Abstract
tRNAs from all 3 phylogenetic domains have a 5-methyluridine at position 54 (T54) in the T-loop. The methyl group is transferred from S-adenosylmethionine by TrmA methyltransferase in most Gram-negative bacteria and some archaea and eukaryotes, whereas it is transferred from 5,10-methylenetetrahydrofolate (MTHF) by TrmFO, a folate/FAD-dependent methyltransferase, in most Gram-positive bacteria and some Gram-negative bacteria. However, the catalytic mechanism remains unclear, because the crystal structure of TrmFO has not been solved. Here, we report the crystal structures of Thermus thermophilus TrmFO in its free form, tetrahydrofolate (THF)-bound form, and glutathione-bound form at 2.1-, 1.6-, and 1.05-Å resolutions, respectively. TrmFO consists of an FAD-binding domain and an insertion domain, which both share structural similarity with those of GidA, an enzyme involved in the 5-carboxymethylaminomethylation of U34 of some tRNAs. However, the overall structures of TrmFO and GidA are basically different because of their distinct domain orientations, which are consistent with their respective functional specificities. In the THF complex, the pteridin ring of THF is sandwiched between the flavin ring of FAD and the imidazole ring of a His residue. This structure provides a snapshot of the folate/FAD-dependent methyl transfer, suggesting that the transferring methylene group of MTHF is located close to the redox-active N5 atom of FAD. Furthermore, we established an in vitro system to measure the methylation activity. Our TrmFO-tRNA docking model, in combination with mutational analyses, suggests a catalytic mechanism, in which the methylene of MTHF is directly transferred onto U54, and then the exocyclic methylene of U54 is reduced by FADH2.
Keywords: modification, X-ray crystallography, TrmFO
Methylation is one of the most common chemical modifications and is found in a broad range of biomolecules, including nucleic acids, proteins, lipids, and small compounds. It is implicated in a variety of cellular processes, such as epigenetics, development, cancer, and bacterial host defense. In RNA, methylated nucleotides appear in most noncoding RNAs and constitute more than half of the posttranscriptional modifications identified so far. tRNAs especially contain >50 different methylated nucleotides, which not only reinforce their L-shaped structure but also improve or switch their molecular recognition (1). The 5-methyluridine at position 54 in the T-loop of tRNA (also designated as T54) is conserved in the 3 domains of life (2). This nucleotide participates in a long-range interaction with the 1-methyladenine at position 58 to maintain the T-loop conformation and stabilize the tRNA L-shape. In thermophilic organisms such as Thermus thermophilus, T54 is further modified to 2-thioribothymidine, which is essential for growth at high temperatures (3, 4). In most Gram-negative bacteria and some archaea and eukaryotes, T54 formation is catalyzed by TrmA, an S-adenosylmethionine (AdoMet)-dependent methyltransferase (MTase) that belongs to the most common MTase family (5–7). The recently-solved crystal structure of Escherichia coli TrmA in complex with a tRNA T-arm analog (8), along with the previous extensive biochemical studies, revealed its mechanisms for catalysis and substrate binding.
Although most MTases use AdoMet as a methyl donor, some other MTases use 5,10-methylenetetrahydrofolate (MTHF) as a 1-carbon donor. It was reported >3 decades ago that in the Gram-positive bacteria Enterococcus faecalis and Bacillus subtilis the biosynthesis of tRNA T54 depends not on AdoMet but on folate (9), and that the purified E. faecalis enzyme catalyzes T54 formation by using MTHF as a 1-carbon donor and FADH2 as a reductant, producing tetrahydrofolate (THF) (10). More recently, bioinformatics searches identified the gene encoding a flavin-dependent MTase, named TrmFO, in the B. subtilis genome (11). Biochemical analyses demonstrated that the purified B. subtilis TrmFO contains a bound FAD and catalyzes T54 formation in a folate/FAD-dependent manner. Sequence analyses revealed that TrmFO orthologs are distributed in most Gram-positive bacteria and some Gram-negative bacteria, including T. thermophilus (12). Notably, the TrmFO and TrmA MTases exhibit a mutually-exclusive phylogenetic distribution. Although Thermotoga maritima TrmFO has been crystallized (13), the mechanism by which TrmFO catalyzes T54 formation in a folate/FAD-dependent manner has remained elusive because of the lack of structural information.
Here, we present the high-resolution crystal structures of T. thermophilus TrmFO, which reveal TrmFO consists of an FAD-binding domain and an insertion domain. Both of these domains share a structural similarity with those of GidA, an enzyme involved in the 5-carboxymethylaminomethylation of tRNA U34. However, a remarkable difference between their domain organizations may account for their distinct biological functions. Furthermore, we have established an in vitro system to evaluate the folate/FAD-dependent methylation activity, which is coupled with the MTHF production by a metabolic serine hydroxymethyltransferase (SHMT) enzyme. Using our in vitro system, we demonstrated that TrmFO catalyzes T54 formation in a folate/FAD-dependent manner. Our TrmFO-tRNA complex model, together with our extensive mutational analyses, allows us to propose a novel catalytic mechanism.
Results and Discussion
Biochemical Characterization.
