Summary
Na+/H+ exchanger regulatory factor (NHERF1) is a signaling adaptor protein comprising two PDZ domains and a C-terminal ezrin-binding (EB) motif. To understand the role of intramolecular interactions in regulating its binding properties, we characterized the complex between the second PDZ domain PDZ2 and the C-terminal 242–358 fragment of NHERF1 using NMR and fluorescence methods. NMR chemical shift and relaxation data implicate 11 C-terminal residues in binding and, together with a thermodynamic analysis of mutant proteins, indicate that the EB region becomes helical when bound to PDZ2. Both specific contacts between PDZ2 and EB as well as non-specific interactions involving a 100-residue flexible linker contribute to stabilizing two structurally distinct closed conformations of NHERF1. The affinity of mutant proteins for an extrinsic ligand is inversely related to the helix-forming propensity of the EB motif. The findings provide a structural framework for understanding how autoinhibitory interactions modulated the binding properties of NHERF1.
Introduction
Na+/H+ exchanger regulatory factor 1 (NHERF1; also known as EBP50) is a multi-domain scaffolding protein localized in the apical membrane of polarized epithelial cells belonging to the Na+-H+ exchanger regulatory factor family of proteins [1, 2]. Members of this protein family interact with a number of tyrosine kinase receptors, G-protein coupled receptors and ion transport proteins [3, 4]. Interactions between NHERF1 and the actin-binding protein ezrin contribute to key cellular signaling events, including intracellular trafficking and assembly of protein complexes involving receptors and ion channels [5, 6]. In addition, NHERF1-ezrin interactions are involved in regulating cell shape and migration, and has been implicated in cancer through interactions with platelet-derived growth factor receptor (PDGF), merlin (NF-2), β-catenin and PTEN [7, 8].
NHERF1 contains two PSD-90/Dlg/ZO-1 homology (PDZ) domains in the N-terminal half of the protein, and a C-terminal domain (CT; residues 242–358) containing a ~14 residues ezrin binding (EB) motif at the extreme C-terminus (Figure 1A). Sequence-based algorithms [9] predict that the 100-residue segment linking PDZ2 and the EB motif is intrinsically disordered. PDZ domains are protein-protein recognition modules that organize diverse macromolecular complexes involved in cell signaling [10]. They recognize peptide motifs of 3–5 residues in length, usually at the C-terminus of the target protein and occasionally at internal locations in β-turns. Although the two PDZ domains of NHERF1 share 57.1% identical amino acids and are predicted to adopt similar structures, they differ in terms of their binding specificities and affinities [3]. For example, the C-terminus of cystic fibrosis transmembrane conductance regulator (CFTR) has a substantially higher affinity for binding PDZ1 than PDZ2 [2, 11, 12]. While the interaction of PDZ1 with the C-terminus of various ligand peptides has been characterized in detail by X-ray crystallography [13, 14], the complex of PDZ2 with target peptides, such as the C-terminus of CFTR, appears inherently more dynamic, and less is known about the functional role of PDZ2. Recent studies suggest that intramolecular interactions between the PDZ2 and CT domains of NHERF1 are important for regulating the binding properties of NHERF1. Li et al. [11] have shown that the interaction of PDZ2 to the C-terminus of CFTR (C-CFTR) can be significantly enhanced in the presence of ezrin, which specifically recognizes the NHERF1 EB motif. A number of kinases have been identified that regulate the binding properties of NHERF1 by phosphorylation of serine residues, including several sites in the flexible portion of the CT domain and one in PDZ2 [15–18].
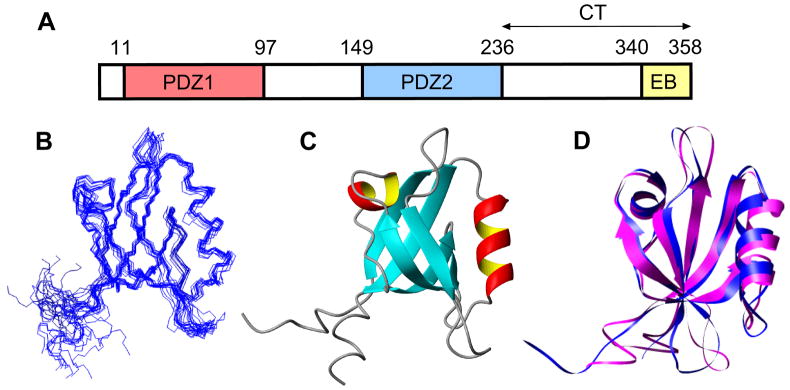
Figure 1. Domain structure of NHERF1 and 3D structure of PDZ domains.
(A) A linear representation of the domain structure of NHERF1. (B) Backbone overlay of NMR solution structures of NHERF1 PDZ2 (12 structures with lowest number of restraint violations). Ramachandran plot analysis of the final structures showed 64%, 30%, 6% and 0% for most favorable, additionally allowed, generously allowed and disallowed regions, respectively (excluding Gly). (C) Ribbon representation of NMR structure of PDZ2. (D) Comparison of the backbone structures of NHERF1 PDZ1 (blue) and NHERF1 PDZ2 (magenta).
To establish a firm structural framework for understanding the ligand binding properties of the PDZ2 domain and its intramolecular interactions with the C-terminus of NHERF1, we determined the NMR solution structure of PDZ2 and characterized its interactions with the C-terminal region of NHERF1. Chemical shift perturbation and relaxation studies revealed the presence of specific intramolecular interactions between PDZ2 and the EB domains. The NMR results indicate that the last 11 residues at the C-terminus of NHERF1 participate in binding and adopt an α-helical conformation when bound to PDZ2. Point mutations in the EB region of the PDZ2-CT fragment to amino acid expected to enhance or disrupt helical secondary structure result in large shifts in the conformational equilibria of the protein between closed (autoinhibited) and open conformations and modulate its binding affinity with respect to C-CFTR.
Results
NMR solution structure of the NHERF1 PDZ2 domain
At the outset of this study high-resolution structural information, a prerequisite for a full understanding its binding properties, was available only for the first, but not the second PDZ domain of NHEF1. Thus, we used NMR to determine the solution structure of the NHERF1 PDZ2 domain (Figure 1). Structure calculations were carried out on the basis of 1707 non-redundant NOE distance constraints, 66 hydrogen bond restraints, and 319 dihedral angle restraints (see Supplemental Data, Table S1). Atomic coordinates and NMR assignments have been deposited in the Protein Data Bank (PDB ID 2jxo) and Biological Magnetic Resonance Bank (BMRB ID 15567), respectively.
