Abstract
We report on a patient with mental retardation, seizures and tall stature with advanced bone age in whom a de novo apparently balanced chromosomal rearrangement 46,XX,t(X;9)(q12;p13.3) was identified. Using array CGH on flow-sorted derivative chromosomes (array painting) and subsequent FISH and qPCR analysis, we mapped and sequenced both breakpoints. The Xq12 breakpoint was located within the gene coding for oligophrenin 1 (OPHN1) whereas the 9p13.3 breakpoint was assigned to a non-coding segment within a gene dense region. Disruption of OPHN1 by the Xq12 breakpoint was considered the major cause of the abnormal phenotype observed in the proband.
Keywords: Translocation, OPHN1, Tall stature, Mental retardation
1. Introduction
A small subset of patients with mental retardation and/or congenital abnormalities present with an apparently balanced de novo chromosomal rearrangement. In most patients it is assumed that the observed phenotypic anomalies are the result of submicroscopic deletions or duplications or alternatively disruption, activation or inactivation of a gene or genes located at or near the breakpoints. Until now, only a limited number of such apparently balanced de novo rearrangements have been investigated to the basepair level. In some instances detailed analysis has led to the identification of disease related genes [2,3,12]. Here we describe the combined application of array painting [8], FISH and real-time quantitative PCR which enabled us to map and sequence the breakpoints of a balanced reciprocal translocation t(X;9)(q12;p13.3) in a girl with mental retardation and tall stature.
2. Materials and methods
2.1. G-banding
Karyotyping was performed on short term lymphocyte cultures from peripheral blood with G-banding. Karyotypes were described according to the guidelines of the ISCN 2005.
2.2. Chromosome flow sorting
Purification of the translocated chromosomes, derivative 9 and X, was carried out using a flow cytometer (MoFlo®, DAKO) as described previously [4,8]. DNA from the flow sorted chromosomes was used as template for rolling-circle amplification (RCA) with Repli-G (Molecular Staging). The amplified DNA was subsequently used as template DNA for array painting, qPCR and sequencing [8].
2.3. Array painting
Using random prime labeling, 500 ng of amplified derivative chromosome 9 and derivative chromosome X DNA was labeled with Cy3 and Cy5 respectively (BioPrime Array CGH Genomic Labeling System, Invitrogen). Repetitive sequences were suppressed with 100 μg Cot-1 DNA (Invitrogen) and 400 μg yeast tRNA. The labeled fragments were resuspended in 60 μl hybridization buffer at 37 °C (50% formamide, 10% dextran sulphate, 0.1% Tween 20, 2× SSC, 10 mM Tris pH 7.4). In-house produced 1 Mb BAC arrays were prehybridized at 37 °C during 1 h using 50 μg Cot-1 DNA (Invitrogen) and 150 μg herring sperm DNA, resuspended in 120 μl hybridization buffer. After removal of the prehybridization mixture, DNA from both derivatives was simultaneously hybridized for 48 h at 37 °C. The slides were washed in 1× PBS/0.05% Tween 20 for 10 min at room temperature, 50% formamide/2× SSC for 30 min at 42 °C and finally 1× PBS/0.05% Tween 20 for 10 min at room temperature. After centrifuge drying, the slides were scanned using a GMS 418 Array Scanner (MWG). The scan images were processed with Imagene software (Biodiscovery) and further analyzed with our in-house developed and freely available software tool arrayCGHbase (http://medgen.ugent.be/arraycghbase/) [13]. Data points were excluded from analysis if one of the following criteria were fulfilled: signal to noise ratio <5; standard deviation of the log2 transformed ratios between triplicates >0.2; only one informative replicate.
2.4. FISH with region specific probes
Fluorescence in situ hybridization (FISH) was performed as described [18]. Locus specific RPCI-BAC probes for chromosome X and 9 were obtained by screening several assemblies (http://genome.ucsc.edu/) of the human genome project. All probes were relocated to the March 2006 Genome assembly (Table 1). BAC probes were labeled either with digoxigenin or biotin and hybridized on patients' metaphase chromosomes.