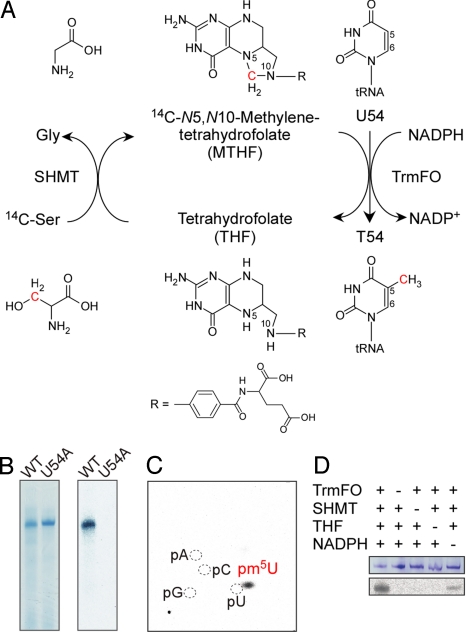
T. thermophilus TrmFO and SHMT were expressed in E. coli and purified to homogeneity (Fig. S1A). The purified TrmFO protein is colored yellow, indicating that oxidized FAD is bound to the protein, as observed in B. subtilis TrmFO (11). To examine its activity, we developed an in vitro assay system in which T. thermophilus SHMT converts [14C]-Ser and THF to glycine and [methylene-14C]-MTHF, and then TrmFO methylates the T. thermophilus tRNAIle transcript by using [methylene-14C]-MTHF and NADPH as the 1-carbon donor and the reductant, respectively (Fig. 1A). Using the in vitro system, we observed 14C incorporation into the tRNAIle transcript but not into the tRNAIle U54A transcript, where U54 is replaced by A (Fig. 1B and Fig. S1B). In addition, we detected [14C]-pm5U formation by 2D TLC after nuclease P1 digestion (Fig. 1C). These results clearly show that TrmFO methylates the C5 atom of U54. No activity was detected in the absence of either SHMT or THF, indicating that the methylene of MTHF serves as a 1-carbon donor (Fig. 1D). The activity was detected in the absence of NADPH, while it was enhanced in the presence of NADPH (Fig. 1D). These results suggest that although TrmFO requires NADPH for its activity, the purified TrmFO used for our experiments contained both the oxidized and reduced forms of flavin. Taken together, our results demonstrate that T. thermophilus TrmFO catalyzes the formation of T54 in the T-loop of tRNA by using MTHF as a 1-carbon donor and NADPH/FAD as a reductant.
Fig. 1.
Methylation activity. (A) Reaction scheme of the in vitro methylation assay. (B) Incorporation of the [14C-methyl] group into the wild-type and U54A T. thermophilus tRNAIle transcripts. After the in vitro methylation reaction, the transcripts were resolved by 10% urea-PAGE. The gels were stained with methylene blue (Left) and autoradiographed (Right). (C) Identification of the methylated position. After the in vitro methylation reaction, the transcript was purified by 10% urea-PAGE, digested with nuclease P1, and analyzed by 2D TLC. (D) After the in vitro methylation reaction in the presence of the indicated component, the transcript was resolved by 10% urea-PAGE. The gels were stained with methylene blue (Upper) and autoradiographed (Lower).
Overall Structure.
We solved the crystal structures of T. thermophilus TrmFO in the THF-free flavoprotein form and in complex with THF at 2.1- and 1.6-Å resolutions, respectively. During the screening of crystallization conditions, we found that the addition of glutathione (GSH) significantly improved the reproducibility of crystals under THF-free conditions, and we solved the structure in complex with GSH at 1.05-Å resolution. The 3 structures are essentially identical, with a rmsd for all of the aligned Cα atoms of <0.3 Å (Fig. S2A), indicating that the binding of either GSH or THF does not induce substantial conformational changes in the enzyme structure. We will hereafter describe the THF complex structure, unless otherwise stated.
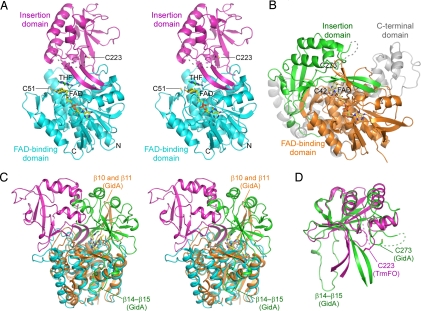
The structure consists of 2 domains, an FAD-binding domain and an insertion domain (Fig. 2A). The FAD-binding domain (residues 1–153 and 314–436) composes a 3-stranded antiparallel β-sheet (β3, β5, and β4), a 6-stranded mixed β-sheet (β6, β2, β1, β7, β16, and β17), 9 α-helices (α1–α5 and α10–α13), and 3 310-helices (η1, η4, and η5). The insertion domain (residues 154–313) is composed of a 2-stranded antiparallel β-sheet (β8 and β15), a 6-stranded antiparallel β-sheet (β10, β11, β12, β13, β9, and β14), 4 α-helices (α6–α9), and 2 310-helices (η2 and η3). The region between α6 and α9 (residues 207–222) of the insertion domain is disordered in the electron density in all 3 of the structures determined here, reflecting the intrinsic flexibility of this region. Consistent with the gel filtration result (Fig. S3), TrmFO exists as a monomer in all 3 crystal forms, indicating that TrmFO might function as a monomer.
Fig. 2.
