACs were pooled at equal DNA amounts and 2–3 μl of this BAC pool DNA was used for a label PCR with either the haptens BIO-2′-deoxyuridine 5′-triphosphate (dUTP), DIG-dUTP, DNP-dUTP, or for direct fluorochrome labeling with Texas-Red-dUTP or TAMRA-dUTP. Three microliters of each labeled BAC pool (approximately 100 ng) and 2 μl (approximately 60 ng) of chromosome painting probe was used per 1 μl hybridization mix.
Multicolor FISH on metaphase chromosomes and on morphologically preserved nuclei, detection of labeled probes by fluorochrome-conjugated antibodies or fluorochrome-conjugated avidin was performed according to protocols described in detail elsewhere (Cremer et al. 2006b). To identify cells in G0- and S-phase, respectively, antibodies against BrdU (Roche) and a monoclonal antibody against the cell-cycle-related nuclear protein pKi67 (mouse-anti-pKi67, Dianova) visualized by an appropriate fluorochrome-coupled secondary antibody was used. Nuclei were counterstained for 5 min with 1 μM ToPro-3 or DAPI (Molecular Probes).
RNA isolation, cDNA and cRNA synthesis, and transcription analysis
Data of the gene expression profiles of stimulated lymphocytes and fibroblasts were based on chip analysis using the human U133A GeneChip (Affymetrix) according to the instructions of the Affymetrix GeneChip Expression Analysis Protocols (Affymetrix, Santa Clara, CA, USA). Briefly, RNA was extracted with Trizol reagent 100 (Invitrogen) according to the manufacturer's instructions. RNA quality was verified by 28S and 18S rRNA integrity on a denaturing gel. Total cDNA was synthesized by using SuperScriptTM II reverse transcriptase (Invitrogen) for the first strand synthesis and E. coli DNA-Polymerase for the second strand synthesis. After reaction, RNA was removed by a RNase H treatment at 37°C for 20 min, followed by in vitro transcription with biotinylated UTP and cytidine triphosphate (CTP) (ENZO). The labeled cRNA was cleaned with the Quiagen's RNeasy Kit (Quiagen 74106). Fifteen micrograms cRNA was fragmentated and hybridized to the human U133A GeneChip (Affymetrix). Staining of the hybridized chips (using Affymetrix fluidics station) was performed using streptavidin phycoerythrin conjugate followed by the addition of biotinylated antibody (Vector Laboratories) and finally treating with streptavidin phycoerythrin conjugate. The arrays were scanned using a Hewlett-Packard GeneArray Scanner 2500.
Microarray data analysis
Analysis was done using MicroArray Suite 5.0 (MAS, Affymetrix) and dCHIP (http://www.dCHIP.org). dChip was used for normalization of the expression data. The Affymetrix U133 array contains 22,283 targets representing 14,500 genes. Intensity values ranged between 40 and 15,509 for the human fibroblasts and between 15 and 9,690 for the human lymphocytes. The normalized data were used to distinguish between expressed and non-expressed genes.
RT-PCR
Verification of expression data obtained from the affymetrix chip was done by a semi-quantitative reverse transcriptase-polymerase chain reaction (RT-PCR); primers for RT-PCR were designed by the software program FastPCR (http://www.biocenter.helsinki.fi/bi/Programs/fastpcr.htm). The reverse primers were chosen from the 3′ UTR and the forward primers from the penultimate exon of the respective gene yielding a length of the amplification product between 200 and 600 bp.
For each primer pair three reactions were set up as follows: 0.5 μM Forward Primer, 0.5 μM Reverse Primer, 250 μM dNTPs, 50 mM KCl, 10 mM Tris–HCl pH 9.0, 1.5 mM MgCl2, 2 μl first strand cDNA and 2.5 U Taq polymerase (Amersham). Cycling conditions were as follows: 30 s at 94°C, 30 s at appropriate primer annealing temperature, 45 s at 72°C; final extension 72°C 5 min. Cycling was stopped for each gene after 15, 22, or 35 cycles. Five microliters of each PCR product was loaded on an ethidium bromide stained gel and visualized under ultraviolet (UV) light. Only BACs where RT-PCR yielded negative results after 35 cycles for all genes located on a BAC clone were included in a pool representing weakly expressed/silent genes. Primers for GAPDH were used as a positive control and reactions without first strand cDNA as negative controls.
Confocal microscopy and 3D reconstruction of image stacks
Nuclei were scanned using a laser scanning confocal microscope (Zeiss LSM 410) for the simultaneous visualization of three fluorochromes or a Leica SPII for the simultaneous visualization of up to five fluorochromes with an axial distance of 150–300 nm between optical sections. Both microscopes were equipped with a 63×/1.4 plan-apochromat oil objective. For each optical section images were collected sequentially for all fluorochromes. Stacks of 8-bit gray scale 2D images were obtained with a pixel size between 40 and 80 nm.
Image processing and quantitative evaluation methods
Confocal images were processed with ImageJ (http://www.rsb.info.nih.gov/ij/). Three-dimensional reconstructions of CT image stacks were performed using Amira 2.3 TGS (http://www.amiravis.com/).
Three-dimensional distance measurement of fluorescence signals to the nuclear border and CT surface
A software was developed for the automatic measurement of the shortest absolute 3D distances of all BAC signals to the surface of their corresponding CT or to the nuclear border (enhanced absolute 3D distances to surfaces, eADS). As a first step the surface of the reference structure (CT, nucleus) is determined by applying a user set threshold: all voxels with intensities below the set threshold are set to zero. The surface of the reference structure contains all voxels with an intensity >0, which are directly adjacent to at least one voxel with an intensity=0. The program measures the shortest 3D distance of all voxels of a BAC signal to the surface of the reference structure by calculating the euclidean distances between the voxels of the BAC signal and voxels of the surface of the reference structure (considering the voxel size in the spatial directions). All signals that are located inside the surface-surrounded volume are assigned as negative distance values, while signals that are located outside of the surface-surrounded volume have positive distance values. All measured distances will be divided into user-defined classes (recommend at least z-voxel size), whereas the relative frequency in every class is dependent on the intensity value of the signal voxels, which are allocated in this class. The mean values of the classes for all evaluated nuclei are plotted as a graph.
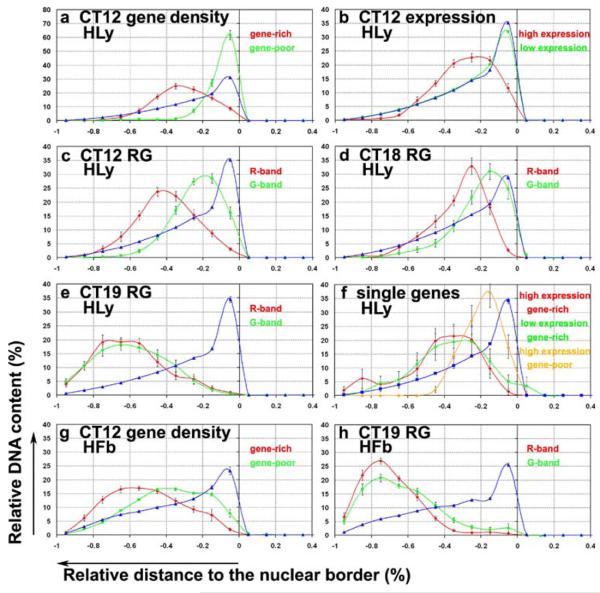
Absolute distances (nm) were applied for distance measurements within CTs, while for evaluations of distance measurements inside the nucleus a normalization was performed to facilitate the comparison of different experiments. Therefore, the maximum distance for each nucleus was acquired and set to 1. Accordingly, all other measured distances of signal-voxels to the surface of a given nucleus were related to the maximum distance and the resulting values (between 0 and 1) were ordered into ten classes by a frequency distribution. The sum of all normalized distances counted for each fluorochrome in a given nucleus was set to 100%. The mean values of the ten classes for all evaluated nuclei were plotted as a graph (see Figs. 3 and 6).
Fig. 3.
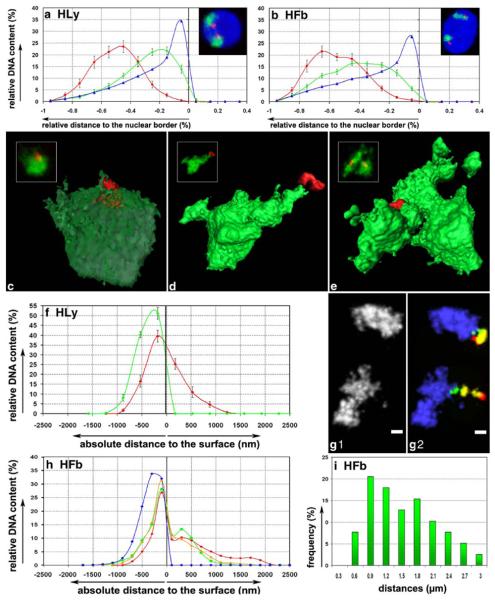
Quantitative 3D evaluation of the normalized distance distributions of differently assigned BAC pools to the nuclear border. The ordinate denotes the normalized sum of voxel intensities for a respective fluorochrome, the abscissa the normalized distance to the nuclear border. Negative values indicate the nuclear inside and “0” the nuclear border. The blue curve denotes the nuclear counterstain in all graphs. The assignment of curves to the respective BAC pools, shown in green, red, and ocher is indicated in each graph. Note the different scaling of the y-axis in a. The apparent small overlap of BAC signals outside of the nucleus is due to the interpolation between distinct values obtained for each class
Fig. 6.
Average absolute distances of differently assigned BAC pools to the CT surface in nuclei of Hly and Hfb. Red dots represent gene-dense regions, R-band-assigned chromatin or highly expressed genes, respectively; green dots represent the respective gene-poor regions, G-band-assigned chromatin or weakly expressed/silent genes hybridized together in the different experiments
Linear regression analysis
For the interpretation of linear regression results the adjusted correlation coefficient of determination AdjR2 was used that takes into account the number of independent variables.