Table 1.
BAC clones used for breakpoint mapping on chromosome X and chromosome 9 with their name, sanger name, chromosome, start position and end position according to the NCBI 36 genome assembly
| Name | Sanger name | Chr | Start (bp) | End (bp) |
|---|---|---|---|---|
| RP11-48L13 | bA48L13 | 9p21.1 | 29493271 | 29639053 |
| RP11-630H9 | bA630H9 | 9p21.1 | 30404961 | 30535678 |
| RP11-133O22 | bA133O22 | 9p13.3 | 33773698 | 33788695 |
| RP11-331F9 | bA331F9 | 9p13.3 | 35581889 | 35670216 |
| RP11-112J3 | bA112J3 | 9p13.3 | 35670117 | 35865495 |
| RP11-327L3 | bA327L3 | 9p13.3 | 35865396 | 35965281 |
| RP11-113A10 | bA113A10 | 9p13.3 | 35965182 | 36088594 |
| RP11-421H8 | bA421H8 | 9p13.3 | 36088495 | 36279930 |
| RP11-84P7 | bA84P7 | 9p13.3 | 36279831 | 36356857 |
| RP11-405L18 | bA405L18 | 9p13.2 | 37453976 | 37496328 |
| RP11-392E22 | bA392E22 | 9p13.2 | 38474287 | 38559167 |
| RP11-274B18 | bA274B18 | 9q21.11 | 70318668 | 70488660 |
| RP4-808O4 | dJ808O4 | Xq12 | 66577626 | 66716658 |
| RP6-201G10 | dA201G10 | Xq12 | 67010325 | 67200935 |
| RP11-516A11 | bA516A11 | Xq12 | 67200836 | 67342790 |
| RP3-360E18 | dJ360E18 | Xq12 | 67342258 | 67474160 |
| RP13-469L5 | bB469L5 | Xq13.1 | 67813008 | 67875523 |
| RP13-178D16 | bB178D16 | Xq13.1 | 67875424 | 67925275 |
| RP11-446J19 | bA446J19 | Xq13.1 | 68510619 | 68634148 |
BAC clones in bold are breakpoint spanning clones (chromosome X) or breakpoint flanking clones (chromosome 9). Clones in italics are clones from the Sanger 1 Mb array.
2.5. Fiber-FISH
Fiber-FISH analysis was performed according to Speleman et al. [16]. Fiber-FISH slides were prepared from a lymphoblastoid cell line of the patient (EBV689) and a normal male control cell line (EBV99) and hybridized with probes RP11-331F9, RP11-112J3 on chromosome 9 and probe RP3-360E18 on chromosome X, respectively flanking and spanning the breakpoint.
2.6. qPCR analysis
Primers for qPCR were designed to amplify products along the sequence interval of interest determined by fiber-FISH. qPCR was performed as described previously [19]. Once the breakpoints were found to lie between two primers, further primers were chosen to amplify fragments in between. Junction fragments were generated by using forward and reverse primers from the different chromosome sequences at either side of the breakpoints.
2.7. Sequencing
The junction fragments were cleaned with exo-SAP (GE Healthcare) and sequenced by using the di-deoxy chain terminator method [15] with the BigDye v3.1 ET terminator cycle sequencing kit from Applied Biosystems. After the sequencing reactions, the products were electrophoresed on a 3100 Genetic Analyzer and analyzed with Sequence Analysis and SeqScape (Applied Biosystems).