Overall structure. (A) Stereoview of the TrmFO-THF complex structure. The FAD-binding and insertion domains are shown in cyan and magenta, respectively. The disordered region is shown as a dashed line. The catalytic Cys residues, Cys-51 and Cys-223, are shown in stick representations. FAD and THF are shown in stick representations with the carbon atoms colored yellow and green, respectively. (B) The crystal structure of C. tepidum GidA (13). The FAD-binding, insertion, and C-terminal domains are colored orange, green, and gray, respectively. The disordered region is shown as a dashed line. The conserved Cys residues, Cys-42 and Cys-273, are shown in stick representations. FAD is shown in a stick representation with the carbon atoms colored yellow. (C) Stereoview of the superposition of TrmFO and GidA, based on the FAD-binding domains. The color code is the same as in A and B. For clarity, the C-terminal domain of GidA is omitted. (D) Superposition of the insertion domains of TrmFO and GidA.
Structural Comparison with GidA.
A Dali search (14) revealed that TrmFO shares structural similarity with the GSH reductase family of flavoproteins. As expected from the sequence homology, TrmFO shares the highest structural similarity with the GidA proteins, including Chlorobium tepidum GidA (15) [Protein Data Bank (PDB) ID code 3CP8; Z-score = 28.5 and rmsd for 256 Cα atoms = 2.3 Å] and E. coli GidA (15) (PDB ID code 3CP2; Z-score = 25.5 and rmsd for 263 Cα atoms = 4.1 Å). GidA is an RNA modification enzyme that associates with MnmE to form an α2β2 heterotetramer and participates in the incorporation of the carboxymethylaminomethyl group onto the C5 atom of U34 in tRNAGlu, tRNALys, and tRNAGln (15, 16). Although the mechanism has not been fully elucidated, GidA may reduce the intermediate Schiff base at the last step of the reaction. GidA consists of 3 domains: an FAD-binding domain, an insertion domain, and a C-terminal domain (Fig. 2B). The FAD-binding and insertion domains of GidA share ≈30% sequence identity with those of TrmFO, while the C-terminal domain, which is responsible for the association with MnmE, is absent in TrmFO (Fig. S4A). C. tepidum GidA has the 2 conserved Cys residues, Cys-42 and Cys-273, which correspond to the catalytic Cys residues of T. thermophilus TrmFO, Cys-51 and Cys-223, respectively, but their catalytic role remains uninvestigated.
The FAD-binding and insertion domains of TrmFO can be individually superposed onto those of C. tepidum GidA (Fig. 2 C and D); the rmsd for 241 Cα atoms of the FAD-binding domains is 2.2 Å, and the rmsd for 115 Cα atoms of the insertion domains is 3.3 Å. However, a structural comparison of TrmFO with GidA revealed that their overall structures are remarkably different, mainly because of their domain orientations, which correspond to their distinct biological functions (Fig. 2C). It also should be noted that GidA behaves as a homodimer, with the interface formed by the FAD-binding and insertion domains (Fig. S5A). In contrast, it seems unlikely that TrmFO forms a similar dimer, because of its different domain arrangements. The insertion domain of TrmFO cannot participate in dimerization with a dimer interface area much smaller than that of GidA. Our gel filtration result also supports the idea that TrmFO exists as a monomer in solution (Fig. S3).
The domain organizations of TrmFO and GidA are stabilized by the characteristic structural elements of each protein, indicating that the domain organizations observed here are not simply the consequences of crystal-packing interactions, but are physiologically relevant. In TrmFO, Tyr-381, Met-393, Gly-398, and Leu-399 of the FAD-binding domain and Ile-163, Leu-165, Met-271, and Val-302 of the insertion domain predominantly contribute to the formation of the domain interface through hydrophobic interactions (Fig. S5B). In addition, the side chains of Arg-263 hydrogen-bond to the main-chain carbonyl of Glu-388. Most of these residues are conserved among the TrmFOs, but are replaced in the GidAs by residues with different physicochemical properties (Fig. S4A); Ile-163, Leu-165, Met-271, Tyr-381, and Leu-399 of T. thermophilus TrmFO are replaced with the hydrophilic Arg-200, Asp-202, Glu-302, Asp-412, and Glu-430 residues in C. tepidum GidA, respectively. Consistent with these observations, the corresponding region of GidA is exposed to the solvent. However, the β14–β15 region in the insertion domain of GidA, which is longer than the corresponding region of TrmFO, makes extensive contacts with the FAD-binding domain (Fig. 2C). Moreover, the GidA-specific β-strands (β10 and β11) form a 4-stranded antiparallel interdomain β-sheet with β12 and β19 to stabilize its domain organization. These observations suggest that TrmFO and GidA originated from a common ancestor, but have acquired distinct functions during evolution, not only by the gain/loss of the C-terminal domain but also by the alteration of the spatial arrangement of the FAD-binding and insertion domains.
FAD Recognition.