Statistic test
To test for significant differences (p≤0.05) in the distribution curve for each fluorochrome, the Wilcoxon signed rank sum test was applied on the average relative distances (ARD) of the evaluated nuclei.
Replication timing
Replication timing measurements have been performed largely as described (Woodfine et al. 2004), but using genome-wide tiling path resolution arrays (Fiegler et al. 2006). Briefly, nuclei in either G1- or S-phase of the cell cycle were flow sorted from a normal lymphoblastoid cell line (HRC575), differentially labeled and co-hybridized to the arrays. The linear ratio of fluorescence intensities for each clone was calculated after appropriate normalization and represent values between 1.0 (late replication) and 2.0 (early replication).
Results
Generation of BAC pools targeting distinct chromatin features
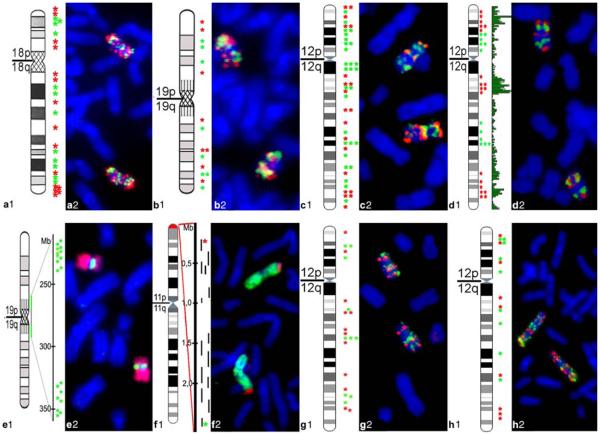
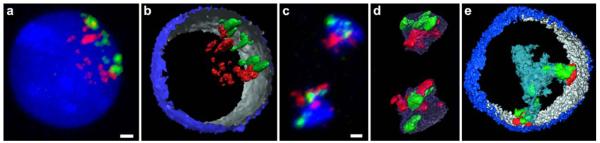
To analyze the spatial arrangement and folding behavior of distinct chromatin segments in human lymphocyte (Hly) and fibroblast (Hfb) nuclei along the entire length of HSA 12, 18, and 19 and of a subtelomeric segment of HSA 11, we used sets of DNA probes addressing distinct chromatin features that were generated from pooled BAC clones selected according to the respective desired chromatin feature (Fig. 1). (1) R- and G-band related clone pools for HSA 12, 18 and 19: These probe sets delineated sequences from central regions of R- or G-band domains, respectively, from either HSA 12, 18, or 19 (Fig. 1a–c); (2) Gene density related clone pools for HSA 11, 12 and 19: The “gene-rich” pool of HSA 12 delineated the three gene-dense regions 12p12.3–12p13.32 (38 genes/Mb; ~10.5 Mb;), 12q13.11–12q13.3 (31 genes/Mb; ~11 Mb), and 12q24.11–12q24.32 (16 genes/Mb; ~16 Mb) represented by 19 BACs, the “gene-poor” pool, the two genepoor regions 12p11.22–12p12.3 (6.5 genes/Mb; ~15 Mb;)\and 12q21.2 –12q21.33 (5 genes/Mb; ~18 Mb) represented by 12 BACs (Fig. 1d). Each probe pool comprised clones from R- as well as G-band domains. The “gene-poor” pool of HSA 19 was represented by 11 BACs from 19p12 and 8 BACs from 19q12 flanking the immediate pericentromeric region over approximately 6 Mb (Fig. 1e). The particular gene-dense and transcriptionally active subtelomeric region located on 11p15.5 was delineated by 14 BACs covering a 2.3 Mb contig (with two small gaps of around 100 and 300 kb) (Fig. 1f). (3)Gene expression related clone pools of HSA 12: For the “high expression” pools, specifically designed for Hly and Hfb, BAC clones—often harboring several genes— were included, in case they contained at least one gene with an expression intensity value >1,000, irrespective of the expression levels of other genes contained in the BAC. The “low expression/silent” gene pools were made up from BAC clones containing only genes with expression intensity values <250 (Fig. 1g–h; for details, see “Materials and methods”).
Fig. 1.
Overview of BAC pools. Panels a1–h1 denote the chromosomal position of each BAC (marked by an asterisk) used in this study with the corresponding idiogram at 850-band resolution. Panels a2–h2 show respective FISH control experiments on metaphase chromosomes (blue). a R/G-band-assigned BAC pool of HSA 18 comprising 12 BACs from G-dark (G) bands (green) and 15 BACs from G-light (R) bands (red). b R/G-band-assigned BAC pool of HSA 19 comprising 8 BACs from G-dark (G) bands (green) and 8 BACs from G-light (R) bands (red). c R/G-band-assigned BAC pool of HSA 12 comprising 28 BACs from G-dark (G) bands (green) and 20 BACs from G-light (R) bands (red). d Gene-density-assigned BAC pool of HSA 12 comprising 19 BACs of gene-dense segments (red) and 12 BACs from gene-poor segments (green). The green bars delineate the gene content along the chromosome. e Gene-density-assigned BAC pool of HSA 19 representing segments of the gene-poor region 19p12–19q12 comprising 19 BACs (green). HSA 19 are painted in red in E2. f BAC pool delineating a gene-dense segment of 11p15.5 comprising 14 BACs. The exact position of each BAC is represented by a black bar in f1; the BAC contig is visualized in red and the painted HSA 11 in green in f2. g–h Expression-assigned BAC pools of HSA 12 in Hly (g) and Hfb (h) comprising 10 (Hly) and 9 (Hfb) BACs with weakly expressed genes (green) and 12 (Hly) and 9 (Hfb) BACs with highly expressed genes (red)
The differently labeled pools were used for 3D FISH experiments together with painting probes for the respective chromosome territories. Tables 2, 3, 4, 5 provide information on band assignment, mean gene density (within 2- and 10-Mb windows), and mean replication timing for each BAC pool. Detailed data for each BAC clone including GC content and levels of transcriptional activity are listed in Tables S1–8.
Table 2.
Overview on features of the used R-/G-band-assigned pools
| 12 R (20 BACs) |
12 G (28 BACs) |
18 R (15 BACs) |
18 G (12 BACs) |
19 R (8 BACs) |
19 G (8 BACs) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene density (2 Mb window) | 16.6 | 8.6 | 7.2 | 4.7 | 32.4 | 30.1 | ||||||
| Gene density (10 Mb window) | 13.4 | 9.4 | 5.7 | 5.0 | 30.9 | 33.5 | ||||||
| Mean replication timing (early-mid-late) | 1.63 (11-7-2) | 1.45 (4-11-13) | 1.55 (2-12-1) | 1.39 (0-5-7) | 1.63 (6-2-0) | 1.56 (4-2-2) | ||||||
| Expression | HLy | HFb | HLy | HFb | HLy | HFb | HLy | HFb | HLy | HFb | HLy | HFb |
| high | 1 | 11 | 1 | 3 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 3 |
| median | 6 | 7 | 1 | 3 | 1 | 3 | 0 | 2 | 4 | 5 | 2 | 3 |
| low | 13 | 2 | 22 | 18 | 12 | 10 | 8 | 6 | 3 | 0 | 5 | 1 |
| unknown | 0 | 0 | 4 | 4 | 1 | 1 | 4 | 4 | 1 | 1 | 1 | 1 |
Mean gene density, replication timing, and expression levels of R/G band assigned BAC pools of HSA 12, 18, and 19. Numbers in brackets indicate the number of BACs with an early, mid, or late replication timing where each fraction comprises one-third of the entire replication range (theoretically between 1.0 and 2.0) observed for the respective chromosome. Data on “expression” classify BACs according to their genes with only low (<300 transcription units), median (300–1,000 transcription units) and high (>1,000 transcription units) expression levels.
Table 3.
Overview on features of the used gene-density-assigned pools of HSA 12 and 19
| 12 rich (19 BACs) |
12 poor (12 BACs) |
19 poor (19 BACs) |
||||
|---|---|---|---|---|---|---|
| R / G bands | 12 / 7 | 2/10 | 0 / 19 | |||
| Gene density (2 Mb window) | 20.3 | 5.2 | 9.3 | |||
| Gene density (10 Mb window) | 16.5 | 5.2 | 9.5 | |||
| Mean replication timing (early-mid-late) |
1.62 (11-6-2) |
1.42 (0-5-7) |
1.38 (2-3-14) |
|||
| Expression | HLy | HFb | HLy | HFb | HLy | HFb |
| high | 1 | 11 | 0 | 1 | 0 | 1 |
| median | 4 | 4 | 0 | 2 | 1 | 0 |
| low | 14 | 4 | 11 | 8 | 8 | 8 |
| unknown | 0 | 0 | 1 | 1 | 10 | 10 |
Band assignment, mean gene density, replication timing, and expression levels of gene-density assigned BAC pools of HSA 12 and 19. Numbers in brackets indicate the number of BACs with an early, mid, or late replication timing where each fraction comprises one-third of the entire replication range (theoretically between 1.0 and 2.0) observed for the respective chromosome. Data on “expression” classify BACs according to their genes with only low (<300 transcription units), median (300–1,000 transcription units) and high (>1,000 transcription units) expression levels.
Table 4.
Overview on features of the used expression-assigned pools of HSA 12
| 12 high in HLy (12 BACs) |
12 low in HLy (10 BACs) |
12 high in HFb (9 BACs) |
12 low in HFb (9 BACs) |
|
|---|---|---|---|---|
| R / G bands | 5 / 7 | 4 / 6 | 6 / 3 | 6 / 3 |
| Gene density (2 Mb window) |
16.5 | 10.5 | 16.6 | 15.7 |
| Gene density (10 Mb window) |
12.6 | 10.5 | 14.3 | 12.3 |
| Mean replication timing (early-mid-late) |
1.70 (10 - 2 - 0) |
1.45 (2 - 3 - 5) |
1.65 (7 - 1 - 1) |
1.51 (2 - 4 - 3) |
Band assignment, mean gene density, and replication timing of expression assigned BAC pools of HSA 12. Numbers in brackets indicate the number of BACs with an early, mid, or late replication timing where each fraction comprises one-third of the entire replication range (theoretically between 1.0 and 2.0) observed for the respective chromosome. BACs are classified according to their gene content with only low (<300 transcription units), and high (transcription units).