Like other PDZ domains, our NMR structure of PDZ2 (Figures 1B and 1C) consists of six β-strands (β1-β6) and two α-helices (α1 and α2). The C-terminal residues, 233–238 (RETDEF), form an additional 1½-turn helix. Superposition of the crystal structure of PDZ1 ([19]; PDB ID 1g9o) with our NMR structure of PDZ2 (PDB ID 2jxo), using Chimera (Figure 1D), yields an rmsd 0.961 Å for backbone atoms of aligned residues (excluding flexible regions). We also compared our NMR structure with a crystal structure of the second PDZ domain of human NHERF1 recently deposited by members of the Structural Genomics Consortium (PDB ID 2ozf). The solution and crystal structures are very similar in terms of secondary structure and backbone conformation, especially in the structural core of the domain (0.827 Å rmsd). The short helix at the C-terminus observed in our NMR structure (NHERF1 residues 150–240) is not present in this crystal structure of the 150–235 fragment [19], but has been observed in other PDZ-containing proteins when the expressed sequence is long enough, such as the crystal structure of PSD-95 PDZ3 (PDB 1be9) [20].
NMR and CD studies of intramolecular PDZ2-CT interactions
Although there are many well-resolved peaks in the heteronuclear single quantum correlation (HSQC) spectrum of the 209-residue PDZ2-CT construct (Figure 2), few sequential connectivities were observed in heteronuclear 3D NMR spectra and only a limited number of inter-residue correlations were observed by nuclear Overhauser effect spectroscopy (NOESY), which can be attributed to the presence of flexible regions and the dynamics of inter-domain interactions giving rise to unfavorable relaxation properties (see below). Thus, it is difficult or impossible to directly solve the NMR structure of intact PDZ2-CT construct.
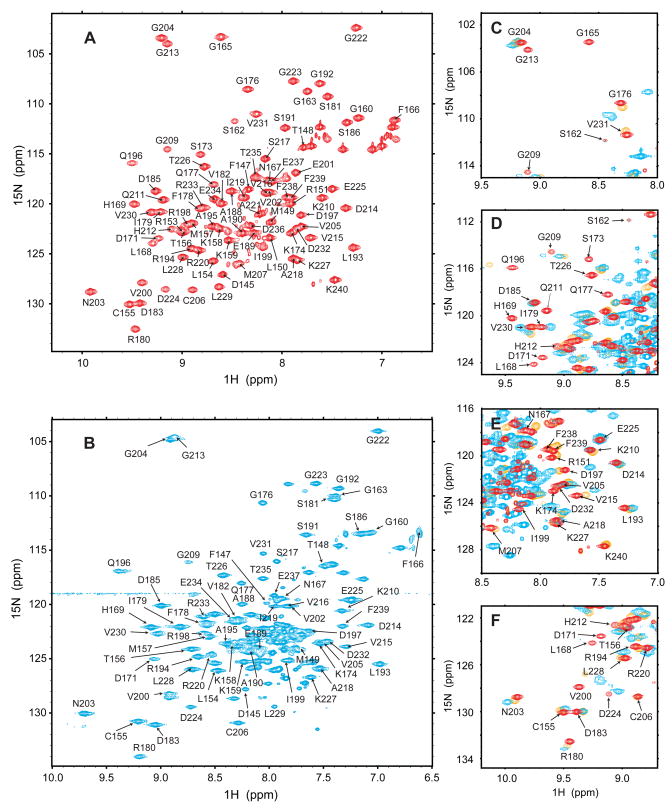
Figure 2. NMR evidence for inter- and intra-molecular interactions between PDZ2 and CT.
(A) HSQC at 15 °C of 15N-labeled PDZ2 in 20 mM HEPES and 150 mM NaCl, pH 7.5. (B) Transverse relaxation-optimized NMR spectrum (TROSY) at 30 °C of PDZ2-CT in 20 mM HEPES and 150 mM NaCl, pH 7.5. (C–F) Expanded regions from HSQC spectra at 20 °C of 15N-labeled PDZ2 (red), equimolar complex of 15N-labeled PDZ2 and unlabeled CT (orange), and PDZ2-CT (cyan) in 20 mM HEPES and 150 mM NaCl, pH 7.5. Assignments for PDZ2 are indicated.
On the other hand, many HN and CO connectivities were observed in HNCO type experiment, which enabled us to assign many of the resonances arising from PDZ2 (residues 150–240) in both the isolated PDZ2 domain and the longer PDZ2-CT construct (residues 150–358). Figure 2A shows the HSQC spectrum of isolated PDZ2, including assignments for 82 residues (from a total of 86 non-proline residues). With the help of HNCO experiments, we assigned the backbone resonances for 67 residues of PDZ2-CT (Figure 2B). In panels C–F, expanded regions of the HSQC spectra of isolated PDZ2 (red), PDZ2-CT (cyan) and an equimolar complex of 15N-labeled PDZ2 with unlabeled CT (orange) are superimposed. As shown in Figure 2B, most of the resolved peaks in the spectrum of PDZ2-CT are from residues in the PDZ2 domain (150–240). The remaining unassigned peaks due to the C-terminal segment form a dense cluster in the center of the HSQC spectrum characteristic of a disordered conformation.
Comparison of Figures 2A and 2B shows that the majority of the resolved and assigned peaks for PDZ2-CT coincide closely with the corresponding peaks of free PDZ2. Moreover, Figures 2C–2F show that there is close agreement between the spectra of PDZ2-CT (cyan) and the non-covalent 1:1 complex of 15N-labeled PDZ2 and unlabeled CT (orange), indicating that both assume a similar overall conformation involving either intra- or intermolecular interaction between PDZ2 and the CT domain. For example, in both cases the N-NH cross-peak of G165 is lost due to line broadening (Figure 2C), and the resonances of Q196, G213 and V215 experience comparable conformational shifts with respect to free PDZ2 (Figures 2D–2F). The possibility that addition of CT causes aggregation was ruled out on the basis of pulsed field gradient (PFG) NMR diffusion measurements on an equimolar mixture of PDZ2 and CT, which are fully consistent with a 1:1 complex between monomers of PDZ2 and CT (see Supplemental Data, Figure S1). Thus, the noncovalent complex between the isolated PDZ2 and CT domains serves as a reliable model for understanding the intramolecular interaction between PDZ2 and the C-terminal domain of NHERF1.