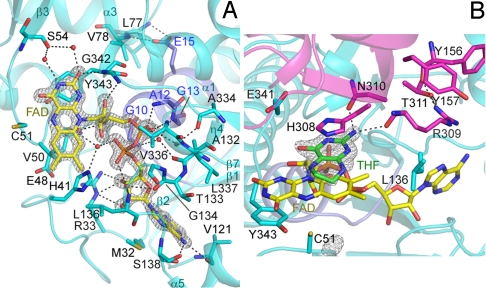
In all 3 of the structures determined here, the FAD is well defined in the electron density (Fig. 3A). The FAD molecule is held by 2 helices (α1 and η4) and 3 loop regions (β1–α1, β2–β3, and β7–α5) through extensive interactions (Fig. 3A). The adenine base is sandwiched between the side chains of Met-32 and Thr-133, while its N6 atom hydrogen-bonds to the Oγ atom of Ser-138 and the main-chain carbonyl group of Val-121. The hydroxyl groups of the ribose moiety hydrogen-bond with the side chain of Arg-33. The pyrophosphate moiety is recognized by the main-chain amide groups of Ala-12 and Val-336 through direct hydrogen bonds and by the main-chain amide or carbonyl of Gly-10, Gly-13, Glu-48, Ala-132, Thr-133, Gly-134, Leu-136, Ala-334, and Leu-337, and the Nε2 atom of His-41 through water-mediated hydrogen bonds. Gly-10, Ala-12, and Gly-13 are part of the GxGxAGxEA signature motif conserved among the GSH reductase family members (Fig. S4B). The invariant Glu-15 in this motif directly hydrogen-bonds with the main-chain amide groups of Leu-77 and Val-78 in α3 to stabilize the local conformation. The ribityl moiety makes van der Waals contacts with the side chain of Val-336, with its hydroxyl groups hydrogen-bonded to the main-chain carbonyl and amide groups of Glu-48 and Gly-342, respectively. The si-face of the flavin ring is held by the side chains of Val-50, Cys-51, and Tyr-343 through van der Waals interactions, while the re-face is accessible to the solvent. The O2 atom of the flavin ring interacts with the main chain amide of Tyr-343 and the carbonyl of Ser-54 through direct and water-mediated hydrogen bonds, respectively. The N3 atom of the flavin ring hydrogen-bonds to the main-chain carbonyl of Ser-54 through a water molecule. The interactions between the protein and FAD, including the water-mediated hydrogen bonds, are preserved in all 3 structures presented here. The residues involved in FAD binding are highly conserved among the TrmFOs (Fig. S4B).
Fig. 3.
Active site of the TrmFO-THF complex. (A) FAD recognition. (B) THF recognition. The color code is the same as in Fig. 1, except that the signature motif of the GSH reductase family is colored blue. Water molecules are shown as red spheres. Hydrogen bonds are shown as dashed lines. The FO − FC omit maps for FAD, contoured at 7σ (A), and THF, contoured at 4σ (B), are shown as gray meshes.
THF Recognition.
In the structure of the THF complex, an electron density for the pteridin moiety of THF is observed (Fig. 3B). The structures of the free-form and the GSH complex lack a corresponding electron density, supporting our assignment of the electron density for THF. Furthermore, we observed substrate inhibition with >2 μM THF in our in vitro assay, suggesting the ability of THF to bind to the active site. No electron density was found for the (p-aminobenzoyl)-glutamate moiety of THF, indicating that these moieties are flexible and thus disordered, but might become fixed upon interactions with the substrate tRNA.
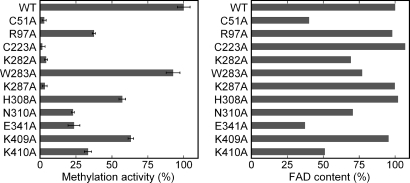
In the THF-bound structure, the pteridin ring is recognized by the strictly conserved residues, His-308, Arg-309, and Asn-310 (Fig. 3B and Fig. S4B). The pteridin ring of THF is sandwiched between the imidazole ring of His-308 and the flavin ring of FAD, with its NA2 atom hydrogen-bonded to the main-chain carbonyl of Arg-309 and the Oδ1 atom of Asn-310. The side chain of His-308 is rotated from its original position in the free-form structure to interact with the pteridin moiety (Fig. S2B). The H308A and N310A mutants showed 57% and 23% activity, compared with that of the wild type, respectively (Fig. 4), which suggests the involvement of these residues in the interactions with the substrate MTHF during catalysis. The invariant Glu-341 resides in the active site, and the E341A mutant showed a decrease in both FAD binding and methylation activity (Fig. 4). Although in the present structure Glu-341 does not directly contact either FAD or THF, these results indicate that Glu-341 is essential for both FAD binding and catalysis. The side chain of Arg-309 contacts the side chains of Leu-136 and Tyr-157 and hydrogen-bonds with the main-chain carbonyl of Tyr-156 and the Oγ1 atom of Thr-311 to stabilize the architecture of the active site. Leu-136, Tyr-157, and Thr-311 are highly conserved among the TrmFOs (Fig. S4B).
Fig. 4.
Methylation activity (Left) and FAD content (Right) of the wild type and mutants. After the in vitro methylation reaction at 60 °C for 20 min, the methylated tRNAIle transcripts were resolved by 10% urea-PAGE and quantified by autoradiography. Because several mutant enzymes showed decreased FAD binding ability, we evaluated the methylation activity by dividing the relative amount of the methylated transcript by the FAD content. The methylation activity of the wild type is expressed as 100% (mean ± SE, n = 3). The FAD content was estimated from the absorbance at 445 nm. The FAD content of the wild type is expressed as 100%.
tRNA Binding Model.