Table 5.
Overview on features of the used single BACs of HSA 12 with inverse features
| BAC gene- rich/high expr |
BAC gene- rich/low expr |
BAC gene- poor/ high expr |
|
|---|---|---|---|
| Gene | ATP5B | NEUROD4 | NAP1L1 |
| R / G bands | R | G1 | R |
| Gene density (2 Mb window) |
37.5 | 23.5 | 5.5 |
| Gene density (10 Mb window) |
24.6 | 28.8 | 2.8 |
| Mean replication timing |
1.80 early | 1.47 median | 1.62 median |
| Expression intensity value |
1,855 | 19 | 1,770 |
Band assignment, mean gene density, replication timing of single BACs and expression levels of selected genes from these BACs
Identification of nuclei in G0- and S-phase
In each 3D FISH experiment we generated light optical serial sections from 20–30 nuclei of Hly and Hfb cells both in G0 and S-phase. Nuclei in S-phase for both cell types were identified by a positive replication pattern after BrdU immunostaining (data not shown). Unstimulated Hly are unambiguously in G0, as they have exited the cell cycle. Hfb nuclei in G0 were identified by a negative immunostaining using an antibody directed against the cell-cycle-related nuclear protein pKi67 (Gerdes et al. 1984). Persistence of the pKi67 epitope for immunostaining after pretreatment including pepsin treatment and heat-denaturation was confirmed in control experiments (Fig. S2). A qualitative and quantitative evaluation, however, revealed on average some decrease of the signal intensity in pepsinized cells and an increase of “pKi67 negative” cells from 49% to 63% in nuclei after pepsin treatment (Fig. S2). Thus, a small percentage of Hfb nuclei assigned as G0 could in fact be cycling.
Distribution of different sequence pools with respect to the nuclear space
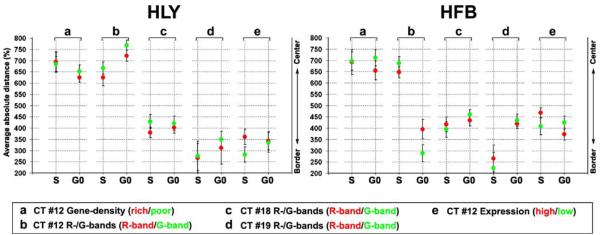
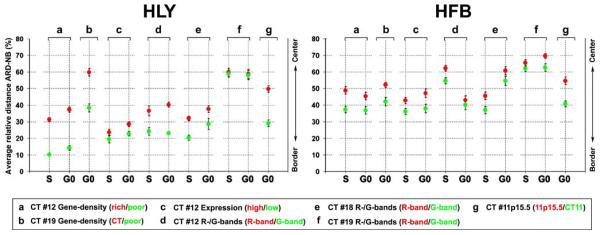
Image stacks were quantitatively evaluated by the eADS program, which provides the smallest (absolute and normalized) 3D distance for each voxel from a BAC signal to the nuclear border (see “Materials and methods”). Table 6 presents for each pool the normalized average relative distances to the nuclear border, further referred to as ARDNB and the relative distance differences (ΔD) of ARD-NB values obtained and compared in one experiment. These results are graphically shown in Fig. 2 and selected examples of the distribution curves are plotted as graphs in Fig. 3.
Table 6.
Average relative distances to the nuclear border (ARD-NB)
| Pool assignment |
Hly/HLb | HFb | ||
|---|---|---|---|---|
| S | G0 | S | G0 | |
|
A CT #12 Gene density |
Drich=31.17% Dpoor=10.13% ΔD=21.04% 24/24(100%) (p<0.001) |
Drich=37.15% Dpoor=14.19% ΔD=22.96% 21/21(100%) (p<0.001) |
Drich=48.76% Dpoor=37.29% ΔD=11.47% 28/30(93%) (p<0.001) |
Drich=45.36% Dpoor=36.81% ΔD=8.55% 24/26(92%) (p<0.001) |
|
B CT #19 Gene density |
--- |
DCT=59.69% Dpoor=38.21% ΔD=21.48% 24/25(96%) (p<0.001) |
--- |
Drich=52.13% Dpoor=42.05% ΔD=10.08% 20/22(91%) (p<0.001) |
|
C CT #12 Expression |
Dhigh=23.66% Dlow=19.3% ΔD=4.36% 15/20(75%) <p=0.027) |
Dhigh=28.53% Dlow=22.79% ΔD=5.74% 16/19(84%) (p<0.001) |
Dhigh=42.71% Dlow=36.02% ΔD=6.69% 17/21(81%) (p=0.001) |
Dhigh=47.16% Dlow=37.88% ΔD=9.28% 15/20(75%) (p<0.001) |
|
D CT #12 R-/G- bands |
DR=36.47% DG=24.2% ΔD=12.27% 13/15(86%) (p<0.001) |
DR=40.1% DG=23.11% ΔD=16.99% 28/28(100%) (p<0.001) |
DR=62.17% DG=54.55% ΔD=7.62% 21/25(84%) (p<0.001) |
DR=42.92% DG=40.06% ΔD=2.86% 12/18(66%) (p=0.081) |
|
E CT #18 R-/G- bands |
DR=31.88% DG=20.38% ΔD=11.5% 28/28(100%) (p<0.001) |
DR=37.5% DG=28.55% ΔD=8.95% 17/19(89%) (p<0.001) |
DR=45.54% DG=36.91% ΔD=8.63% 22/25(88%) (p<0.001) |
DR=60.64% DG=54.61% ΔD=6.03% 16/23(70%) (p=0.004) |
|
F CT #19 R-/G- bands |
DR=59.75% DG=58.92% ΔD=0.83% 13/22(59%) (p=0.436) |
DR=58.78% DG=58.03% ΔD=0.75% 13/23(56%) (p=0.537) |
DR=65.38% DG=61.91% ΔD=3.47% 17/24(71%) (p=0.015) |
DR=69.58% DG=62.63% ΔD=6.95% 23/23(100%) (p<0.001) |
|
G CT #11 11p15.5 |
--- |
Dp15.5=49.69% DCT11=28.99% ΔD=20.7% 22/23(95%) (p<0.001) |
Dp15.55=54.52% DCT11=40.65% ΔD=13.87% 17/19(89%) (p<0.001) |
|
Average relative distances to the nuclear border (ARD-NB) of differently assigned BAC pools. The two top lines in each panel denote ARD-NB values, where Drich and Dpoor denote gene-density-assigned pools (rows A–B), Dhigh and Dlow expression-assigned pools (row C), DR and DG R- and G-band-assigned pools (rows D–F) and Dp15.5 and DCT11 11p15.5 and the entire CT11 delineating pool (row G); ΔD denotes the distance differences between the respective pairs. The two bottom lines indicate the number and percentage of individual nuclei within a series with DR>DG, Drich>Dpoor, Dhigh>Dlow and Dp15.5>DCT11 and significance. Grey shaded boxes indicate non-significant differences (p>0.05).
Fig. 2.
Average relative distances of differently assigned BAC pools to the nuclear border (ARD-NB) of Hly and Hfb. Red dots represent gene-dense regions, highly expressed genes, or R-band-assigned chromatin respectively; green dots represent the respective gene-poor regions, weakly expressed/silent genes, or G-band-assigned chromatin and CT11 hybridized together in the different experiments. The differently assigned pairs of pools compared within one experiment show for all series smaller ARD-NBs for the green dots compared to the respective red dots
For each chromosome studied and in both the spherical nuclei of Hly and the flat ellipsoidal nuclei of Hfb, BAC pools assigned to gene-dense segments, highly expressed genes, and R-bands were all found, at different levels of significance, at a more interior nuclear position in contrast to the more peripheral location of pools defined for genepoor segments, weakly expressed/silent genes, and G-band assigned chromatin. Consistent differences in the radial nuclear distributions of BAC pool signals between cycling (S-phase) and quiescent (G0-phase) cells were not observed.
Distance differences (ΔD) were most pronounced between the gene-dense and gene-poor assigned probe pools from HSA 12 and 19 and the gene-dense segment of 11p15.5 compared to the entire CT11 (Table 6 and Fig. 2, columns A, B, G). Figure 4 shows examples of 3D FISH experiments with the gene-density-related HSA 12 and HSA 19 BAC pools on human lymphocytes, which demonstrate the polarity of HSA 12 assigned gene-dense and gene-poor sequence pools both with regard to the nucleus (Fig. 4a,b) and the HSA 12 territories (Fig. 4c,d). Figure 4e illustrates the distinct orientation of the gene-poor region 19p12–19q12 with regard to the remaining HSA 19 territory.
Fig. 4.
Multicolor 3D FISH in HLy with gene-density-assigned BAC pools of chromosomes 12 and 19. a Maximum Z-projection of the nuclear DNA counterstain (blue), the gene-rich (red) and the gene-poor (green) BAC pool of HSA12 illustrate the distinct nuclear distribution of these BAC pools. b 3D reconstruction of the same nucleus shown in (a) using TGS Amira 3.1.1. Part of the nuclear border is indicated by reconstruction of the counterstain periphery (outside blue; inside silver-grey). c Maximum Z-projection of chromosome paint 12 (blue), the gene-rich (red) and the gene-poor (green) BAC pool of chromosome 12 exemplifying a typical intra-territorial distribution. d 3D reconstruction of the same CTs using TGS Amira 3.1.1. e 3D reconstruction using TGS Amira 3.1.1 of painted CTs 19 (light blue) together with the BAC pool delineating the gene-poor segments of 19p12–19q12 (green) and centromeres of CT 19 (red). Part of the nuclear border is again indicated by reconstruction of the counterstain periphery (outside dark blue; inside silver-grey). Bars correspond to 1 μm
In comparison to the gene-density-assigned pools in Hly, ΔD values were considerably smaller obtained between highly and weakly expressed/silent genes of HSA 12 in Hly (Table 6, Fig. 2 left, column C). Notably, these sequence pools included BACs from R- and G-bands, and from regions with different gene density.