3. Results
3.1. Clinical findings
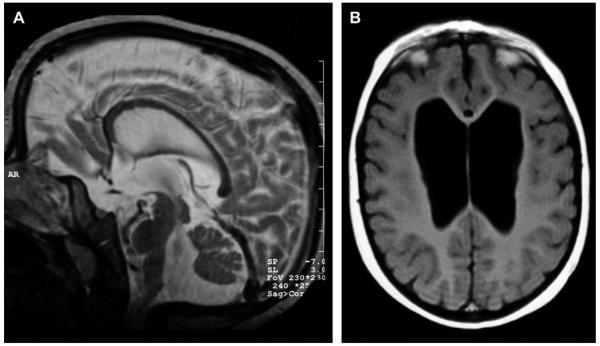
The proband was born at 38 weeks gestation after an uncomplicated pregnancy. The girl was the first child from healthy non-consanguineous parents. Birth weight was 2580 g (P10), length 45.5 cm (P10 = 47 cm) and head circumference 33 cm (P10–P25). The postnatal course was uneventful. The girl had a delayed neuromotor development with sitting at 11 months and walking at 23 months. She developed a first epileptic insult at the age of 10 months for which she was treated with phenobarbital. The seizures were characterized by upward deviation of the eyes, anteflexion of the head and flexion of the legs. The electroencephalogram did not show signs of hypsarythmia. A first evaluation at the age of 14.5 months revealed a weight of 13.9 kg (P90 = 12 kg), length of 82 cm (P90 = 81 cm) and head circumference of 47.5 cm (P50=–P90). There was no clear facial dysmorphism at that age. Just intermittent strabismus, a receding frontal hairline and mild hypotelorism were noted. Diagnostic work-up because of the seizures revealed a de novo balanced reciprocal translocation between the long arm of the X chromosome and the short arm of chromosome 9 (46,XX,t(X;9)(q12;p13)). An MRI of the brain showed moderately enlarged cerebral ventricles, as well as an enlarged fourth ventricle, with the frontal horn of the lateral ventricles most prominently enlarged. Bilateral hypoplasia of the head of the caudate nucleus contributed to the balloon-like appearance of the frontal horn. There was vermian hypoplasia, associated with an infracerebellar CSF collection continuous with the fourth ventricle. The brain stem had a relatively small appearance (Fig. 1). Around the age of 3 years the anti-epileptic treatment was switched from phenobarbital to valproate. At the age of 5 years she started in special education school. Psychometric evaluation at that age revealed an IQ score of 60. On follow-up she developed an accelerated growth with shift of all parameters at or above the 90th centile. According to the bone age atlas of Greulich & Pyle, her bone age at the age of 8 years was between 11 and 12 years. On the last physical examination she was 113/12 years old. Weight was 69.4 kg (+4.6 sd) (BMI: 28.5), length 155.7 cm (+1.38 sd) and head circumference 54.2 cm (+1.47 sd). She had a friendly and affectionate personality. Her speech was not so easy to understand. When talking, she tended to hold her head in an oblique position, thus looking mainly with the left eye. Dysmorphic facial features included prominent incisors and relatively large ears (Fig. 2). On the trunk a supernumerary nipple on both sides was observed. A mild extension deficit was present in the elbows and knees. In the upright position, she was holding her knees in mild flexion. Her gait was slow and unsteady. Her feet were small and flat.
Fig. 1.
Neuroradiological findings (A): Sagittal spin-echo T2 image shows vermian and brain stem hypoplasia, with infracerebellar CSF collection and enlarged fourth ventricle. (B) Axial spin-echo T1 image shows enlarged lateral ventricles, with the frontal horns most prominently enlarged.
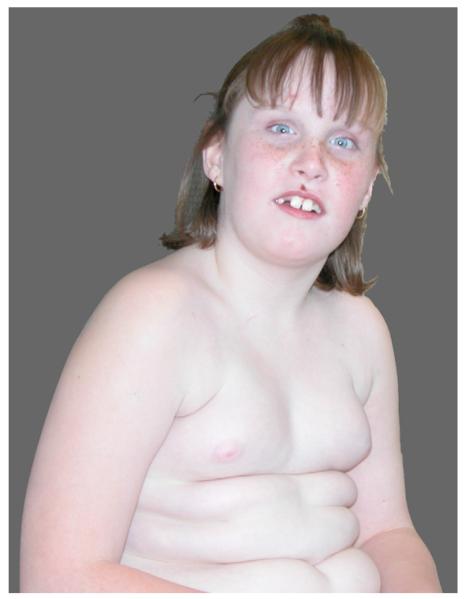
Fig. 2.