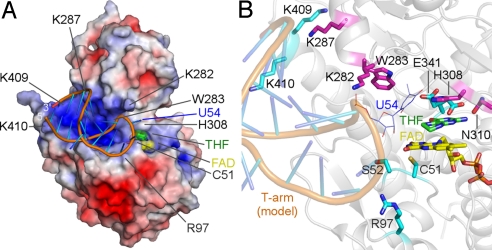
A positively-charged, concave surface is created near the bound flavin ring by the highly-conserved basic residues, including Arg-97, Lys-282, Lys-287, Lys-409, and Lys-410 (Fig. 5A). The side chains of these residues could interact with the phosphate backbone of the substrate tRNA. The target base U54 must be flipped out from the T-loop of the tRNA for modification, because it is originally buried inside the T-loop, where it forms a reverse-Hoogsteen base pair with A58. Indeed, the crystal structure of TrmA in complex with a 19-nt tRNA T-arm analog revealed that TrmA recognizes a flipped-out U54 (8). Because TrmFO is likely to recognize U54 in a similar manner, we manually docked the T-arm analog in the TrmA-RNA structure onto the TrmFO-THF structure (Fig. 5). In this docking model, the T-arm analog could be accommodated by the positively-charged, concave surface of TrmFO without serious steric hindrances. On the basis of the positions of the T-arm, a full-length yeast tRNAPhe (PDB ID code 1EHZ) could also be docked onto the model without any clashes.
Fig. 5.
tRNA docking model. (A) Model of the TrmFO-RNA complex. The 19-nt T-arm analog in the TrmA-RNA structure (8) is docked onto the TrmFO-THF structure. The electrostatic surface potential was calculated by using APBS (29). (B) Close-up view of the potential tRNA binding site. The residues that seem to be involved in substrate binding and catalysis are shown in stick representations. The residues in the FAD-binding and insertion domains are colored cyan and magenta, respectively.
In this model, the side chains of Arg-97, Lys-282, Lys-287, Lys-409, and Lys-410 are located in the proximity of the backbone phosphate groups of the modeled tRNA, and the side chain of the conserved Trp-283 is close to the target base U54 (Fig. 5). To validate our docking model, we prepared Ala mutants of Arg-97, Lys-282, Trp-283, Lys-287, Lys-409, and Lys-410 and examined their in vitro methylation activity (Fig. 4). The K282A and K287A mutants exhibited almost no activity, indicating that Lys-282 and Lys-287 are critical for tRNA binding. The R97A, K409A, and K410A mutants showed decreased activity, indicating the importance of these residues for tRNA binding. The W283A mutant displayed slightly decreased activity, suggesting that Trp-283 may interact with the base or ribose moiety of the substrate tRNA. Taken together, these mutational results support our docking model. However, the structure of the TrmFO-RNA complex is required to fully understand the precise mechanism of tRNA recognition by TrmFO.
Catalytic Mechanism.
The present structure of the TrmFO-THF complex provides structural insight into hydride transfer from FADH2 to the target uridine in a folate/FAD-dependent MTase. Modeling of MTHF onto the THF complex structure indicates that the methylene group linking N5 and N10 of MTHF, which should be transferred onto C5 of U54, is located in the vicinity of the redox-active N5 of FAD. Furthermore, our docking model of the TrmFO-RNA complex suggests that the substrate tRNA could bind at the active site, with U54 close to the bound THF (Fig. 5). Taken together, we propose a catalytic mechanism in which MTHF and the tRNA substrate are simultaneously bound at the active site, where the methylene of MTHF is directly transferred onto C5 of U54, and then the resultant exocyclic methylene at C5 of U54 is reduced by FADH2 to form T54 (Fig. S6). Given our biochemical data indicating the requirement of NADPH for the activity, FAD is likely to be reduced to FADH2 by NADPH before MTHF binding. Like the pteridin moiety of THF, the nicotinamide ring of NADPH might bind between the imidazole ring of His-308 and the flavin ring of FAD.
In addition to TrmA and TrmFO, the MTases acting on the C5 of uridine include 2 classes of thymidylate synthase, folate-dependent ThyA (17, 18) and folate/FAD-dependent ThyX (19), which both methylate dUMP to form dTMP for DNA synthesis. ThyA uses MTHF as both a 1-carbon donor and a reductant, producing dihydrofolate, whereas ThyX uses MTHF as a 1-carbon donor and FADH2 as a reductant, producing THF. Previous biochemical and crystallographic studies revealed that, despite their distinct catalytic mechanisms, both TrmA (7, 8, 20) and ThyA (17, 18) in common form an essential covalent intermediate between the thiol group of a Cys residue and C6 of the substrate uridine, increasing the nucleophilicity of C5 of the uridine to facilitate the methylation. In addition, biochemical studies suggested that ThyX forms a covalent intermediate between the hydroxyl of a Ser residue and C6 of the substrate uridine (21). Thus, it is possible that TrmFO uses a similar strategy to activate the target U54. In the TrmFO structures, the invariant Cys-51 is located in the vicinity of the flavin ring (Fig. 5B). The C51A mutant indeed exhibited almost no activity (Fig. 4), suggesting the possibility that the thiol of Cys-51 attacks C6 of U54 of the substrate tRNA to form a covalent intermediate during catalysis.