A comparison of the ARD-NB values obtained for the R- and G-band-assigned sequence pools from HSA 12, 18, and 19 in Hly revealed a wide variation of ΔD values in Hly ranging from an apparently lacking difference for CT19 (ΔD <1%) to significant differences for CT12 and CT18 (Table 6 and Fig. 2 left, columns D, E, and F). The direct comparison of the ARD-NB values of all R-band-assigned sequence pools with each other (red dots in Fig. 2 left, columns D–F), and for all G-band-assigned pools (green dots in Fig. 2 left, columns D–F) also revealed a remarkable range of values in Hly and Hfb (Table 6 and Fig. 2 right, columns D–F). These data indicate that the assignment to either a G- or R-band per se does not implicate a distinct positioning with regard to the nuclear center or periphery.
The impact of different chromatin features on nuclear position assessed by regression analysis
To directly compare and weight the impact of different local chromatin features on nuclear positioning we plotted the normalized ARD-NB value of each sequence pool described above against its mean gene density, GC content, replication timing, and average expression level to assess the correlation between nuclear position and these different parameters (Fig. 5). The mean gene density for a given BAC pool was assessed from the number of genes found in 2- and 10-Mb windows, respectively, around each BAC sequence (http://www.ncbi.nlm.nih.gov). The mean replication timing was calculated by averaging the replication timing values of BACs constituting each pool. In cases, where a replication timing value was not known for a given BAC, data from the nearest neighbor BAC were substituted (see Supplementary material, Tables S1–8). Values for replication timing were obtained by comparative hybridizations of S vs G1 phase genomic DNA and genomic DNA from various stages of S-phase prepared from lymphoblastoid cells after flow sorting (see “Materials and methods”).
Fig. 5.
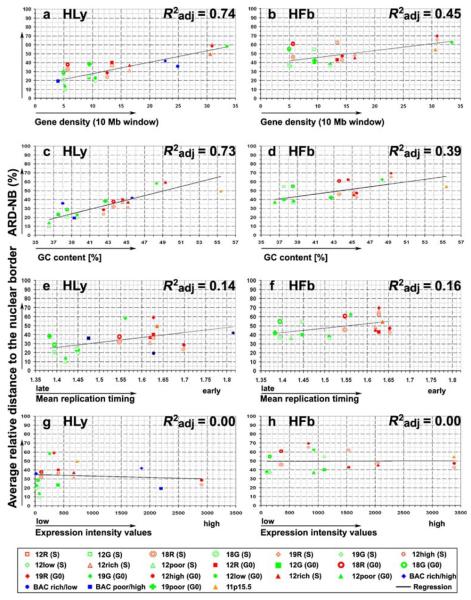
Correlation analysis of the average radial position of chromatin with distinct chromatin features. The ordinate denotes in all graphs the ARDNB values assessed for the different BAC pools in Hly and Hfb. The abscissa denotes the different chromatin features defined for the respective BAC pools, namely, the average gene density (a, b), GC content (c–d), replication timing (e–f), and expression intensity values (g–h). Symbols for each BAC pool are explained in the legend at the bottom. The regression line for each graph was determined by the adjusted correlation coefficient. While a high correlation from the plotted values and the regression curve could be shown for ARD-NB to gene density and GC content, this correlation was much lower with replication timing and expression activity
The mean expression level of a sequence pool was determined by averaging the expression intensity values of each (one) gene on a given BAC clone that showed the highest transcriptional activity on this BAC. Accordingly, average expression intensities for a given BAC pool are a maximum estimate. The average GC content was derived from sequence data provided in (http://www.ncbi.nlm.nih.gov). Independent of its original designation regression analysis for the collectivity of all BAC pools revealed a high correlation between the average radial nuclear position and mean gene density obtained for both 10- and 2-Mb windows around each BAC of a given BAC pool (Fig. 5a,b), with 2-Mb windows resulting in slightly lower values (data not shown). The same close correlation was found for the GC content represented by each BAC pool (Fig. 5c,d).
In contrast, the correlation with the mean replication timing of the different BAC pools was much less pronounced (Fig. 5e,f). A caveat should be added with regard to this result: replication timing values derived from lymphoblastoid cells may not exactly reflect replication timing values for Hly and HFb and some discrepancies have in fact been observed (our own unpublished observation). As overall DNA replication timing profiles, however, were found to be remarkably similar for HSA 22 in both lymphoblastoid cells and fibroblasts (White et al. 2004), we tentatively assumed that replication timing values for HSA 11, 12, 18, and 19 obtained for lymphoblastoid cells are at least fairly representative for Hfb. Finally, a positive correlation between the nuclear position and expression levels represented by the different BAC pools could not be established (Fig. 5g,h).
The data described above consistently support the conclusion that different average radial nuclear positions depend on regional gene density and not on band assignment or expression levels. To corroborate this conclusion further, we compared the radial positions of three BACs in Hly. One BAC contains the highly expressed gene ATP5B located in the highly gene-dense region of the R-band located at 12q13.13, the second BAC contains the highly expressed gene NAP1L1 in the gene-poor immediate environment of the R-band located at 12q21.2, where as the third BAC contains the weakly expressed/silent gene NEUROD4 within a gene-dense G1 band located at 12q13.2. In spite of a drastic difference in their expression levels, ATP5B and NEUROD4 were both found in the nuclear interior consistent with their location in gene-dense regions, whereas the highly expressed NAP1L1 gene was localized at the nuclear periphery consistent with its location in a gene poor region (Table 7 and Fig. 3f).
Table 7.
Nuclear positioning of single genes with inverse features in the nucleus
| Gene assignment | ARD-NB | Frequency and significance of distinct nuclear positioning | ||||
|---|---|---|---|---|---|---|
| BAC (A) | BAC (B) | BAC (C) | ||||
|
BAC (A) gene-rich high expression ATP5B |
41.95% | BAC (A) | --- | (A)/(B) 12/20 (60%) n.s. (p=0.154) |
(A)/(C) 20/20 (100%) s. (p<0.001) |
|
|
BAC (B) gene-rich low expression NEUROD4 |
35.85% | BAC (B) | (B)/(A) 8/20 (40%) n.s. (p=0.154) |
--- | (B)/(C) 16/20(80%) s. (p<0.001) |
|
|
BAC (C) gene-poor high expression NAP1L1 |
19.26% | BAC (C) | (C)/(A) 0/20 (0%) s. (p<0.001) |
(C)/(B) 4/20 (20%) s. (p<0.001) |
--- | |
Nuclear positioning and spatial relation of BACs represent single genes with inverse features.
The table shows the ARD-NB and the ratio between the different BACs, indicating their frequency and significance of a more internal position of the genes compared within 20 nuclei. Grey shaded boxes indicate non-significant differences (p>0.05).
Distribution of different sequence pools with respect to individual CTs
Next we addressed the issue of distribution differences of the various sequence pools with respect to individual CTs. The painted CTs 11, 12, 18, and 19 showed subregions with different signal intensities and highly variable shapes (compare Fig. 8),and it should be noted that small chromatin protrusions expanding from territory surfaces may not be outlined by chromosome painting. Such protrusions, however, can be visualized as BAC signals located outside of the thresholded CT periphery. Thresholded light optical images of CTs also reveal invaginations enlarging the threshold surface. Using the eADS program we measured the smallest absolute distances of each voxel assigned to a given BAC pool to the CT surface irrespective of its position inside or outside the CT border.
Fig. 8.
Spatial arrangement of a 2.3 Mb contig of the gene-dense region 11p15.5. a–b Quantitative evaluation of the 11p15.5 segment in the nucleus compared to the entire CT 11, both in Hly (a) and Hfb (b). The abscissa denotes the normalized distances to the nuclear rim, the ordinate the normalized sum of voxel intensities for CT11 (green) and the 11p15.5 contig (red). c–e 3D reconstructions and maximum projections (inserts) of CT11 (green) and the 11p15.5 contig (red) in Hly (c) and Hfb (d–e). c A rather compact conformation of CT 11 with the contig smoothly extending from the CT surface. d Contig, which protrudes completely from its CT. e Contig mainly found in the CT interior. f Quantitative evaluation of the 11p15.5 distribution with regard to the CT surface in HLy. The abscissa denotes the absolute distance distribution to the CT surface in nanometers, the ordinate the normalized sum of voxel intensities for CT11 (green) and 11p15.5 contig (red). g1 Maximum projection of CT11; g2 maximum projection of CT11 (blue) and the contig in Hfb, where the most telomeric clone is delineated in red, the intermediate clones in yellow, and the most centromeric clone in green. The images exemplify two contig conformations, one with a coiled structure and one with a conformation of a >2 μm extended structure. Note that the paint (g1) does not delineate the contig. h Quantitative evaluation of the 11p15.5 distribution with regard to the CT 11 surface (blue) in Hfb, with distinctly labeled BACs of the contig as outlined in (g). The abscissa denotes the absolute distance distribution to the CT surface in nanometer, the ordinate the normalized sum of voxel intensities for CT11 (blue), and the different clones of 11p15.5 contig (green, yellow, red). i Distance distributions between the geometrical centers of the most telomeric (red) and the most centromeric (green) clones of the 11p15.5 contig
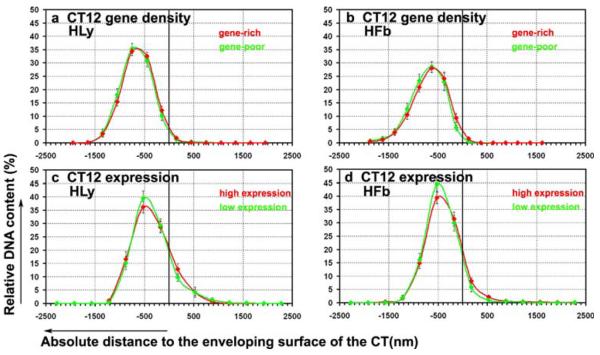
The results of distance measurements for the different BAC pools are listed in detail in Table 8 and mean values are graphically shown in Fig. 6. Selected examples of the distribution curves are plotted as graphs in Fig. 7. Negative and positive values indicate positions inside and outside the CT surface, respectively. These analyses yielded the following results: the mean distances of BAC signals from a given pool to the CT surface, obtained from different experiments, varied considerably (Table 8 and Fig. 6). However, the spatial arrangements of two sequence pools directly compared in one experiment for the same CTs were very similar with regard to the CT surface (Fig. 7). Moderately significant differences (p<0.025) were only observed for R-band- and G-band-assigned sequence pools in CTs 12 of Hfb (G0) and for sequence pools comprising either highly expressed or weakly expressed/silent genes in CTs 12 of both Hly and Hfb S-phase nuclei (Table 8). Unexpectedly, highly expressed genes were often located slightly more toward the CT 12 interior compared to weakly expressed/silent genes (Table 8, row E). For all chromosomes studied most signals (>92% on average) were located within the segmented border of each CT. From the small percentage of BAC signals found outside the CT border, the large majority was noted within a shell of 400 nm and all remaining signals within a distance <1 μm (Fig.7).