The far-UV circular dichroism (CD) spectrum of the isolated CT (Figure 3B) exhibits a strong negative band at 205 nm and a weak shoulder near 220 nm, consistent with a largely disordered conformation with little regular secondary structure (~10% α-helix and ~15% β-sheet, estimated using the program K2d; www.embl-heidelberg.de/~andrade/k2d.html). These results are consistent with secondary structure prediction based on the sequence (Supplemental Figure S6), which indicates that most of the CT region is likely to be disordered with only marginal helix forming tendencies in two short segments near the C-terminus (residues 327–336 and 348–355). In contrast, the spectra of the PDZ2 domain (residues 150–240) and the PDZ2-CT construct (residues 150–358) show a pronounced negative band at 222 nm indicative of helical conformation (Figure 3A).
Figure 3. Secondary structure and stability of PDZ2, CT and PDZ2-CT.
CD spectra of (A) PDZ2 at 15 °C (solid line) and PDZ2-CT at 20 °C (dashed line); and (B) CT at 15 °C. (C) Urea unfolding of PDZ2 and PDZ2-CT monitored by using CD at 222 nm at 15 °C. The lines represent a two-state fit of the data for PDZ2 (Cm=1.25±0.33 M, m = 1.41±0.13 kcal mol−1 M−1; ΔG0=1.76±0.46 kcal mol−1) and PDZ2-CT (Cm=4.18±0.03 M, m = 1.6±0.11 kcal mol−1 M−1; ΔG0=6.69±0.15 kcal mol−1), respectively.
To determine the energetic contribution of intramolecular interactions between the PDZ2 and CT domains, we measured the protein stability of PDZ2 and PDZ2-CT by using the CD signal at 222 nm to monitor the changes in helical secondary structure content as a function of urea concentration. As shown in Figure 3C, the isolated PDZ2 domain undergoes an immediate increase in the CD signal upon addition of urea and loses most of its helical secondary structure in a transition centered near 1.2 M indicative of a marginally stable structure. In contrast, the cooperative unfolding transition for PDZ2-CT is shifted up to much high urea concentration (4.2 M). The unfolding data were initially fitted using a unimolecular two-state equilibrium unfolding model (see Methods), which is adequate to describe the data for PDZ2 and the main transition for PDZ2-CT. The corresponding thermodynamic parameters (see Figure 3 caption) indicate that inclusion of the C-terminal segment results in a major increase in structural stability (ΔΔG = 4.93 kcal/mol).
Residues in PDZ2 involved in binding to CT
To characterize the surface on PDZ2 involved in recognition of the EB motif, we recorded HSQC spectra on a 1.46 mM solution of 15N-labeled PDZ2 before and after addition of unlabeled CT (to final concentrations of 0.032 mM). The spectral changes observed (cf. Figure 2) are dominated by exchange-broadening. The resulting decrease in peak intensity is especially pronounced (~50%) for G165, F166, L168, H169, Q177 and Q211, which are thus implicated in binding (Figure 4). Except for Q211, these residues are on strand β2 or adjacent regions of the β-sheet. The residues homologous to G165, F166, L168 have been reported to be involved in PDZ1 binding to other target peptides [13, 14]. Q211 in a loop N-terminal to helix α2 (Figure 4B) is the only residue outside the β-sheet undergoing a significant loss of peak intensity. All other residues experience a smaller, relatively uniform, decrease in peak intensity (26 ± 8 %), which can be accounted for by the decrease in the overall rotational correlation time of the PDZ2-CT complex relative to that of the isolated, more compact, PDZ2 domain. PFG diffusion data (Supplemental Figure S1) confirm that the spectral changes observed are due to formation of a stoichiometric complex between PDZ2 and CT rather than aggregation.
Figure 4. Residues on PDZ2 interacting with the EB region.
(A) Fractional intensity change in percent (complex/free) from the 15N relaxation dispersion reference spectra (without CPMG sequences) recorded on free PDZ2 (1.51 mM) and PDZ2 (1.46 mM) in the presence of CT (0.032 mM). Unassigned residues (Y164, S170, and K172) and those with overlapping peaks in the spectrum of the complex (A190, I199, V202, and M207) are not shown. (B) Ribbon diagram of the NMR structure of PDZ2 with a surface plot in the background. Residues with more than 50% intensity change are shown as ball-and-stick in red. S162, the phosphorylation site in the PDZ2 alone, is shown as ball-and-stick in yellow. (C) Ribbon diagram surface plot of the NMR structure of PDZ2 showing homologous side chains in PDZ1 involved in interactions with the peptide DEQL [19].
Residues in CT involved in binding to PDZ2
To determine which residues in CT (NHERF1 residues 242–358) are involved in binding to PDZ2 (residues 150–240), we recorded 1H-15N HSQC of uniformly 15N-labeled CT (15N CT) with increasing amounts of unlabeled PDZ2 (14N PDZ2). The spectrum of free 15N CT (Figure 5A) shows a narrow range of chemical shifts, especially in the proton dimension (7.8 – 8.9 ppm), confirming that CT adopts a largely disordered conformation. Increasing concentrations of 14N PDZ2, approaching that of 15N CT (0.53 mM), result in a major loss in intensity for a small number of peaks while the majority are unaffected (Figure 5A). After subtracting the spectrum of the complex from that of free 15N CT, the difference spectrum shows only 15 remaining peaks (Figure 5B). The remarkable observation that a majority of the cross peaks cancel out in the difference spectra indicates that line broadening rather than chemical shift perturbation is the dominant effect and is consistent with low-affinity, but specific, interactions between PDZ2 and CT. Thus, the peaks in the difference spectrum must arise from the residues on CT involved in interactions with PDZ2. By recording 15N edited NOE-HSQC and TOC-HSQC spectra of free CT, we were able to assign the majority of these peaks to the last 11 residues at the C-terminus of CT (Figure 5B). The assignments were further confirmed by HNCACB and HN(CO)CACB spectra of 13C and 15N labeled CT.
Figure 5. NMR evidence that PDZ2 binds the C-terminal EB region in a helical conformation.
1H-15N HSQC spectra of 15N-labeled CT (242–358). (A) The superimposed spectra of HSQC of 15N-labeled CT (red) and HSQC of 15N-labeled CT (0.53 mM)/unlabeled PDZ2 (0.53 mM) equimolar complex (black); (B) The difference spectrum obtained by subtracting HSQC of 15N CT from that of equimolar 15N-labeled CT/unlabeled PDZ2 complex (red: positive contours; black: negative contours). The spectra of free and complex were normalized based on the intensities of the peak with an arrow in (A). (C) Chemical shift changes due to binding of 15N-labeled CT to unlabeled PDZ2 derived from relaxation dispersion measurements (Figure S3), including 10 of the last 11 residues at the C-terminal of NHERF1. (D) C-terminal helix of the NHERF1 peptide (346–358) as observed in the crystal structure of the FERM-NHERF1 complex [25]. The side chains are shown in ball-and-stick representation. F355 and L358 (red) are involved in binding to PDZ2 and FERM. M346, W348, and L354 (green) are involved in binding to FERM only. K350, K351, and S356 (blue) are important in the binding to PDZ2 only.