Clinical photograph of the proband at the age of 8 years showing obesity and mild facial dysmorphism with prominent maxillary incisors, relatively large ears and high forehead.
3.2. Molecular cytogenetic findings
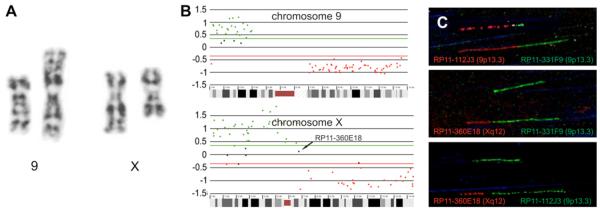
Partial G-banded karyotypes of the apparently balanced chromosomal rearrangement are shown in Fig. 3A. The karyotype could be described as 46,XX,t(X;9)(q12;p13). Following chromosome sorting and rolling circle amplification, array painting was performed (Fig. 3B). Together with FISH analysis using region specific probes (Table 1), the breakpoint could be localized in BAC RP11-360E18 on chromosome Xq12 and in between BACs RP11-331F9 and RP11-112J3 on chromosome 9p13.3 (Fig. 3B). The breakpoints were fine mapped using fiber FISH and could be further localised in RP11-360E18 and RP11-331F9 to an accuracy of ~20 kb (Fig. 3C).
Fig. 3.
Derivative chromosome 9 and derivative chromosome X after G-banding. (A) array CGH profile after array painting for chromosome X and chromosome 9. (B) Fiber-FISH results with BAC clones RP11-112J3 (9p13.3, red), RP11-360E18 (Xq12, red) and BAC clones RP11-331F9 (9p13.3, green) and RP11-112J3 (9p13.3, green) (C).
Consecutive qPCRs allowed us to narrow down the breakpoint on chromosome X between 67,404,215 bp and 67,407,331 bp (3116 bp) and on chromosome 9 between 35,656,338 bp and 35,656,776 bp (438 bp). Subsequently, using appropriate primers on both derivatives, both breakpoints were amplified and sequenced (Fig. 4).
Fig. 4.
Sequence of both breakpoints. A for derivative 9 and B for derivative X. The nucleotides in bold represent chromosome 9 sequence, the underlined nucleotides represent primer sequence used for cloning, the adenine between brackets indicates one basepair that is lost either from chromosome 9 or chromosome X.
4. Discussion
We report on a girl with mental retardation (IQ = 60), seizures and tall stature with advanced bone age. Congenital abnormalities of the internal organs were not observed. Only a mild craniofacial dysmorphism with strabismus, a receding frontal hairline and mild hypotelorism was noted. On the trunk, a supernumerary nipple on both sides was present. Brain MRI revealed a mild dilatation of the ventricular system. Karyotyping was performed and revealed an apparently balanced translocation: 46,XX,t(X;9)(q12;p13). Breakpoints were sequenced after flow-sorting and array painting in order to determine their genomic position.