Table 8.
Absolute distances to the chromosome territory (CT) surface
| Pool assignment |
Hly/HLb | HFb | ||
|---|---|---|---|---|
| S | G0 | S | G0 | |
|
A CT #12 Gene density |
Drich=−694nm Dpoor=−685nm ΔD=−9nm 12/21(57%) (p=0.465) |
Drich=−625nm Dpoor=−652nm ΔD=+27nm 13/24(54%) (p=0.353) |
Drich=−692nm Dpoor=−697nm ΔD=+5nm 9/18(50%) (p=0.966) |
Drich=−655nm Dpoor=−712nm ΔD=+57nm 14/26(54%) (p=0.178) |
|
B CT #12 R-/G- bands |
DR=−623.55nm DG=−667.26nm ΔD=+43.71nm 7/18(39%) (p=0.196) |
DR=−721.8nm DG=−768.5nm ΔD=+46.7nm 11/25(44%) (p=0.197) |
DR=−647.11nm DG=−686.65nm ΔD=+39.39nm 10/30(33%) (p=0.109) |
DR=−395.2nm DG=−289.2nm ΔD=−105nm 14/19(74%) (p<0.002) |
|
C CT #18 R-/G- bands |
DR=−380.4nm DG=−429.8nm ΔD=+49.4nm 11/26(42%) (p=0.195) |
DR=−400.6nm DG=−420.2nm ΔD=+19.6nm 10/20(50%) (p=0.498) |
DR=−416nm DG=−394.1nm ΔD=−21.9nm 16/30(53%) (p=0.422) |
DR=−434.2nm DG=−460.1nm ΔD=+25.9nm 9/20(45%) (p=0.756) |
|
D CT #19 R-/G-bands |
DR=−266.37nm DG=−275.76nm ΔD=+9.39nm 10/20(50%) (p=0.756) |
DR=−311.7nm DG=−349.2nm ΔD=+37.5nm 12/27(44%) (p=0.524) |
DR=−265.1nm DG=−223.29nm ΔD=−41.81nm 12/20(60%) (p=0.729) |
DR=−418.75nm DG=−433.96nm ΔD=+15.21nm 11/23(48%) (p=0.338) |
|
E CT #12 expression |
Dhigh=−360nm Dlow=−282nm ΔD=−78nm 17/24(70%) (p=0.025) |
Dhigh=−343nm Dlow=−335nm ΔD=−8nm 12/25(48%) (p=0.893) |
Dhigh=−468nm Dlow=−408nm ΔD=−60nm 16/23(70%) (p=0.02) |
Dhigh=−373nm Dlow=−424nm ΔD=+51nm 10/21(47%) (p=0.102) |
Average absolute distances of differently assigned BAC pools to the CT surface. The two upper lines in each row denote the mean absolute distances to the territory surface determined for each differently assigned BAC pool in nm, where negative values indicate a position inside the CT. Drich and Dpoor denote gene density assigned pools (row A), Dhigh and Dlow expression-assigned pools (row E) and DR and DG R- and G-band-assigned pools (rows B–D). ΔD denotes the absolute differences between the respective pairs. The bottom lines denote the number and percentage of individual nuclei within a series with Drich>Dpoor, Dhigh>Dlow, and DR>DG and significance. Grey shaded boxes indicate non-significant differences (p>0.05).
Fig. 7.
Quantitative 3D evaluation for the distance distributions of differently assigned BAC pools to their territory surface. The abscissa denotes the absolute distance to the CT surface in nanometers. Negative values represent a position inside, positive values a position outside of the segmented CT surface, the black bar displays the surface of the segmented CT. The ordinate shows the normalized sum of intensities in the voxels for a respective fluorochrome. a CT12 gene-density BAC pool in Hly (G0), red curve represents gene-rich BACs, green curve gene-poor BACs. b CT12 gene-density BAC pool in Hfb (G0), red curve represents gene-rich BACs, green curve gene-poor BACs. c CT12 expression assigned BAC pool in HLy (G0), red curve represents BACs containing highly expressed genes, green curve BACs weakly expressed/silent genes. d CT12 gene-expression-assigned BAC pool in quiescent Hfb (G0), red curve represents BACs containing highly expressed genes, green curve BACs containing weakly expressed/silent genes. Note the indiscriminate distribution of the BAC pools regarding the position in their corresponding CT in all curves
These results argue against the hypothesis that gene dense or highly expressed chromatin segments typically loop out widely from the respective territory as it has been described for a number of specific highly expressed genes (for review, see Bartova and Kozubek 2006; Foster and Bridger 2005). Two previous reports (Gilbert et al. 2004; Mahy et al. 2002) described single cosmids or PACs from 11p15.5 to be frequently located >1.5 μm outside the CT 11 border. These clones are part of a particularly gene-dense and highly expressed segment of several megabases within 11p15.5. We visualized this segment by a BAC contig of 2.35 Mb covering this region except for two small gaps, (see Fig. 1f and Table S7). This contig comprises approximately 80 genes, many of them highly expressed in fibroblasts and to a less extent in lymphocytes (Table S7).
In agreement with our data obtained for HSA 12, 18, and 19, this gene-dense contig was positioned distinctly toward the nuclear interior compared to the entire CT 11 both in Hly and in Hfb (Fig. 2, column G and Fig. 8a,b). Figure 8c–e demonstrates a pronounced variability of 3D arrangements of the contig, ranging from compact structures partly hidden in the territory interior or exposed at the territory periphery to finger-like protrusions expanding from the CT 11 surface. Maximum distances of signal voxels were noted up to 1.2 μm outside of the segmented CT 11 in Hly nuclei (Fig. 8f) and up to 2.3 μm in Hfb nuclei (Fig.8h), where the most telomeric clone was delineated in red and the most centromeric clone in green. Figure 8g exemplifies two CTs 11 of a Hfb nucleus, one with an extended protrusion of the contig and the other representing a rather compact structure. Quantitative measurements of BAC signals in 20 Hly and 20 Hfb nuclei using the eADS program revealed around 60% of all BAC signals located inside segmented HSA 11 CTs in Hly (Fig. 8f).
In Hfb, only 46% of signals assigned to the most telomeric clone (red) were found inside CT 11, while this amount was around 60% for the remaining signals (Fig. 8h). Distance measurements between the geometrical centers of the most telomeric and the most centromeric BAC revealed for the majority a distance range between 0.9 and 1.8 μm with maximum distances up to 3 μm (Fig. 8i). These data suggest a highly variable structure and potentially large extension of this specific chromatin segment.
Summary of results
We found sequence pools addressing gene-dense regions, highly expressed genes or R-band-assigned chromatin of different chromosomes both in lymphocyte and fibroblast nuclei, all localized more toward the nuclear interior compared to their chromosomal counterparts addressing gene-poor regions, weakly expressed/silent genes and G-band-assigned chromatin. Regression analyses, however, demonstrated that the radial nuclear position for each sequence pool could closely be attributed only to its gene density within a window of several megabases and to GC content. In contrast, replication timing seems to be of minor importance. Finally, expression levels or the assignment to either a G- or R-band per se do not implicate a distinct positioning with regard to the nuclear center or periphery.
Radial positional differences of sequence pools at the nuclear level were also reflected at the level of individual chromosome territories. The polarized distribution of gene-dense and gene-poor chromatin segments within a single CT argues for a considerable structural transformation of interphase CTs in comparison to mitotic chromosomes. However, gene-dense or highly expressed sequences were not generally found preferentially at the CT surface, but were distributed throughout the territory space with little expansions from territory surfaces. As a particular case, our study confirms previous reports of an extended structure of a segment on 11p15.5.
Discussion
Linking nuclear architecture with local chromatin features
A variety of local chromatin features, including gene density, transcriptional activity, replication timing, local enrichments of repetitive sequences, and GC content have been linked with radial arrangements of higher order chromatin patterns observed in several cell types of humans and other species (for recent reviews, see Bartova and Kozubek 2006; Foster and Bridger 2005; Parada et al. 2004; Pederson 2004). So far, however, it has remained elusive, whether these features contribute rather indiscriminately to higher order chromatin arrangements, or whether one of these features has a dominant impact. Other features could then reflect only their interdependence with this predominant feature or contribute synergistically (or anti-synergistically) to a cell-type-specific nuclear topography to enhance nuclear functions. While the weight of previous studies already hints to the relevant impact of gene density for radial nuclear chromatin arrangements (see below), a systematic study was lacking so far, which directly compared and weighted different features of local chromatin arrangements.
In this study we aimed, in particular, to reveal whether local gene densities per se and/or other features associated with gene density by means of co-evolution and/or functional relationship have major effects on radial nuclear chromatin arrangements. At a functional level, we aimed to elucidate the possible impact of replication timing and gene expression levels on higher order chromatin arrangements.
Local gene density dominates radial chromatin arrangement in the nucleus
A gene-density-correlated nuclear architecture was initially described for entire CTs in spherically shaped nuclei of lymphocytes (Boyle et al. 2001; Cremer et al. 2001; Croft et al. 1999). Evidence for a gene density-related positioning of chromosomal sub-domains came from observations that translocated parts of derivative chromosomes largely retain their position observed for the normal chromosome (Cremer et al. 2003; Croft et al. 1999), from studies comparing homologous segments between different species with karyotypic rearrangements (Neusser et al. 2007; Tanabe et al. 2002) and from studies of single-gene loci vs non-genic sequences (Lukasova et al. 2004; Scheuermann et al. 2004).