To confirm that the loss in peak intensity is due to exchange broadening associated with complex formation, we conducted a series of relaxation-dispersion experiments on 15N CT in the absence and presence of 14N PDZ2, using a constant-time version [21] of the Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence [22]. The 15N R2 relaxation dispersion profiles for residues at the C-terminal end of CT are presented in Supplemental Data (Figure S2). K350, K351, F355, S356, and L358 are the only residues in CT that show a significant change in the relaxation dispersion profiles upon addition of PDZ2. This observation is fully consistent with the line-broadening effects seen in Figure 5A. The parameters estimated by fitting the data to the general two-site exchange equation are given in Supplemental Data (Table S2). Global fitting of the relaxation dispersion curves for 10 C-terminal residues yielded an exchange rate, kex, of 2,300 ± 300 s−1, which is consistent with the fast exchange limit for most residues while those with large chemical shift differences approach the intermediate exchange regime. The corresponding chemical shift changes for CT residues 348–358 of CT (except N357, whose signal was too weak) in the presence of PDZ2 are shown in Figure 5C.
The observed NMR spectral changes (Figures 5) show that a majority of the amino acids among the last 11 residues of CT are involved in interactions with PDZ2 domain. The number and range of residues involved in binding is inconsistent with an extended conformation of the peptide ligand, as found in other PDZ-peptide complexes, in which only 3–5 residues participate in binding [10]. However, our findings can be readily explained if these 11 residues assume an α-helical conformation when bound to PDZ2 (Figure 5D). Additional support for this hypothesis was obtained by using a synthetic peptide corresponding to residues 347–358 of CT as a ligand for PDZ2. In the total correlation spectrum (TOCSY) of this peptide (3.3 mM) in the presence of PDZ2 (0.29 mM), compared to that of free peptide (2.3 mM), residues K350, K351, N352, E353, F355, S356, L358 exhibit measurable chemical shift perturbations and changes in line-width for NH and Hα resonances (Supplemental Figure S3). L358 shows especially prominent changes in line shape, which is consistent with the relaxation dispersion evidence that this residue experiences a large change in chemical shift upon binding (Figure 5). The patterns observed for this 12-residue peptide are fully consistent with the line-broadening and relaxation dispersion effects observed for the complex with full-length CT, and thus indicate that residues outside the C-terminal EB motif are unlikely to participate in intimate contacts with PDZ2.
Mutational analysis of PDZ2-EB interactions
As a further test of our hypothesis that the EB motif assumes a helical conformation upon interaction with PDZ2, we prepared a series of PDZ2-CT variants in which Glu353, a residue on the solvent-exposed side of the predicted EB helix (Figure 5D), was replaced by residues expected to stabilize the helix (Ala) or disrupt helix formation (Pro, Gly or Asp), as does a Gly substitution at an adjacent surface residue, Asn352. For comparison, we prepared a L358D variant in which the most critical residue recognized by the PDZ domain is replaced by a charged side chain in order to disrupt binding. Figure 6 illustrates the effects of mutations on the urea-induced conformational equilibrium, which includes both local and global unfolding transitions. The observed changes in intrinsic fluorescence (dominated by Tyr164 and Trp348) vs. urea concentration (Supplemental Data, Figure S4) indicate a complex unfolding mechanism involving partially unfolded states populated at intermediate urea concentrations. We used a three-state unfolding mechanism to globally fit to the combined set of fluorescence emission spectra vs. urea concentration for each protein, following procedures outlined in [23]. The global fitting parameters (Cm and m-values for each transition) listed in Table 1 define the populations of folded, intermediate and unfolded states vs. urea concentration (Figures 6A and 6B).
Figure 6. Effect of mutations in the EB region of NHERF1 on the three-state unfolding equilibrium of PDZ2-CT.
observed by global analysis of the fluorescence emission spectra vs. urea concentration (Supplemental Data, Figure S4). In panels (A) and (B) the populations of the three predominant equilibrium states, N (solid), I (dashed) and U (dash-dot), are plotted vs. urea concentrations for PDZ-CT and three variants. (C) Intrinsic tyrosine fluorescence spectra of the I-state (normalized relative to the emission maximum of the N-state) for WT PDZ2-CT and three variants. (D) Bar graph showing the effect of mutations on the free energy of the N ⇔ I transition (Δ ΔG1, hatched) and the I ⇔ U unfolding transition (Δ ΔG2). Free energy changes were calculated at the Cm1/2 values of WT PDZ2-CT, using the transition parameters in Table 1.
Table 1.
Effect of point mutations in the EB regions on the fluorescence-detected three-state unfolding equilibrium of PDZ2-CTa
| Cm1 (M) | m1 (kcal mol−1M−1) | ΔΔG1b (kcal mol−1) | Cm2 (M) | m2 (kcal mol−1M−1) | ΔΔG2b (kcal mol−1) | flIrel | [α] | |
|---|---|---|---|---|---|---|---|---|
| WT | 1.80 (0.07) | 0.56 (0.05) | 0 | 3.84 (0.01) | 1.70 (0.01) | 0 | 0.51 | 1.51 |
| N352G | 2.50 (0.05) | 0.47 (0.04) | 0.33 (0.09) | 3.77 (0.01) | 1.87 (0.01) | −0.13 (0.02) | 0.00 | 0.4 |
| E353A | 1.96 (0.08) | 0.87 (0.14) | 0.14 (0.15) | 3.64 (0.01) | 1.66 (0.01) | −0.33 (0.02) | 0.73 | 1.9 |
| E353G | 3.85 (0.28) | 0.41 (0.06) | 0.85 (0.26) | 3.85 (0.03) | 1.82 (0.01) | 0.02 (0.06) | 0.07 | 0.63 |
| E353P | 3.56 (0.21) | 0.49 (0.05) | 0.86 (0.22) | 3.56 (0.03) | 1.61 (0.02) | −0.45 (0.04) | 0.16 | 0.51 |
| E353D | 3.68 (0.09) | 0.86 (0.04) | 1.61 (0.21) | 3.85 (0.03) | 1.82 (0.01) | 0.24 (0.05) | 0.30 | 0.62 |
| L358D | 3.22 (0.21) | 0.41 (0.07) | 0.58 (0.22) | 3.54 (0.02) | 1.68 (0.01) | −0.50 (0.04) | 0.04 | 1.38 |
Cm1/2 and m1/2 represent the equilibrium transition parameters obtained by global fitting of a three-state unfolding equilibrium to fluorescence spectra of wild-type and mutant forms of the PDZ2-CT fragment NHERF-1 (in 20 mM potassium phosphate, pH 7.5, 1 mM DTT, at 15 °C). Standard errors are shown in parentheses. flIrel represents the relative fluorescence intensity for the emission of Tyr164 at 302 nm in the I-state relative to that in the N-state. For comparison, [α] indicates the α-helix content of the EB region (residues 348–358) predicted using Agadir (http://www.embl-heidelberg.de/Services/serrano/agadir/agadir-start.html).
mutational free energy change measured at the midpoint concentration of the transition for the wild type.