By analogy with the catalytic mechanisms of ThyA (17, 18) and ThyX (22, 23), the molecular details of the catalytic mechanism of TrmFO can be inferred as follows (Fig. S6). The thiol of Cys-51 attacks C6 of U54 to form a covalent binary intermediate. The imidazolidine ring of MTHF opens to form an iminium ion at its N5. The activated C5 of U54 attacks the N5 methylene of MTHF to form a covalent ternary intermediate. A general base abstracts the proton at C5 of U54, resulting in the breakdown of the covalent ternary intermediate. The resultant exocyclic methylene at C5 of U54 is reduced by hydride transfer from FADH2 to complete the reaction. The invariant Ser-52, which resides next to Cys-51, might act as a general base to abstract the proton at C5 of U54. In the ThyA reaction, the exocyclic methylene at C5 of dUMP is reduced by the hydrogen at C6 of THF. In this model, C5 of U54 could be located closer to the N5 of FAD rather than the C6 of THF. Therefore, our proposed mechanism provides a possible structural explanation for the reduction of the exocyclic methylene of U54 by the hydrogen, not from the C6 of THF, but from the N5 of FADH2 in the TrmFO reaction. The notion is consistent with our hypothesis that a Cys-51-U54 covalent intermediate is formed during catalysis, because the FAD N5 atom is located close to Cys-51.
The crystal structures of ThyX complexed with dUMP (24) and complexed with the substrate analog BrdUMP (25) revealed that the substrate uridine base stacks with the flavin ring of FAD. However, the catalytic mechanism for the ThyX reaction remains elusive, because no structure of a ThyX-folate complex has been solved. Our present findings suggest the possibility that dUMP and MTHF simultaneously bind at the active site of ThyX during catalysis, which is compatible with the proposed model for T. maritima ThyX (22, 23). Although we cannot exclude the possibility that TrmFO and ThyX methylate the uridine base by different mechanisms because their structures are dissimilar, it seems reasonable that the substrate uridine and MTHF simultaneously bind at the active site for the methylene transfer reaction.
GSH Binding and Its Biological Implications.
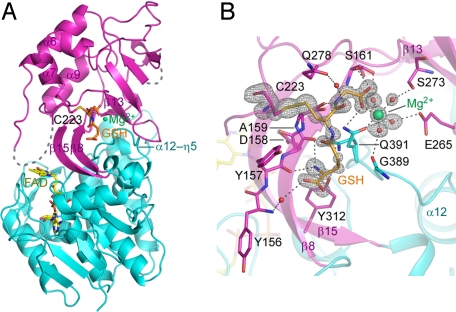
We found that the addition of GSH greatly improved the quality of the crystals, and we solved the GSH-bound structure at 1.05-Å resolution. The structure shows that GSH indeed binds to the insertion domain of the protein (Fig. 6A) and its electron density is well defined (Fig. 6B). GSH is bound in the pocket formed by 2 β-strands (β8 and β13) and 3 loop regions (α6–α7, β13–α9, and α12–η5). GSH is recognized by the main chains of Tyr-156, Tyr-157, Asp-158, and Ala-159 and the side chains of Ser-161, Gln-278, Tyr-312, and Gln-391, through direct or water-mediated hydrogen bonds (Fig. 6B). Notably, the side chain of the invariant Cys-223 forms a disulfide bond with the thiol group of GSH. The electron density also reveals a bound Mg2+ in the vicinity of GSH, based on its coordination geometry, peak heights, bond lengths, and crystallization conditions (Fig. 6B). This Mg2+ is octahedrally coordinated by the carboxylate group of the GSH glycine moiety, the Oε1 of Gln-391, and 4 water molecules. The side chains of Ser-161, Glu-265, and Ser-273 and the main-chain carbonyl of Gly-389 hold the 4 water molecules through hydrogen-bonding interactions. In the structures of the free form and the THF complex, which were crystallized in the absence of GSH, no electron density for either GSH or Mg2+ is visible in the corresponding region, supporting our assignment.
Fig. 6.
Structure of the TrmFO-GSH complex. (A) Overall structure. GSH is shown in a stick representation with carbon atoms colored orange. A Mg2+ ion and water molecules are shown as green and red spheres, respectively. Hydrogen bonds are shown as dashed lines. The color code is the same as in Fig. 1. (B) GSH recognition. Hydrogen bonds are shown as dashed lines. The FO − FC omit map for Cys-223, GSH, Mg2+, and Mg2+-coordinating water molecules, contoured at 5.5σ, is shown as a gray mesh.
Tyr-157, Asp-158, Ala-159, Cys-223, Gln-278, and Gln-391 are highly conserved among the TrmFOs (Fig. S4B), suggesting that GSH binding might be biologically relevant. We examined the effect of GSH on the activity and found that the activity was decreased to 10% in the presence of 0.1 mM GSH. Furthermore, the C223A mutant exhibited almost no activity (Fig. 4). These results indicate the importance of the thiol group of Cys-223 for the catalysis. Although Cys-223 is located >20 Å away from the flavin ring (Fig. 6A), upon tRNA binding a large conformational change might occur to allow Cys-223 to participate in the catalysis. Alternatively, upon tRNA binding, TrmFO might form a homodimer so that Cys-223 is located close to the active site of the adjacent monomer. Because the binding mode between TrmFO and GSH is reminiscent of a protein–protein interaction, it could also be imagined that GSH binding mimics an interaction between TrmFO and its unknown protein partner. Notably, SHMT from organisms possessing TrmFO contains a conserved YGGCE motif (the chemical structure of GSH can mimic the latter 3 residues). In the crystal structure of T. thermophilus SHMT (PDB ID code 2DKJ), the invariant Cys-64 in the motif is located at the end of an α-helix around the active site, with its thiol group buried inside the protein molecule (Fig. S7). It seems possible that a structural rearrangement could enable Cys-64 to participate in association with TrmFO. Thus, it is tempting to speculate that TrmFO and SHMT form a channeling complex to facilitate T54 formation. It should be noted that most of the cellular MTHF is likely to be used for dTMP formation by ThyA or ThyX, and thus it might be reasonable that the MTHF generated by SHMT is directly used by TrmFO, through channeling complex formation.