The situation has been less clear for flat ellipsoid nuclei of fibroblasts. Several studies essentially described a chromosome size rather than a gene-density-correlated radial CT arrangement in nuclei of quiescent fibroblasts (Bolzer et al. 2005; Cremer et al. 2001; Croft et al. 1999), while a switch to a more gene-density-correlated arrangement was reported in cycling cells (Bridger et al. 2000). Bolzer et al. 2005 showed the clustering of Alu sequences as a marker for gene-dense regions in the interior of Hfb nuclei, suggesting that both size and gene-density-correlated features of chromatin arrangements co-exist in human fibroblast nuclei. A recent study of Neusser et al. (see concomitant paper in this issue, Neusser et al. 2007) compared the nuclear topology of orthologous chromosome segments in fibroblast nuclei of different primate species comprising evolutionary translocations, fusions, and fissions. These experiments provided further evidence for a polarized and gene-density-correlated arrangement of chromosome subregions within a territory in flat-ellipsoidal nuclei. In particular, observations in a primate species (Wolf's guenon) with a rather homogeneously sized chromosome complement argue against a functional role of chromosome-size-dependent spatial chromatin arrangements and for geometrical constrains in flat-ellipsoidal fibroblast nuclei. In this study we demonstrated also for human Hfb nuclei, that individual CTs show in fact a gene-density-correlated orientation of chromatin at the subchromosomal level in cycling and quiescent cells, substantiating local gene density as a dominant parameter of the polarized CT architecture also for this cell type.
Only few studies so far directly addressed the issue of the nuclear topography of chromosomal G- and R-bands. G-band-assigned chromatin constitutes the mid-replicating chromatin rim typically found beneath the nuclear envelope (Sadoni et al. 1999). However, our data do not support evidence that most G-bands (defined on an 850-band resolution level) are necessarily linked with a peripheral position. With increasing length of mitotic chromosomes small R-bands appear within larger blocks of G-dark bands (Lehrer et al. 2004), and it is not clear how this feature of mitotic chromosomes translates into the 3D architecture of chromatin located at or close to the nuclear envelope.
We could not find an overall correlation between radial nuclear positioning of genes and their transcriptional status. The influence of transcriptional activity on gene positioning in the nucleus has been a controversial issue (for review, see Bartova and Kozubek 2006; Kosak and Groudine 2004; Lanctot et al. 2007; Parada et al. 2004). Lukasova et al. 2002 and Scheuermann et al. 2004 suggested an interior nuclear position of transcribed genes, compared to non-transcribed genes in a given cell type. As local gene density was not considered in these studies, these observations may rather reflect gene density than the transcriptional activity of a studied gene per se. A repositioning toward the nuclear interior has been described for single genes, such as the CFTR gene, in cell types, where it is actively transcribed (Zink et al. 2004). While specific genes may in fact show such a dynamic behavior, our data do not substantiate gene repositioning as a common phenomenon. Highly expressed genes located in a gene-poor environment can be well found at the nuclear periphery, as exemplified recently for the β-globin gene (Brown et al. 2006) and in the present study for the NAP1L1 gene.
Analysis of gene repositioning in living cells is mostly done by the GFP-lac repressor/lac operator system that introduces heterologous binding sites for fluorescently labeled proteins at the gene of interest. For instance (Chuang et al. 2006; Tumbar and Belmont 2001) used a fusion of the acidic activation domain (AAD) of the viral protein VP16 and the DNA-binding domain of the lac repressor in conjunction with an integrated array of lac operators and analyzed chromatin movement during transcriptional activation in mammalian cells. The targeted locus moved from the periphery toward the nuclear interior after mitosis in VP16 AAD-expressing living cells, and long-range movements >2 μm of the locus were observed. Although, undoubtedly, important insights into the relationships between chromatin movement and gene regulation have come from the use of this technique, the physiological relevance of studying very large arrays of tandemly repeated genes has to be addressed.
Interphase chromosomes show a high plasticity
Our data support a high plasticity of interphase CTs. The topography of chromatin in CTs can clearly deviate from the straight linear arrangement of metaphase chromosomes and does not support a previous claim that chromosome structure during interphase is largely maintained from metaphase chromosomes (Claussen 2005; Lemke et al. 2002). Evidence for a high plasticity of interphase chromosomes was recently provided (Simonis et al. 2006; Wurtele and Chartrand 2006; Zhao et al. 2006). They used an advancement of the chromosome conformation capture (3C) method called 4C (chromosome conformation capture-on-chip, Simonis et al. 2006, or circular chromosome conformation capture, Zhao et al. 2006, respectively). By recircularization of the template, an inverse PCR and subsequent sequencing or microarray techniques a genome-wide identification of sequences cross-linking with a specific genomic locus became possible. A predominance of cis-interactions of the mouse β-globin locus on MMU 7 was observed, yet, the interacting loci could be tens of megabases away from the β-globin locus (Simonis et al. 2006). The observation of mostly cis-interactions emphasizes at the same time a territorial organization of chromosomes that imposes some constraint on interchromosomal interactions.
The observation of a gene density-driven folding of CT subdomains raises the question of the size range over which local differences of gene density are transferred as a polarized nuclear arrangement of gene-dense and gene-poor segments within a CT. Murmann et al. (2005) analyzed the spatial position of six genes in different cell types with normal karyotypes and in cell lines where these genes were involved in different translocations. They suggested that the regional gene density within a 2-Mb window around a gene of interest was a good predictor for the positioning of a genomic locus within the cell nucleus, while we found a still better overall correlation within 10-Mb windows around the selected genomic regions. In line with these observations is a recent study of Shopland et al. 2006, reporting on the structural organization of a 4.3-Mb contig of mouse chromosome MMU 14 with four clusters of genes separated by gene deserts that were preferentially aligned with the nuclear periphery. The organization of chromosomes in alternating segmental clusters of low and high gene density comprising windows of at least several megabases (see, e.g., supplementary Fig. S1) can facilitate such a polarized folding structure of individual CTs within these windows. Recent comparisons between human and mouse revealed that the synteny of gene-rich clusters with broadly expressed genes has been conserved (Singer et al. 2005). As sharing of common regulatory elements is not a common feature for most co-expressed gene clusters (Sproul et al. 2005), their conservation might be influenced by constraints of the folding capacity.
The chromosome territory periphery does not provide a special compartment for transcription
Early studies (Dietzel et al. 1999; Kurz et al. 1996) and even recent observations (Clemson et al. 2006) provided evidence for active genes positioned at the periphery of chromosome territories. However, the common acceptance of concept was challenged, when active genes were also found inside CTs (for review, see Cremer and Cremer 2006; Cremer et al. 2006a; Gilbert et al. 2005; Kosak and Groudine 2004; van Driel et al. 2003; Williams 2003) consistent with the upcoming chromosome territory-interchromatin compartment (CT-IC) model that postulates a sponge-like architecture of CTs with interconnected chromatin domains permeated by a network of IC channels (Cremer and Cremer 2001, see supplementary figure therein). Here, the only topological requirement for transcription is the position of genes at the surface of chromatin domains lining the IC. Our data also argue against a special importance of the CT periphery, as we did not observe an overall preferential position of gene-dense DNA segments or transcriptionally active genes at or outside the CT periphery.
Technical issues of chromosome painting can contribute to variable and sometimes discrepant results concerning the position of specific loci inside or outside a given CT. These include differences of fixation procedures and a possible bias in amplification efficiency of chromosome painting probes. The suppression of interspersed repetitive sequences (contributing >40% in the human genome; Waterston et al. 2002) by Cot-1 DNA during FISH results in an incomplete visualization of CTs even by optimized protocols. In addition, the subjective choice of a threshold for confocal image stacks may result in the visual loss of fine DNA structures for a given territory and affect the location of its border. Thus, absolute distance measurements of a single gene or locus to the thresholded border of a CT, although a commonly used approach, should be interpreted with caution. To diminish this problem, we focused on comparative measurements of differently labeled targets with regard to a given CT border. In this case relative differences are maintained independently of the inevitably arbitrary determination of this border in chromosome painting experiments.
For some genes or small chromatin segments, however, impressive distances of >2 μm from their CT border have been shown that cannot easily be explained just by threshold setup, but have to be assumed as expanded structures (for review, see Foster and Bridger 2005). Our study provides further evidence to this phenomenon. In Hfb nuclei we observed that the highly gene-dense region located in 11p15.5 expanded up to >2 μm outside of the thresholded CT border, a value which clearly exceeded the maximum distance (<1 μm), which we observed between any HSA 12, 18, and 19 BAC and the respective thresholded CT borders. While resolution of 3D-FISH and confocal microscopy does not provide information on the degree of decondensation of small subsegments within this region, the maximum extension found for the entire contig points to a compaction factor of at least 1:300, which is an order of magnitude beyond the compaction factor of about 1:30 assumed for an extended 30-nm fiber (for review, see Cremer et al. 2004).
All segments previously described as looped-out structures represent gene-dense chromosome regions with either tissue-specific or ubiquitously increased gene expression such as the MHC cluster (Volpi et al. 2000), the EDC cluster (Williams et al. 2002), the Hoxb complex (Chambeyron and Bickmore 2004; Chambeyron et al. 2005), the β-globin locus (Brown et al. 2006; Ragoczy et al. 2003), or the 11p15.5 region (Gilbert et al. 2004; Mahy et al. 2002). Yet, the underlying mechanisms or preconditions that predispose these regions to form such extended structures are still elusive. Gene density alone is probably not a sufficient indicator, as we could not detect pronounced chromatin extensions for segments of HSA 12, even though their gene density within 2- and 10-Mb windows are the same as or even higher than gene densities of the MHC and Hoxb cluster or the EDC complex (data not shown). For gene-dense segments involved in frequent chromatin protrusions an open chromatin conformation was reported (Gilbert et al. 2004). However, an open chromatin conformation was also described for the two gene-dense regions of HSA 12 (Gilbert et al. 2004; see Fig. 3 therein), for which we could not demonstrate chromatin protrusions in our present study. Finally, looping out of the EDC and the MHC locus has been linked with high transcriptional activation. However, for the β-globin locus a looping out was already observed before transcriptional induction (Ragoczy et al. 2003). We observed highly extended structures of CTs 19 in Hly that obviously link gene-dense chromatin in the nuclear interior with the (peri)centromeric region at the nuclear envelope.