Truncation variant of PDZ2-CT comprising residues 150–343 of NHERF-1.
The mutations have a profound effect on the transition between the N- and I-states populated at low and intermediate denaturant concentrations, whereas the main unfolding transition centered around 3.8 M urea is less affected by the mutations. The local fitting parameters (intercepts of the fluorescence signal vs. urea concentration at a given wavelength) allow us to extract the intrinsic fluorescence spectra for each state (Figure S4). It is fortuitous that the PDZ2-CT fragment of NHERF1 contains only two fluorescent residues, Tyr164 in PDZ2 and Trp348 in the EB region. Sigmoidal transitions indicative of cooperative unfolding are seen only at wavelengths below about 340 nm where Tyr emission contributes to the spectrum. The data at higher wavelengths dominated by Trp emission show a monotonous increase, indicating that Trp348 is in a similar, solvent-exposed, environment in all conformational states. Subtraction of the fitted U-state spectrum from the spectra of the N- and I-states thus reveals the contribution of Tyr164, which is highly sensitive to mutation of residues in the EB region (Figure 6C). Figure 6D shows the effect of mutations on the free energies associated with each of the two unfolding transitions, ΔΔG1 and ΔΔG2, evaluated at the respective midpoints of the wild-type (Table 1).
Affinity of PDZ2-CT for C-CFTR
To understand the impact of intramolecular structural rearrangements on the functional binding properties of NHERF1, we used the cytoplasmic domain of CFTR as a representative ligand. CFTR contains a C-terminal PDZ recognition motif (DTRL) known to interact with both PDZ domains of NHERF1 [2, 11, 24]. We used surface plasmon resonance (SPR) to measure binding of C-CFTR (residues 1411–1480 at the C-terminus of CFTR) to several PDZ2-CT variants (Supplemental Data, Figure S5). The dissociation constant we measured for the wild-type protein (2.0 ± 0.2 μM) is 2-fold higher than the value (1.1 ± 0.08 μM) measured by Cushing et al. using fluorescence anisotropy [12]. This difference is in part due to the presence of autoinhibitory interactions with CT. By comparison, the E353P mutation, which interferes with helix formation in the EB region, binds C-CFTR with 4-fold higher affinity (Kd = 0.48 ± 0.06 μM), and the C-terminal L358D mutation also enhances binding (Kd = 1.6 ± 0.1 μM). In contrast, the helix-stabilizing E353A mutation gives rise to a noticeable decrease in affinity for C-CFTR (Kd = 2.5 ± 0.5 μM). Thus, the affinity of PDZ2-CT variants for the CFTR ligand peptide is inversely related to the helix-forming propensity of the EB motif (Table 1), which supports our hypothesis that intramolecular interactions between PDZ2 and a helix at the C-terminus compete with the binding of extrinsic ligands.
Discussion
NHERF1 interacts with ion channels and receptors through its tandem PDZ domains, which recognize specific PDZ-binding motifs with the consensus sequence D(S/T)X(V/I/L) (X denoting any amino acid residue). A mechanism involving competing intramolecular interactions is consistent with the fact that the same motif is also found at the C-terminus of NHERF1 (FSNL). The dynamic nature and low affinity of the PDZ2-CT interaction make this system not only unsuitable for X-ray crystallographic analysis, but also pose major challenges for structure determination by conventional NMR methods. Nevertheless, we were able to resolve and assign a majority of the peaks due to PDZ2 in the presence of covalently or non-covalently bound CT (Figure 2B). The lack of major changes in chemical shift indicates that the overall structure of PDZ2 is preserved in the complex. By measuring the changes in peak intensity for individual residues in 15N-labeled PDZ2 due to line broadening upon addition of unlabeled CT, we were able to identified key residues involved in recognition of the C-terminus (Figure 4). These residues, located mainly on the β2 strand and adjacent loops, form a contiguous binding surface (Figure 4B). The interface is similar, but not identical to that observed in the complex of other PDZ domains with extended peptides, which often make more extensive contacts with the α2 helix [13, 14, 20].
Our NMR and CD spectra (Figures 2, 3 and 5) indicate that the CT domain, expressed either as an isolated fragment or covalent linked to PDZ2, is highly flexible and largely disordered in solution, as predicted on the basis of its sequence, using a disorder prediction algorithm [9]. The following observations provide evidence for specific intramolecular interactions between PDZ2 and the C-terminal EB region of NHERF1: (1) the PDZ2-CT fragment is much more stable than the isolated PDZ2 domain (Figure 3C); (2) adding a small amount of unlabeled CT to an 15N-labeled sample of PDZ2 causes a major decrease in intensity for selected residues on β2 and adjacent loops; (3) addition of unlabeled PDZ2 to 15N-labeled CT causes major changes in peak intensity and relaxation dispersion profiles (Figures 5); (4) a 12-residue peptide with a sequence corresponding to the EB region of NHERF1 exhibits changes in line-width and chemical shifts on addition of unlabeled PDZ2 (Supplemental Figure S3); (5) mutation of residues in the EB at or near the C-terminus cause major shifts in the conformational equilibrium between alternative folded states and modulate the fluorescence of Tyr164 in the PDZ2 peptide binding groove (Figure 6).
In all other PDZ-peptide complexes reported previously, the peptide ligand assumes an extended conformation and inserts itself between the α2 helix and β2 strand in an antiparallel arrangement with respect to β2 [10, 14, 20]. The binding groove accommodates a maximum of five residues for an extended ligand peptide with the last three residues at the C-terminus playing the principal role in PDZ-ligand recognition. In contrast, our NMR studies (Figure 5) show that PDZ2 interacts with a much longer segment spanning 11 residues at the C-terminus of NHERF1, including L358, F355, S356, K351, K350 and W348 (listed in decreasing order with respect to relaxation effects). The fact that L358 experiences particularly large spectral perturbations confirms that this C-terminal residue plays a key role in PDZ2-peptide recognition, as previously found in the case of the complex between NHERF1 PDZ1 and CFTR [13]. Our results are clearly inconsistent with binding in an extended conformation, which would extend far beyond the length of the binding groove on PDZ2. On the other hand, since the axial distance between adjacent amino acids is shorter in an α-helix (1.5 Å) compared to β-sheet or extended conformation (~3.5 Å), an α-helix of 11–12 residues covers the same length as a 5-residue extended peptide, and can thus be accommodated in the peptide binding pocket of PDZ2. Moreover, the perturbations show a periodic pattern with larger changes in relaxation parameters for every 3rd or 4th residue (Figure 5). To our knowledge, this is the first direct structural evidence for a peptide interacting with a PDZ domain in an α-helical conformation. The distribution of contact residues on PDZ2 in its complex with CT (Figure 4B) suggests that the helical EB domain is accommodated in the groove between β2 and α2. A previous attempt to model this complex resulted in a different orientation for the C-terminal helix and a more exposed N-terminal end [18]. This preliminary docking model has to be revised in light of the NMR data obtained in the present study.