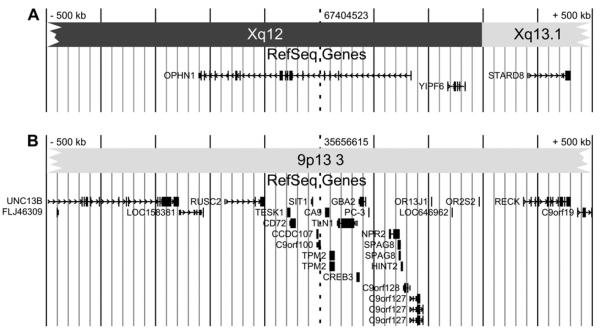
On Xq12, the breakpoint interrupts the 390 kb large oligophrenin 1 gene (OPHN1) (Fig. 5). The OPHN1 gene was identified through the breakpoint analysis of a female patient with a t(X;12) translocation [2]. OPHN1 has 25 exons and encodes a Rho-GTPase-activating protein. The Rho proteins are important mediators of intracellular signal transduction affecting cell migration, cell morphogenesis and synapse maturation [3]. The oligophrenin protein regulates cytoskeletal dynamics through Rho-GTPase modulation and is specifically involved in dendritic spine morphogenesis [9]. Of all the genes involved in X-linked mental retardation, six encode regulators or effectors of Rho-GTPase proteins, suggesting an important role of the Rho signalling pathway in cognitive functions [21]. Oligophrenin expression is highest in the developing central nervous system, and more precisely in the neuronal and glial cells [7]. In adult life, oligophrenin expression is enriched in the hippocampus, the olfactory bulb, and the Purkinje cell layer of the cerebellum.
Fig. 5.
Genomic context of the breakpoints on chromosome X (A) and chromosome 9 (B) with the RefSeq genes (UCSC Genome Browser on March 2006 Assembly) in a 500 kb interval upstream and downstream of the breakpoint. The dotted line indicates the breakpoint.
Although OPHN1 mutations were first described to cause a rather non-specific form of mental retardation [3], several publications have now highlighted the recognizable phenotype with neurological abnormalities and mild facial dysmorphism [1,5,21]. Hypoplasia of the cerebellum is frequently observed on brain imaging studies and seem to result in dysmetria, adiadochokinesia, and oculomotor problems (nystagmus, strabismus, external ophthalmoplegia) in the affected individuals [6,17]. Ataxia is rarely observed. Seizures are commonly reported. The expression of oligophrenin 1 in the craniofacial skeleton, especially at the level of the mandible [21] may explain the facial dysmorphism seen in some patients with the “oligophrenin 1 syndrome”. The craniofacial dysmorphism is more pronounced in older patients and includes a long face with prominent chin, hypotelorism, deep-set eyes with prominent supraorbital ridges, long tubular nose, short philtrum, and thin upper lip. Macrocephaly and tall stature have also been reported in several families [14,21], In accordance with its X-linked recessive inheritance pattern, the “oligophrenin 1 syndrome” phenotype is usually encountered in affected males, with female carriers only showing a rather mild and non-specific phenotype (learning difficulties and strabismus) unless they carry an X;autosome translocation as is illustrated by our patient. In females with an X;autosome translocation, X inactivation initially occurs at random but is followed by cellular selection, favouring the cells without a partial autosome inactivation. Accordingly, nearly 95% of females with a balanced X;autosome translocation show a skewed X-inactivation pattern with the normal X chromosome inactivated in almost all cells [11]. This explains that females bearing an X;autosome translocation are as severely affected as males.
The chromosome 9p13 breakpoint is localised within a non-coding segment on chromosome 9. A position effect on the expression of the flanking genes cannot be ruled out. Position effects due to chromosomal aberrations, leading to altered expression of genes have been described up to 1.3 Mb of the breakpoint [10,20]. This possibility was not further investigated because the major part of the abnormal phenotype in our patient could be explained by the disruption of the OPHN1 gene.
So far, only a few constitutional, apparently balanced, translocations have been mapped at the sequence level, mostly due to the huge amount of experimental laboratory. However, the information on responsible genes obtained through above described efforts is of the utmost importance. The strategy followed in this study is an example of a clear cut way to facilitate the analysis of balanced translocations, enabling to narrow down the translocation breakpoints in only a limited number of experiments.
Although technically challenging, this study further illustrates the feasibility for rapid breakpoint mapping and sequencing, using a combined approach of chromosome flow sorting (or alternatively microdissection), linear DNA amplification and array CGH. It can be anticipated that similar studies on larger series of de novo apparently balanced translocations in mentally retarded patients will lead to the discovery of several new genes as a cause for mental retardation.