Materials and Methods
Sample Preparation.
Cloning, expression, and purification of T. thermophilus TrmFO and SHMT are described in SI Text.
Crystallography.
Crystallization, data collection, and structural determination of T. thermophilus TrmFO are described in SI Text and Table S1.
Measurements of tRNA Methylation Activity of TrmFO by an in Vitro Assay System.
To measure the TrmFO activity radioisotopically, we developed an in vitro assay system. l- [U-14C]-serine (1.85 GBq/mmol) and THF were purchased from GE Healthcare and Sigma, respectively. T. thermophilus tRNAIle wild-type and U54A transcripts were prepared as described (26). This assay system is a semiquantitative method, because the rate-limiting factor of the methyl-transfer reaction may be the concentration of [methylene-14C]-MTHF, which is generated from [14C]-Ser by SHMT. The methylation activity was inhibited in the presence of high concentrations of THF, suggesting that excess amounts of THF probably serve as an analog of MTHF. Our pilot experiments showed that the optimal concentrations of THF are 1–10 μM for 1.8 μM TrmFO. Thus, all experiments were carried out under conditions in which the enzyme concentrations were nearly equal to the THF concentration. The reaction mixture (50 μL), consisting of 1.8 μM TrmFO, 2.3 μM SHMT, 1 μM THF, 1 mM NADPH, 28 mM 2-mercaptoethanol, 5 μL of [14C]-Ser, and 1.7 μM transcript, was incubated at 60 °C for 20 min. The incorporation of the [14C-methyl] group into the transcript was quantified after 10% urea-PAGE, using a Fuji Photo Film BAS2000 imaging analyzer system, as described (27). We confirmed the linearity of the reaction at least up to 30 min under our assay conditions. We thus evaluated the methylation activity of the wild-type and mutant enzymes based on the amount of the methylated transcript after a 20-min reaction. To examine the effect of GSH on the methylation activity, a reaction mixture (50 μL) supplemented with 0.1 mM GSH was used.
Nucleotide Analysis by 2D TLC.
The 14C-labeled tRNAIle transcript was purified by 10% urea-PAGE and then completely digested with nuclease P1. After the addition of standard nucleotides, the sample was spotted onto a TLC plate (Merck), which was developed as described (28). The incorporation of the [14C-methyl] group was monitored with a Fuji Photo Film BAS2000 imaging analyzer. Standard nucleotides were located by UV irradiation.
Measurements of UV-Visible Absorption Spectra.
After dialysis, the protein concentration was quantified with a Bio-Rad protein assay kit, using BSA as the standards, and absorption spectra (300–500 nm) were measured with a Shimazu UV-1800 UV-visible spectrophotometer.
Supplementary Material
Acknowledgments.
We thank the staff at the Photon Factory Advanced Ring (Tsukuba, Japan) and SPring-8 (Hyogo, Japan) for technical help during data collection and Dr. Luc Bonnefond for critical reading of the manuscript. This work was supported by a grant-in-aid for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science, and Technology (to O.N.), a grant-in-aid for Science Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology (to H.H.), grants-in-aid for science research from the Ministry of Education, Culture, Sports, Science, and Technology (to H.N, R.I., H.H., and O.N.), and Mitsubishi Foundation grants (to O.N.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structural factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3G5Q, 3G5R, and 3G5S).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901330106/DCSupplemental.