In the crystal structure of a C-terminal fragment of NHERF1 (339–358) bound to the radixin FERM domain (structurally unrelated to PDZ), the C-terminal 11 residues of NHERF1 form a 3-turn amphipathic α-helix [25]. Based on our NMR results, the majority of residues strongly affected by binding to PDZ2 (L358, F355, K351, and K350) are located on the hydrophobic side of the helix. By measuring the effect of mutations on affinity, Terawaki, et al. found that residue M346 also contributes to binding in the case of the FERM domain [25]. In contrast, our data indicate that only the helical part of the EB region (residues 348–358) is involved in the binding to PDZ2. Thus, while both PDZ2 and the FERM domain recognize the EB region in an α-helical conformation, the binding interface with these structurally unrelated binding partners is not identical.
Morales, et al. published biochemical and cell biological evidence for intramolecular interactions between the PDZ domains and C-terminus of NHERF1 [8]. They reported that mutations near the C-terminus (F355P, F355R and a C-terminal deletion) abolished the intramolecular contact between PDZ2 and CT, which they attribute to loss of α-helical structure at the C-terminus. They also showed that intramolecular interactions were only moderately perturbed by another set of mutations (S356A, L358F, and L354I/358F), which are expected to disrupt some of the canonical PDZ-peptide contacts, but not a putative α-helix. Although highly suggestive, the results from this mutagenesis study are inconclusive with respect to the exact conformation of the C-terminal peptide when bound to PDZ2, since proline substitution of an amino acid is expected to disrupt both α-helical as well as β-sheet structure. Furthermore, Pro or Arg substitutions at position 355 involve drastic changes in the size and shape of a side chain that is also predicted to contact the PDZ domain in the canonical extended binding motif.
Our NMR and mutational studies offer more detailed structural insights and provide direct support for the hypothesis that the EB region of NHERF1 adopts a helical conformation when engaged in intramolecular contacts with PDZ2. Especially compelling is our finding that amino acid changes at an exposed site of the putative helix cause significant shifts in the conformational transition preceding the main unfolding transition (Figure 6, Table 1). Mutations that destabilize the C-terminal helix (N352G, E353P and E353G) or remove a key contact with PDZ2 (L358D) shift the midpoint of the first transition (Cm1) toward higher urea concentrations and are associated with positive ΔΔG1 (Figure 6D). In contrast, the mutations cause only small shifts in the second (major) unfolding transition and give rise to small or negative ΔΔG2. As a result, the maximum population of the I-state (Figures 6A and 6B) is considerably lower for destabilizing mutations (~40%) compared to wild-type and the helix-stabilizing E353A variant (~75%). Thus, contrary to expectations, disruption of the C-terminal helix or its contacts with the PDZ2 domain stabilizes the native state relative to the I-state populated at intermediate urea concentrations. Interestingly, the variation in population of the I-state correlates with its fluorescence properties; the proteins with well-populated I-states (wild-type and E353A) have substantially enhanced tyrosine emission spectra (Figure 6C).
Our equilibrium denaturation experiments (Figure 6) show that unfolding of PDZ2-CT occurs in several steps involving at least two alternative folded states in addition to fully unfolded conformations. For the wild-type protein and all mutants studied, a three-state unfolding model fully accounts for all of the fluorescence data vs. urea concentration and yields a robust set of fitting parameters characterizing the thermodynamic and spectral properties of the populated states (Table 1, Figure S4). The second transition centered around 3.8 M urea is relatively insensitive to mutation of C-terminal residues, and thus represents the cooperative unfolding of the PDZ2 domain as well as any parts of the linker segment (residues 242–347) it may be in contact with. In contrast, the conformational transition at lower urea concentrations is very sensitive to point mutations in the EB region (except for E353A, which supports helix formation). The free energy perturbations, ΔΔG1, due to mutations that interfere with helix formation or remove critical contacts with PDZ2 (L358D) are all positive (Figure 6D). Thus, mutations that disrupt binding to the PDZ domain all result in a significant stabilization of the native state relative to intermediate and unfolded states. These surprising observations can be explained in terms of the mechanism schematized in Figure 7, which features two partially folded states, Iopen and IEB, in addition to the native (N) and fully unfolded (U) states. The N ⇔ Iopen ⇔ U branch is the dominant unfolding pathway for the disruptive mutants, which exhibit two distinct unfolding transitions. The wild type protein and the helix-stabilizing E353A variant can assume an alternative folded state, IEB, in which PDZ2 engages the helical EB motif (IEB and Iopen are unresolved in our equilibrium unfolding measurements). IEB dominates the population at intermediate urea concentrations (~1.5–3.5 M), and is thus less structured (in terms of solvent-accessible surface area) than the N-state. This suggests that portions of the linker segment (240–347) are structured in N and become solvent-exposed in IEB. Mutations that block formation of the C-terminal helix or its contact with PDZ2 shift the conformational equilibrium from IEB towards Iopen and N, which accounts for the observed stabilization of the N-state at the expense of the less structured intermediates.
Figure 7. Schematic diagram of the various conformational states populated in the unfolding equilibrium of PDZ2-CT.
, including the native state (N) populated in the absence of denaturant, a state with the C-terminal EB helix interacting with PDZ2 (IEB), an open state in which only the PDZ domain is folded (Iopen), and the fully unfolded ensemble (U).