Acknowledgements
We are grateful to the patient and her family for their cooperation. The authors wish to thank the Mapping Core and Map Finishing groups of the Wellcome Trust Sanger Institute for initial clone supply and verification, we also would like to thank Marie-Louise Duits for providing us with the MRI images of the patient. This work was made possible by grant G.0200.03 from the FWO and GOA grant 12051203 from the Ghent University. BLN and NPC are supported by the Wellcome Trust.
KB is a Research Assistant and JV a Postdoctoral Researcher of the Research Foundation – Flanders (FWO – Vlaanderen). This study is supported, in part, by the Fund for Scientific Research, Flanders, with a mandate fundamental clinical research to GRM.
This text presents research results of the Belgian program of Interuniversity Poles of attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming (IUAP).
References
- 1.Bergmann C, Zerres K, Senderek J, Rudnik-Schoneborn S, Eggermann T, Hausler M, Mull M, Ramaekers VT. Oligophrenin 1 (OPHN1) gene mutation causes syndromic X-linked mental retardation with epilepsy, rostral ventricular enlargement and cerebellar hypoplasia. Brain. 2003;126:1537–1544. doi: 10.1093/brain/awg173. [DOI] [PubMed] [Google Scholar]
- 2.Bienvenu T, Der-Sarkissian H, Billuart P, Tissot M, Des Portes V, Bruls T, Chabrolle JP, Chauveau P, Cherry M, Kahn A, Cohen D, Beldjord C, Chelly J, Cherif D. Mapping of the X-breakpoint involved in a balanced X;12 translocation in a female with mild mental retardation. Eur. J. Hum. Genet. 1997;5:105–109. [PubMed] [Google Scholar]
- 3.Billuart P, Bienvenu T, Ronce N, des Portes V, Vinet MC, Zemni R, Crollius H. Roest, Carrie A, Fauchereau F, Cherry M, Briault S, Hamel B, Fryns JP, Beldjord C, Kahn A, Moraine C, Chelly J. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;392:923–926. doi: 10.1038/31940. [DOI] [PubMed] [Google Scholar]
- 4.Carter NP. Bivariate chromosome analysis using a commercial flow cytometer. Methods Mol. Biol. 1994;29:187–204. doi: 10.1385/0-89603-289-2:187. [DOI] [PubMed] [Google Scholar]
- 5.Chabrol B, Girard N, N'Guyen K, Gerard A, Carlier M, Villard L, Philip N. Delineation of the clinical phenotype associated with OPHN1 mutations based on the clinical and neuropsychological evaluation of three families. Am. J. Med. Genet. 2005;138:314–317. doi: 10.1002/ajmg.a.30882. [DOI] [PubMed] [Google Scholar]
- 6.des Portes V, Boddaert N, Sacco S, Briault S, Maincent K, Bahi N, Gomot M, Ronce N, Bursztyn J, Adamsbaum C, Zilbovicius M, Chelly J, Moraine C. Specific clinical and brain MRI features in mentally retarded patients with mutations in the Oligophrenin-1 gene. Am. J. Med. Genet. 2004;124:364–371. doi: 10.1002/ajmg.a.20422. [DOI] [PubMed] [Google Scholar]
- 7.Fauchereau F, Herbrand U, Chafey P, Eberth A, Koulakoff A, Vinet MC, Ahmadian MR, Chelly J, Billuart P. The RhoGAP activity of OPHN1, a new F-actin-binding protein, is negatively controlled by its amino-terminal domain. Mol. Cell. Neurosci. 2003;23:574–586. doi: 10.1016/s1044-7431(03)00078-2. [DOI] [PubMed] [Google Scholar]
- 8.Fiegler H, Gribble SM, Burford DC, Carr P, Prigmore E, Porter KM, Clegg S, Crolla JA, Dennis NR, Jacobs P, Carter NP. Array painting: a method for the rapid analysis of aberrant chromosomes using DNA microarrays. J. Med. Genet. 2003;40:664–670. doi: 10.1136/jmg.40.9.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govek EE, Newey SE, Akerman CJ, Cross JR, Van der Veken L, Van Aelst L. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nat. Neurosci. 2004;7:364–372. doi: 10.1038/nn1210. [DOI] [PubMed] [Google Scholar]