Conclusions
The high impact of regional gene density on the radial position of chromatin begs the question of its biological meaning. What are the functional reasons for the enrichment of gene-dense chromatin in the nuclear interior and of gene-poor chromatin at the nuclear envelope and what are the mechanisms that initiate and maintain such a polarized organization? The spatial proximity of genes in the nuclear interior may facilitate their temporal transcriptional coordination irrespective of their location on different chromosomes (Osborne et al. 2004; Shopland et al. 2003; Spilianakis et al. 2005) and may help to establish a dynamic chromatin topography, which optimally suits these requirements. An increased freedom of movement in an open chromatin conformation has been described for a number of the gene-dense and transcriptionally active regions (Gilbert et al. 2004). A number of mechanisms have been delineated that bind gene-poor chromatin to the nuclear lamina through a complex interplay between nuclear envelope proteins and DNA-binding proteins (for review, see Gruenbaum et al. 2005; Maraldi et al. 2005).
Interestingly, no mechanisms are presently known that would fix gene-dense chromatin in the nuclear interior. It has long been observed that gene density is correlated to GC content (for review, see Eyre-Walker and Hurst 2001). This correlation is much stronger in warm-blooded compared to cold-blooded vertebrates (Saccone et al. 2002). High GC levels in gene-dense regions of warm-blooded vertebrates possibly emerged as a result of selective pressure for thermostability or bending properties of the DNA molecule (Federico et al. 2006; Vinogradov 2003) to mediate the stable maintenance of an open conformation of gene-dense regions. The enrichment of Alu (SINE) repeats in gene-dense regions could also be relevant in this context. Interestingly, “recent” Alus show rather a preference for insertion into AT-rich DNA segments, and only older Alus show a progressively stronger bias toward GC-rich DNA (for review, see Eyre-Walker and Hurst 2001), pointing that the enrichment of Alus in GC-rich DNA is the result of a positive selection. Different explanations have been offered, but the high GC content (>50%) of Alu repeats itself might also act to favor their co-existence with gene-dense chromatin regions.
The existence of transcriptionally highly active genes also in the gene-poor environment of the nuclear periphery argues for an impact of other factors such as the switching of replication timing, ultra-conserved DNA elements (Bejerano et al. 2004), and local chromatin unfolding in addition to the influence of local gene density. Finally, the observation of gene-dense chromatin confirmed in a variety of different cell types and species so far does not implicate that such an arrangement is mandatory and does not exclude the existence of specific cell types with an opposite nuclear distribution.
Supplementary Material
Acknowledgments
We gratefully acknowledge very helpful discussions and technical support from Christian Lanctôt, Stefan Müller, Heiner Albiez, and Boris Joffe from our group. We thank C. Cremer for supporting JvH from his funding. We are grateful to Thomas Ried, NCI, Bethesda, MD, for generously providing us BAC-DNA from clones of chromosomes 12, 18, and 19. This work was supported by the Wilhelm-Sanderstiftung (2001.079.2) to TC and MRS and by the EU (3D Genome, LSHG-CT-2003-503441) to TC. HF and NPC were supported by the Wellcome Trust.
Abbreviations
- 3D
three-dimensional
- 2D
two-dimensional
- ARD-NB
average relative distances to the nuclear border
- CT
chromosome territory
- CT-IC
chromosome territory-interchromatin compartment
- (ΔD)
relative distance difference
- (e)ADS
(enhanced) Absolute 3D distances to surface
- G-band
giemsa dark band
- Hfb
human fibroblasts
- Hly
human (B- and T-) lymphocytes
- HSA
homo sapiens chromosome
- M-FISH
Multicolor fluorescence in situ hybridization
- R-band
giemsa light (reverse) band
- RIDGEs
regions of increased gene expression
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00412-007-0098-4) contains supplementary material, which is available to authorized users.
Contributor Information
Katrin Küpper, Department of Biology II, Anthropology and Human Genetics, Ludwig Maximilians University, Munich, Germany.
Alexandra Kölbl, Department of Biology II, Anthropology and Human Genetics, Ludwig Maximilians University, Munich, Germany.
Dorothee Biener, Department of Biology II, Anthropology and Human Genetics, Ludwig Maximilians University, Munich, Germany.
Sandra Dittrich, Department of Biology II, Anthropology and Human Genetics, Ludwig Maximilians University, Munich, Germany.
Johann von Hase, Kirchhoff Institute for Physics, University of Heidelberg, Heidelberg, Germany.
Tobias Thormeyer, Department of Biology II, Anthropology and Human Genetics, Ludwig Maximilians University, Munich, Germany.
Heike Fiegler, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Cambridge, UK.
Nigel P. Carter, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Cambridge, UK
Michael R. Speicher, Institute of Medical Biology and Human Genetics, Medical University of Graz, Graz, Austria
Thomas Cremer, Department of Biology II, Anthropology and Human Genetics, Ludwig Maximilians University, Munich, Germany.
Marion Cremer, Department of Biology II, Anthropology and Human Genetics, Ludwig Maximilians University, Munich, Germany, e-mail: Marion.Cremer@lrz.uni-muenchen.de.
References
- Alexandrova O, Solovei I, Cremer T, David CN. Replication labeling patterns and chromosome territories typical of mammalian nuclei are conserved in the early metazoan Hydra. Chromosoma. 2003;112:190–200. doi: 10.1007/s00412-003-0259-z. [DOI] [PubMed] [Google Scholar]
- Bartova E, Kozubek S. Nuclear architecture in the light of gene expression and cell differentiation studies. Biol Cell. 2006;98:323–336. doi: 10.1042/BC20050099. [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Bridger JM, Boyle S, Kill IR, Bickmore WA. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol. 2000;10:149–152. doi: 10.1016/s0960-9822(00)00312-2. [DOI] [PubMed] [Google Scholar]
- Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, Voute PA, Heisterkamp S, van Kampen A, Versteeg R. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- Claussen U. Chromosomics. Cytogenet Genome Res. 2005;111:101–106. doi: 10.1159/000086377. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci USA. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig JM, Bickmore WA. Chromosome bands—flavours to savour. BioEssays. 1993;15:349–354. doi: 10.1002/bies.950150510. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur J Histochem. 2006;50:223–272. [PubMed] [Google Scholar]
- Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001b;9:541–567. doi: 10.1023/a:1012495201697. [DOI] [PubMed] [Google Scholar]
- Cremer M, Kupper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol. 2003;162:809–820. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Kupper K, Dietzel S, Fakan S. Higher order chromatin architecture in the cell nucleus: on the way from structure to function. Biol Cell. 2004;96:555–567. doi: 10.1016/j.biolcel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories—a functional nuclear landscape. Curr Opin Cell Biol. 2006a;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Cremer M, Weierich C, Solovei I. Epigenetics protocols database: multicolor 3D-FISH in vertebrate cells. edited by the epigenome network of excellence. 2006b. http://www.epigenome-noe.net/researchtools/protocols.php.