The mechanism outlined in Figure 7 also accounts for the observed effect of mutations on the tyrosine fluorescence spectra (Figure 6C). For WT and E353A, the tyrosine emission band in the non-native intermediates (comprising both Iopen and IEB) is nearly as high as that in the N-state (Figure S4), indicating that Tyr164 is buried in both IEB and N. On the other hand, solvent-quenching of Tyr164 fluorescence in Iopen explains the low I-state fluorescence of the N352G, E353G, E353P, E353D and L358D variants in which IEB is unstable and Iopen is well populated. Mechanisms with fewer states cannot fully account for all observations. For example, if IEB were the predominant state in the absence of denaturant, as initially proposed [8, 11], disruptive mutations in the C-terminal PDZ-binding motif would greatly destabilize the native state and shift the equilibrium towards partially or fully unfolded states (Iopen or U). In contrast, the disruptive changes in the EB motif consistently stabilize the folded state N, indicating that PDZ2-EB interactions are unfavorable in the absence of denaturant and are replaced by energetically more favorable interactions involving the flexible linker region.
The ligand binding properties of mutant NHERF1 constructs are fully consistent with the proposed mechanism (Figure 7) if we postulate that the peptide binding groove of PDZ2 is accessible for interaction with extrinsic ligand peptides only in Iopen. This explains our observation that the affinity of PDZ2 for C-CFTR increases when we disrupt its autoinhibitory interactions with the EB motif (either be mutating the important C-terminal Leu or by blocking helix formation). Further corroborating evidence comes from the fluorescence of Tyr164 (Figure 6C), which reports on the solvent accessibility of a key residue in the ligand binding grove of PDZ2. Our observation of strong tyrosine fluorescence bands under native conditions and those favoring formation of IEB (Figure 6C) indicates that Tyr164 is buried in a non-aqueous environment in both N and in IEB, but exposed to the quenching solvent in Iopen. Since tight binding of PDZ ligand peptides involves intimate contact with the Tyr164 side chain [13, 14], it is likely that not only IEB, but also the N-state is unable to bind extrinsic peptide ligands. Whether this reflects competing interactions involving the flexible linker region or structural changes within the PDZ2 domain remains to be explored. The latter possibility is in line with recent kinetic evidence that binding a high-affinity peptide ligand to the 2nd PDZ domain of tyrosine phosphatase PTP-BL is accompanied by structural rearrangements at the interface between the α2 helix and β2 strand [26].
Conclusions
While biological mechanisms often rely on strong and highly specific molecular interactions, weak interactions can also have an impact. For example, the dynamic nature of weak interactions can be advantageous for subtle regulation of binding affinities in signal transduction. However, compared to high-affinity binding, it is more challenging to obtain detailed structural insight into weakly interacting systems. Here, we demonstrate that NMR chemical shift and relaxation dispersion measurements are well suited for characterizing weak intermolecular interactions and can be used to detect perturbations even in the presence of low amounts of a binding partner. Using this approach, we were able to identify the residues from PDZ2 and CT involved in mutual interdomain interactions in the closed (autoinhibited) state of NHERF1 in which the C-terminal EB region adopts an α-helical conformation. Thermodynamic analysis of PDZ2-CT variants with mutations in the EB motif reveals a complex unfolding equilibrium involving three structurally distinct folded states in addition to the fully unfolded state. Only one of these, Iopen, binds a physiological ligand of NHERF1, whereas the binding site on PDZ2 is occluded in the more stable folded states, IEB and N, due to intramolecular interactions with CT. These dynamic contacts involving the C-terminal EB helix or the intervening flexible linker region have a profound effect on the overall structure of NHERF1 and modulate the binding properties of its domains, and thus are critical for understanding its functions as a signaling adaptor. Together with the fact that the linker sequence contains several potential phosphorylation sites, these observations suggest a novel functional role for intrinsically disordered protein segments in regulating the balance between intra- and inter-molecular ligand interactions.
Methods
Recombinant NHERF1 constructs, including PDZ2 (150–240), CT (242–358), and PDZ2-CT (150–358), were expressed in E. coli using the pET151/D-TOPO vector (Invitrogen, Inc., Carlsbad, CA) with a 6xHis-tag at the N-terminus and purified according to [11]. The 6xHis tag was removed using TEV protease. The cleavage left 7 additional residues (GIDPFTM) at the N-terminus of the purified proteins. Uniformly labeled (15N and/or 13C) samples of proteins were prepared from bacteria grown in M9 media supplemented with 15N NH4Cl and/or 13C glucose as the only nitrogen and carbon sources, respectively. Mutants in the EB region were introduced into PDZ2-CT using the QuikChange method and expressed in the pET19b vector (Novagen EMD, Gibbstown, NJ). Mutant proteins were purified as above, but without cleavage of the 6xHis-tag.
NMR data were collected on a Bruker DMX-600 spectrometer equipped with a 5-mm x,y,z-shielded pulsed-field gradient triple-resonance probe (Bruker Biospin GmbH, Rheinstetten, Germany). For structure determination, peptide studies, and relaxation measurements, data were collected at 15°C. For other experiments, the conditions are given in the figure captions. Typically, NMR samples contained ~1 mM protein in 20 mM HEPES, 150 mM NaCl, pH of 7.5, and contained 0.5 mM dithiothreitol (DTT), unless mentioned otherwise. Additional information on NMR data acquisition, relaxation dispersion experiments, processing and structure calculations can be found in Supplemental Data.
CD spectra on NHERF1 PDZ2, CT and PDZ2-CT in 20 mM potassium phosphate buffer at pH 7.5 were acquired (2 nm bandwidth, 1 mm optical path) on an Aviv 62A spectrometer (Aviv Biomedical, Inc., Lakewood, NJ). Fluorescence spectra were recorded on a PTI fluorometer (Birmingham, NJ). Urea-induced unfolding transitions were measured by recording the ellipiticity at 222 nm (2 nm bandwidth, 2 mm optical path) or fluorescence emission scans from 285–420 nm (276 nm excitation) on a series of protein samples (in 20 mM potassium phosphate buffer with 1 mM DTT and pH 7.5) prepared by mixing aliquots of equimolar stock solutions of native and fully unfolded (10 M urea) proteins.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM056250 to H.R., grants from the American Cancer Society (IRG-92-027-09) and the W.W. Smith Charitable Trust to Z.B., and a grant from the National Cancer Institute (CA06927) and an appropriation by the Commonwealth of Pennsylvania to the Fox Chase Cancer Center. Critical support was provided by the Biotechnology, DNA Sequencing, Molecular Modeling, and Spectroscopy Support Facilities of the Fox Chase Cancer Center. We thank Drs. J. Chernoff, R.L. Dunbrack and E.A. Golemis for helpful comments on the manuscript.
Footnotes
Supplemental Data
Supplemental Data including additional details on NMR methods and structural statistics, NMR diffusion measurements, 15N R2 relaxation dispersion data, NMR analysis of a synthetic EB peptide bound to PDZ2, equilibrium unfolding studies and SPR binding data, are available online at xxx.