- 10.Kleinjan DJ, van Heyningen V. Position effect in human genetic disease. Hum. Mol. Genet. 1998;7:1611–1618. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- 11.Mattei MG, Mattei JF, Ayme S, Giraud F. X-autosome translocations: cytogenetic characteristics and their consequences. Hum. Genet. 1982;61:295–309. doi: 10.1007/BF00276593. [DOI] [PubMed] [Google Scholar]
- 12.McMullan TW, Crolla JA, Gregory SG, Carter NP, Cooper RA, Howell GR, Robinson DO. A candidate gene for congenital bilateral isolated ptosis identified by molecular analysis of a de novo balanced translocation. Hum. Genet. 2002;110:244–250. doi: 10.1007/s00439-002-0679-5. [DOI] [PubMed] [Google Scholar]
- 13.Menten B, Pattyn F, De Preter K, Robbrecht P, Michels E, Buysse K, Mortier G, De Paepe A, van Vooren S, Vermeesch J, Moreau Y, De Moor B, Vermeulen S, Speleman F, Vandesompele J. arrayCGHbase: an analysis platform for comparative genomic hybridization microarrays. BMC Bioinformatics. 2005;6:124. doi: 10.1186/1471-2105-6-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philip N, Chabrol B, Lossi AM, Cardoso C, Guerrini R, Dobyns WB, Raybaud C, Villard L. Mutations in the oligophrenin-1 gene (OPHN1) cause X linked congenital cerebellar hypoplasia. J. Med. Genet. 2003;40:441–446. doi: 10.1136/jmg.40.6.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speleman F, Delattre O, Peter M, Hauben E, Van Roy N, Van Marck E. Malignant melanoma of the soft parts (clear-cell sarcoma): confirmation of EWS and ATF-1 gene fusion caused by a t(12;22) translocation. Mod. Pathol. 1997;10:496–499. [PubMed] [Google Scholar]
- 17.Tentler D, Gustavsson P, Leisti J, Schueler M, Chelly J, Timonen E, Anneren G, Willard HF, Dahl N. Deletion including the oligophrenin-1 gene associated with enlarged cerebral ventricles, cerebellar hypoplasia, seizures and ataxia. Eur. J. Hum. Genet. 1999;7:541–548. doi: 10.1038/sj.ejhg.5200320. [DOI] [PubMed] [Google Scholar]
- 18.Van Roy N, Laureys G, Van Gele M, Opdenakker G, Miura R, van der Drift P, Chan A, Versteeg R, Speleman F. Analysis of 1;17 translocation breakpoints in neuroblastoma: implications for mapping of neuroblastoma genes. Eur. J. Cancer. 1997;33:1974–1978. doi: 10.1016/s0959-8049(97)00319-5. [DOI] [PubMed] [Google Scholar]
- 19.Vandesompele J, De Paepe A, Speleman F. Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal. Biochem. 2002;303:95–98. doi: 10.1006/abio.2001.5564. [DOI] [PubMed] [Google Scholar]
- 20.Velagaleti GV, Bien-Willner GA, Northup JK, Lockhart LH, Hawkins JC, Jalal SM, Withers M, Lupski JR, Stankiewicz P. Position effects due to chromosome breakpoints that map approximately 900 Kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am. J. Hum. Genet. 2005;76:652–662. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zanni G, Saillour Y, Nagara M, Billuart P, Castelnau L, Moraine C, Faivre L, Bertini E, Durr A, Guichet A, Rodriguez D, des Portes V, Beldjord C, Chelly J. Oligophrenin 1 mutations frequently cause X-linked mental retardation with cerebellar hypoplasia. Neurology. 2005;65:1364–1369. doi: 10.1212/01.wnl.0000182813.94713.ee. [DOI] [PubMed] [Google Scholar]