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel S, Schiebel K, Little G, Edelmann P, Rappold GA, Eils R, Cremer C, Cremer T. The 3D positioning of ANT2 and ANT3 genes within female X chromosome territories correlates with gene activity [in process citation] Exp Cell Res. 1999;252:363–375. doi: 10.1006/excr.1999.4635. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD. The evolution of isochores. Nat Rev Genet. 2001;2:549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- Federico C, Scavo C, Cantarella CD, Motta S, Saccone S, Bernardi G. Gene-rich and gene-poor chromosomal regions have different locations in the interphase nuclei of cold-blooded vertebrates. Chromosoma. 2006;115:123–128. doi: 10.1007/s00412-005-0039-z. [DOI] [PubMed] [Google Scholar]
- Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Scott CE, Smith J, Vetrie D, Gorman P, Tomlinson IP, Carter NP. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- Fiegler H, Redon R, Andrews D, Scott C, Andrews R, Carder C, Clark R, Dovey O, Ellis P, Feuk L, Hunt P, Kalaitzopoulos D, Larkin J, Montgomery L, Perry GH, Plumb BW, Porter K, Rigby RE, Rigler D, Valesia A, Langford C, Humphray SW, Scherer SW, Lee C, Hurles ME, Carter NP. Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res. 2006;16:1566–1574. doi: 10.1101/gr.5630906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster HA, Bridger JM. The genome and the nucleus: a marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma. 2005;114:212–229. doi: 10.1007/s00412-005-0016-6. [DOI] [PubMed] [Google Scholar]
- Francke U. Digitized and differentially shaded human chromosome ideograms for genomic applications. Cytogenet Cell Genet. 1994;65:206–218. doi: 10.1159/000133633. [DOI] [PubMed] [Google Scholar]
- Furey TS, Haussler D. Integration of the cytogenetic map with the draft human genome sequence. Hum Mol Genet. 2003;12:1037–1044. doi: 10.1093/hmg/ddg113. [DOI] [PubMed] [Google Scholar]
- Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Gilchrist S, Bickmore WA. Chromatin organization in the mammalian nucleus. Int Rev Cytol. 2005;242:283–336. doi: 10.1016/S0074-7696(04)42007-5. [DOI] [PubMed] [Google Scholar]
- Grimwood J, Gordon LA, Olsen A, Terry A, Schmutz J, Lamerdin J, Hellsten U, Goodstein D, Couronne O, Tran-Gyamfi M, Aerts A, Altherr M, Ashworth L, Bajorek E, Black S, Branscomb E, Caenepeel S, Carrano A, Caoile C, Chan YM, Christensen M, Cleland CA, Copeland A, Dalin E, Dehal P, Denys M, Detter JC, Escobar J, Flowers D, Fotopulos D, Garcia C, Georgescu AM, Glavina T, Gomez M, Gonzales E, Groza M, Hammon N, Hawkins T, Haydu L, Ho I, Huang W, Israni S, Jett J, Kadner K, Kimball H, Kobayashi A, Larionov V, Leem SH, Lopez F, Lou Y, Lowry S, Malfatti S, Martinez D, McCready P, Medina C, Morgan J, Nelson K, Nolan M, Ovcharenko I, Pitluck S, Pollard M, Popkie AP, Predki P, Quan G, Ramirez G, Rash S, Retterer J, Rodriguez A, Rogers S, Salamov A, Salazar A, She X, Smith D, Slezak T, Solovyev V, Thayer N, Tice H, Tsai M, Ustaszewska A, Vo N, Wagner M, Wheeler J, Wu K, Xie G, Yang J, Dubchak I, Furey TS, DeJong P, Dickson M, Gordon D, Eichler EE, Pennacchio LA, Richardson P, Stubbs L, Rokhsar DS, Myers RM, Rubin EM, Lucas SM. The DNA sequence and biology of human chromosome 19. Nature. 2004;428:529–535. doi: 10.1038/nature02399. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Habermann FA, Cremer M, Walter J, Kreth G, von Hase J, Bauer K, Wienberg J, Cremer J, Cremer T, Solovei I. Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res. 2001;9:569–584. doi: 10.1023/a:1012447318535. [DOI] [PubMed] [Google Scholar]
- Holmquist G, Gray M, Porter T, Jordan J. Characterization of Giemsa dark-and light-band DNA. Cell. 1982;31:121–129. doi: 10.1016/0092-8674(82)90411-1. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Groudine M. Form follows function: the genomic organization of cellular differentiation. Genes Dev. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- Kurz A, Lampel S, Nickolenko JE, Bradl J, Benner A, Zirbel RM, Cremer T, Lichter P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol. 1996;135:1195–1205. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture and the regulation of gene expression. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Lehrer H, Weise A, Michel S, Starke H, Mrasek K, Heller A, Kuechler A, Claussen U, Liehr T. The hierarchically organized splitting of chromosome bands into sub-bands analyzed by multicolor banding (MCB) Cytogenet Genome Res. 2004;105:25–28. doi: 10.1159/000078005. [DOI] [PubMed] [Google Scholar]
- Lemke J, Claussen J, Michel S, Chudoba I, Muhlig P, Westermann M, Sperling K, Rubtsov N, Grummt UW, Ullmann P, Kromeyer-Hauschild K, Liehr T, Claussen U. The DNA-based structure of human chromosome 5 in interphase. Am J Hum Genet. 2002;71:1051–1059. doi: 10.1086/344286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasova E, Kozubek S, Kozubek M, Falk M, Amrichova J. The 3D structure of human chromosomes in cell nuclei. Chromosome Res. 2002;10:535–548. doi: 10.1023/a:1020958517788. [DOI] [PubMed] [Google Scholar]
- Lukasova E, Kozubek S, Falk M, Kozubek M, Zaloudik J, Vagunda V, Pavlovsky Z. Topography of genetic loci in the nuclei of cells of colorectal carcinoma and adjacent tissue of colonic epithelium. Chromosoma. 2004;112:221–230. doi: 10.1007/s00412-003-0263-3. [DOI] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol. 2002;159:753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraldi NM, Squarzoni S, Sabatelli P, Capanni C, Mattioli E, Ognibene A, Lattanzi G. Laminopathies: involvement of structural nuclear proteins in the pathogenesis of an increasing number of human diseases. J Cell Physiol. 2005;203:319–327. doi: 10.1002/jcp.20217. [DOI] [PubMed] [Google Scholar]
- Misteli T. Spatial positioning; a new dimension in genome function. Cell. 2004;119:153–156. doi: 10.1016/j.cell.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Murmann AE, Gao J, Encinosa M, Gautier M, Peter ME, Eils R, Lichter P, Rowley JD. Local gene density predicts the spatial position of genetic loci in the interphase nucleus. Exp Cell Res. 2005;311:14–26. doi: 10.1016/j.yexcr.2005.07.020. 311:14–26. [DOI] [PubMed] [Google Scholar]
- Neusser M, Schubel V, Koch A, Cremer T, Mueller S. Comparative analysis of the three-dimensional genome architecture in interphase nuclei of primates. Chromosoma. 2007 doi: 10.1007/s00412-007-0099-3. DOI 10.1007/s00412-007-0099-3. [DOI] [PubMed] [Google Scholar]
- Nusbaum C, Zody MC, Borowsky ML, Kamal M, Kodira CD, Taylor TD, Whittaker CA, Chang JL, Cuomo CA, Dewar K, FitzGerald MG, Yang X, Abouelleil A, Allen NR, Anderson S, Bloom T, Bugalter B, Butler J, Cook A, DeCaprio D, Engels R, Garber M, Gnirke A, Hafez N, Hall JL, Norman CH, Itoh T, Jaffe DB, Kuroki Y, Lehoczky J, Lui A, Macdonald P, Mauceli E, Mikkelsen TS, Naylor JW, Nicol R, Nguyen C, Noguchi H, O'Leary SB, O'Neill K, Piqani B, Smith CL, Talamas JA, Topham K, Totoki Y, Toyoda A, Wain HM, Young SK, Zeng Q, Zimmer AR, Fujiyama A, Hattori M, Birren BW, Sakaki Y, Lander ES. DNA sequence and analysis of human chromosome 18. Nature. 2005;437:551–555. doi: 10.1038/nature03983. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Parada LA, Sotiriou S, Misteli T. Spatial genome organization. Exp Cell Res. 2004;296:64–70. doi: 10.1016/j.yexcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Pederson T. The spatial organization of the genome in mammalian cells. Curr Opin Genet Dev. 2004;14:203–209. doi: 10.1016/j.gde.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Postberg J, Alexandrova O, Cremer T, Lipps HJ. Exploiting nuclear duality of ciliates to analyse topological requirements for DNA replication and transcription. J Cell Sci. 2005;118:3973–3983. doi: 10.1242/jcs.02497. [DOI] [PubMed] [Google Scholar]
- Ragoczy T, Telling A, Sawado T, Groudine M, Kosak ST. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 2003;11:513–525. doi: 10.1023/a:1024939130361. [DOI] [PubMed] [Google Scholar]
- Saccone S, Federico C, Bernardi G. Localization of the gene-richest and the gene-poorest isochores in the interphase nuclei of mammals and birds. Gene. 2002;300:169–178. doi: 10.1016/s0378-1119(02)01038-7. [DOI] [PubMed] [Google Scholar]
- Sadoni N, Langer S, Fauth C, Bernardi G, Cremer T, Turner BM, Zink D. Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments. J Cell Biol. 1999;146:1211–1226. doi: 10.1083/jcb.146.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh Y, Laemmli UK. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell. 1994;76:609–622. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Scheuermann MO, Tajbakhsh J, Kurz A, Saracoglu K, Eils R, Lichter P. Topology of genes and nontranscribed sequences in human interphase nuclei. Exp Cell Res. 2004;301:266–279. doi: 10.1016/j.yexcr.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol. 2003;162:981–990. doi: 10.1083/jcb.200303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase J, Stein S, Vincent S, Molloy KR, Kreth G, Cremer C, Bult CJ, O'Brien TP. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol. 2006;174:27–38. doi: 10.1083/jcb.200603083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006 doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Singer GA, Lloyd AT, Huminiecki LB, Wolfe KH. Clusters of co-expressed genes in mammalian genomes are conserved by natural selection. Mol Biol Evol. 2005;22:767–775. doi: 10.1093/molbev/msi062. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Sproul D, Gilbert N, Bickmore WA. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet. 2005;6:775–781. doi: 10.1038/nrg1688. [DOI] [PubMed] [Google Scholar]
- Tanabe H, Muller S, Neusser M, von Hase J, Calcagno E, Cremer M, Solovei I, Cremer C, Cremer T. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc Natl Acad Sci USA. 2002;99:4424–4429. doi: 10.1073/pnas.072618599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Belmont AS. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat Cell Biol. 2001;3:134–139. doi: 10.1038/35055033. [DOI] [PubMed] [Google Scholar]
- van Driel R, Fransz PF, Verschure PJ. The eukaryotic genome: a system regulated at different hierarchical levels. J Cell Sci. 2003;116:4067–4075. doi: 10.1242/jcs.00779. [DOI] [PubMed] [Google Scholar]
- Versteeg R, van Schaik BD, van Batenburg MF, Roos M, Monajemi R, Caron H, Bussemaker HJ, van Kampen AH. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13:1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. DNA helix: the importance of being GC-rich. Nucleic Acids Res. 2003;31:1838–1844. doi: 10.1093/nar/gkg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, Trowsdale J, Sheer D. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- White EJ, Emanuelsson O, Scalzo D, Royce T, Kosak S, Oakeley EJ, Weissman EJ, Gerstein M, Groudine M, Snyder M, Schubeler D. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc Natl Acad Sci USA. 2004;101:17771–17776. doi: 10.1073/pnas.0408170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RR. Transcription and the territory: the ins and outs of gene positioning. Trends Genet. 2003;19:298–302. doi: 10.1016/S0168-9525(03)00109-4. [DOI] [PubMed] [Google Scholar]
- Williams RR, Broad S, Sheer D, Ragoussis J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp Cell Res. 2002;272:163–175. doi: 10.1006/excr.2001.5400. [DOI] [PubMed] [Google Scholar]
- Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP. Replication timing of the human genome. Hum Mol Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- Wurtele H, Chartrand P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended chromosome conformation capture methodology. Chromosome Res. 2006;14:445–477. doi: 10.1007/s10577-006-1075-0. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Singh Sandhu K, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- Zink D. The temporal program of DNA replication: new insights into old questions. Chromosoma. 2006;115:273–287. doi: 10.1007/s00412-006-0062-8. [DOI] [PubMed] [Google Scholar]
- Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, Rosenecker J, Schindelhauer D. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.