References
- 1.Weinman EJ, Steplock D, Wang Y, Shenolikar S. Characterization of a protein cofactor that mediates protein kinase A regulation of the renal brush border membrane Na(+)-H+ exchanger. J Clin Invest. 1995;95:2143–2149. doi: 10.1172/JCI117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall RA, Ostedgaard LS, Premont RT, Blitzer JT, Rahman N, Welsh MJ, Lefkowitz RJ. A C-terminal motif found in the beta2-adrenergic receptor, P2Y1 receptor and cystic fibrosis transmembrane conductance regulator determines binding to the Na+/H+ exchanger regulatory factor family of PDZ proteins. Proc Natl Acad Sci U S A. 1998;95:8496–8501. doi: 10.1073/pnas.95.15.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinman EJ, Hall RA, Friedman PA, Liu-Chen LY, Shenolikar S. The association of NHERF adaptor proteins with g protein-coupled receptors and receptor tyrosine kinases. Annu Rev Physiol. 2006;68:491–505. doi: 10.1146/annurev.physiol.68.040104.131050. [DOI] [PubMed] [Google Scholar]
- 4.Guggino WB, Stanton BA. New insights into cystic fibrosis: molecular switches that regulate CFTR. Nat Rev Mol Cell Biol. 2006;7:426–436. doi: 10.1038/nrm1949. [DOI] [PubMed] [Google Scholar]
- 5.Mahon MJ, Donowitz M, Yun CC, Segre GV. Na(+)/H(+ ) exchanger regulatory factor 2 directs parathyroid hormone 1 receptor signalling. Nature. 2002;417:858–861. doi: 10.1038/nature00816. [DOI] [PubMed] [Google Scholar]
- 6.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 7.Georgescu MM, Morales FC, Molina JR, Hayashi Y. Roles of NHERF1/EBP50 in cancer. Curr Mol Med. 2008;8:459–468. doi: 10.2174/156652408785748031. [DOI] [PubMed] [Google Scholar]
- 8.Morales FC, Takahashi Y, Momin S, Adams H, Chen X, Georgescu MM. NHERF1/EBP50 head-to-tail intramolecular interaction masks association with PDZ domain ligands. Mol Cell Biol. 2007;27:2527–2537. doi: 10.1128/MCB.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 10.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Dai Z, Jana D, Callaway DJ, Bu Z. Ezrin controls the macromolecular complexes formed between an adapter protein Na+/H+ exchanger regulatory factor and the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2005;280:37634–37643. doi: 10.1074/jbc.M502305200. [DOI] [PubMed] [Google Scholar]
- 12.Cushing PR, Fellows A, Villone D, Boisguerin P, Madden DR. The relative binding affinities of PDZ partners for CFTR: a biochemical basis for efficient endocytic recycling. Biochemistry. 2008;47:10084–10098. doi: 10.1021/bi8003928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karthikeyan S, Leung T, Ladias JA. Structural basis of the Na+/H+ exchanger regulatory factor PDZ1 interaction with the carboxyl-terminal region of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2001;276:19683–19686. doi: 10.1074/jbc.C100154200. [DOI] [PubMed] [Google Scholar]
- 14.Karthikeyan S, Leung T, Ladias JA. Structural determinants of the Na+/H+ exchanger regulatory factor interaction with the beta 2 adrenergic and platelet-derived growth factor receptors. J Biol Chem. 2002;277:18973–18978. doi: 10.1074/jbc.M201507200. [DOI] [PubMed] [Google Scholar]
- 15.Hall RA, Spurney RF, Premont RT, Rahman N, Blitzer JT, Pitcher JA, Lefkowitz RJ. G protein-coupled receptor kinase 6A phosphorylates the Na(+)/H(+) exchanger regulatory factor via a PDZ domain-mediated interaction. J Biol Chem. 1999;274:24328–24334. doi: 10.1074/jbc.274.34.24328. [DOI] [PubMed] [Google Scholar]
- 16.He J, Lau AG, Yaffe MB, Hall RA. Phosphorylation and cell cycle-dependent regulation of Na+/H+ exchanger regulatory factor-1 by Cdc2 kinase. J Biol Chem. 2001;276:41559–41565. doi: 10.1074/jbc.M106859200. [DOI] [PubMed] [Google Scholar]
- 17.Raghuram V, Hormuth H, Foskett JK. A kinase-regulated mechanism controls CFTR channel gating by disrupting bivalent PDZ domain interactions. Proc Natl Acad Sci U S A. 2003;100:9620–9625. doi: 10.1073/pnas.1633250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Poulikakos PI, Dai Z, Testa JR, Callaway DJ, Bu Z. Protein kinase C phosphorylation disrupts Na+/H+ exchanger regulatory factor 1 autoinhibition and promotes cystic fibrosis transmembrane conductance regulator macromolecular assembly. J Biol Chem. 2007;282:27086–27099. doi: 10.1074/jbc.M702019200. [DOI] [PubMed] [Google Scholar]
- 19.Karthikeyan S, Leung T, Birrane G, Webster G, Ladias JA. Crystal structure of the PDZ1 domain of human Na(+)/H(+) exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains. J Mol Biol. 2001;308:963–973. doi: 10.1006/jmbi.2001.4634. [DOI] [PubMed] [Google Scholar]
- 20.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 21.Tollinger M, Skrynnikov NR, Mulder FA, Forman-Kay JD, Kay LE. Slow dynamics in folded and unfolded states of an SH3 domain. J Am Chem Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 22.Loria JP, Rance M, Palmer AG. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc. 1999;121:2331–2332. [Google Scholar]
- 23.Latypov RF, Cheng H, Roder NA, Zhang J, Roder H. Structural characterization of an equilibrium unfolding intermediate in cytochrome c. J Mol Biol. 2006;357:1009–1025. doi: 10.1016/j.jmb.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Raab RW, Schatz PJ, Guggino WB, Li M. Peptide binding consensus of the NHE-RF-PDZ1 domain matches the C-terminal sequence of cystic fibrosis transmembrane conductance regulator (CFTR) FEBS Lett. 1998;427:103–108. doi: 10.1016/s0014-5793(98)00402-5. [DOI] [PubMed] [Google Scholar]
- 25.Terawaki S, Maesaki R, Hakoshima T. Structural basis for NHERF recognition by ERM proteins. Structure. 2006;14:777–789. doi: 10.1016/j.str.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Gianni S, Walma T, Arcovito A, Calosci N, Bellelli A, Engstrom A, Travaglini-Allocatelli C, Brunori M, Jemth P, Vuister GW. Demonstration of long-range interactions in a PDZ domain by NMR, kinetics, and protein engineering. Structure. 2006;14:1801–1809. doi: 10.1016/j.str.2006.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.