References
- 1.Björk GR. Genetic dissection of synthesis and function of modified nucleosides in bacterial transfer RNA. Prog Nucleic Acid Res Mol Biol. 1995;50:263–338. doi: 10.1016/s0079-6603(08)60817-x. [DOI] [PubMed] [Google Scholar]
- 2.Jühling F, et al. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe K, Oshima T, Nishimura S. CD spectra of 5-methyl-2-thiouridine in tRNA-Met-f from an extreme thermophile. Nucleic Acids Res. 1976;3:1703–1713. doi: 10.1093/nar/3.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shigi N, Suzuki T, Tamakoshi M, Oshima T, Watanabe K. Conserved bases in the TPsi C loop of tRNA are determinants for thermophile-specific 2-thiouridylation at position 54. J Biol Chem. 2002;277:39128–39135. doi: 10.1074/jbc.M207323200. [DOI] [PubMed] [Google Scholar]
- 5.Ny T, Björk GR. Cloning and restriction mapping of the trmA gene coding for transfer ribonucleic acid (5-methyluridine)-methyltransferase in Escherichia coli K-12. J Bacteriol. 1980;142:371–379. doi: 10.1128/jb.142.2.371-379.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urbonavičius J, et al. Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol Microbiol. 2008;67:323–335. doi: 10.1111/j.1365-2958.2007.06047.x. [DOI] [PubMed] [Google Scholar]
- 7.Kealey JT, Gu X, Santi DV. Enzymatic mechanism of tRNA (m5U54)methyltransferase. Biochimie. 1994;76:1133–1142. doi: 10.1016/0300-9084(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 8.Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J. Structure of a TrmA-RNA complex: A consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc Natl Acad Sci USA. 2008;105:6876–6881. doi: 10.1073/pnas.0802247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delk AS, Romeo JM, Nagle DP, Jr, Rabinowitz JC. Biosynthesis of ribothymidine in the transfer RNA of Streptococcus faecalis and Bacillus subtilis. A methylation of RNA involving 5,10-methylenetetrahydrofolate. J Biol Chem. 1976;251:7649–7656. [PubMed] [Google Scholar]
- 10.Delk AS, Nagle DP, Jr, Rabinowitz JC. Methylenetetrahydrofolate-dependent biosynthesis of ribothymidine in transfer RNA of Streptococcus faecalis. Evidence for reduction of the 1-carbon unit by FADH2. J Biol Chem. 1980;255:4387–4390. [PubMed] [Google Scholar]
- 11.Urbonavičius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria-evolutionary implications. Nucleic Acids Res. 2005;33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urbonavicius J, Brochier-Armanet C, Skouloubris S, Myllykallio H, Grosjean H. In vitro detection of the enzymatic activity of folate-dependent tRNA (Uracil-54,-C5)-methyltransferase: Evolutionary implications. Methods Enzymol. 2007;425:103–119. doi: 10.1016/S0076-6879(07)25004-9. [DOI] [PubMed] [Google Scholar]
- 13.Cicmil N. Crystallization and preliminary X-ray crystallographic characterization of TrmFO, a folate-dependent tRNA methyltransferase from Thermotoga maritima. Acta Crystallogr F. 2008;64:193–195. doi: 10.1107/S1744309108003825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite version 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer S, Scrima A, Versees W, Wittinghofer A. Crystal structures of the conserved tRNA-modifying enzyme GidA: Implications for its interaction with MnmE and substrate. J Mol Biol. 2008;380:532–547. doi: 10.1016/j.jmb.2008.04.072. [DOI] [PubMed] [Google Scholar]
- 16.Yim L, Moukadiri I, Björk GR, Armengod ME. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli. Nucleic Acids Res. 2006;34:5892–5905. doi: 10.1093/nar/gkl752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finer-Moore JS, Santi DV, Stroud RM. Lessons and conclusions from dissecting the mechanism of a bisubstrate enzyme: Thymidylate synthase mutagenesis, function, and structure. Biochemistry. 2003;42:248–256. doi: 10.1021/bi020599a. [DOI] [PubMed] [Google Scholar]
- 18.Stroud RM, Finer-Moore JS. Conformational dynamics along an enzymatic reaction pathway: Thymidylate synthase, “the movie.”. Biochemistry. 2003;42:239–247. doi: 10.1021/bi020598i. [DOI] [PubMed] [Google Scholar]
- 19.Myllykallio H, et al. An alternative flavin-dependent mechanism for thymidylate synthesis. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 20.Gu X, Santi DV. Covalent adducts between tRNA (m5U54)-methyltransferase and RNA substrates. Biochemistry. 1992;31:10295–10302. doi: 10.1021/bi00157a017. [DOI] [PubMed] [Google Scholar]
- 21.Leduc D, et al. Functional evidence for active site location of tetrameric thymidylate synthase X at the interphase of three monomers. Proc Natl Acad Sci USA. 2004;101:7252–7257. doi: 10.1073/pnas.0401365101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gattis SG, Palfey BA. Direct observation of the participation of flavin in product formation by thyX-encoded thymidylate synthase. J Am Chem Soc. 2005;127:832–833. doi: 10.1021/ja0432214. [DOI] [PubMed] [Google Scholar]
- 23.Agrawal N, Lesley SA, Kuhn P, Kohen A. Mechanistic studies of a flavin-dependent thymidylate synthase. Biochemistry. 2004;43:10295–10301. doi: 10.1021/bi0490439. [DOI] [PubMed] [Google Scholar]
- 24.Mathews II, et al. Functional analysis of substrate and cofactor complex structures of a thymidylate synthase-complementing protein. Structure (London) 2003;11:677–690. doi: 10.1016/s0969-2126(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 25.Sampathkumar P, et al. Structure of the Mycobacterium tuberculosis flavin dependent thymidylate synthase (MtbThyX) at 2.0-Å resolution. J Mol Biol. 2005;352:1091–1104. doi: 10.1016/j.jmb.2005.07.071. [DOI] [PubMed] [Google Scholar]
- 26.Takeda H, et al. The substrate specificity of tRNA (m1G37) methyltransferase (TrmD) from Aquifex aeolicus. Genes Cells. 2006;11:1353–1365. doi: 10.1111/j.1365-2443.2006.01022.x. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto H, et al. Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus. J Biol Chem. 2004;279:49151–49159. doi: 10.1074/jbc.M408209200. [DOI] [PubMed] [Google Scholar]
- 28.Keith G. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie. 1995;77:142–144. doi: 10.1016/0300-9084(96)88118-1. [DOI] [PubMed] [Google Scholar]
- 29.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.