Abstract
Excess copper is poisonous to all forms of life, and copper overloading is responsible for several human pathologic processes. The primary mechanisms of toxicity are unknown. In this study, mutants of Escherichia coli that lack copper homeostatic systems (copA cueO cus) were used to identify intracellular targets and to test the hypothesis that toxicity involves the action of reactive oxygen species. Low micromolar levels of copper were sufficient to inhibit the growth of both WT and mutant strains. The addition of branched-chain amino acids restored growth, indicating that copper blocks their biosynthesis. Indeed, copper treatment rapidly inactivated isopropylmalate dehydratase, an iron-sulfur cluster enzyme in this pathway. Other enzymes in this iron-sulfur dehydratase family were similarly affected. Inactivation did not require oxygen, in vivo or with purified enzyme. Damage occurred concomitant with the displacement of iron atoms from the solvent-exposed cluster, suggesting that Cu(I) damages these proteins by liganding to the coordinating sulfur atoms. Copper efflux by dedicated export systems, chelation by glutathione, and cluster repair by assembly systems all enhance the resistance of cells to this metal.
Keywords: copA, Escherichia coli, glutathione, hydrogen peroxide, Suf
Copper is universally toxic. Indian childhood cirrhosis and Tyrolean infantile liver cirrhosis are human pathologic processes that result from high environmental exposure (1, 2), whereas Wilson disease is a copper-overloading genetic disorder that stems from the dysfunction of a copper pump that normally keeps levels within tolerable limits (3–6). Microbes are particularly vulnerable to metal poisoning because they cannot control their extracellular environment, and so studies into the mechanisms of metal toxicity have often exploited them as model systems.
Escherichia coli elaborates 3 homeostatic systems that try to maintain low levels of intracellular copper. CopA is a P-type ATPase that pumps copper from the cytoplasm into the periplasm; it is a homologue of the ATP7B protein that is defective in Wilson disease (7). CueO is an oxidase that is believed to oxidize periplasmic Cu(I) to Cu(II), possibly to slow its adventitious entry into the cytoplasm (8, 9). Both CopA and CueO are strongly induced when cells are exposed to low levels of copper (7, 10–12). Higher levels additionally activate the Cus system, which pumps copper from the periplasm back to the extracellular environment (13). Mutants that lack these systems stop growing when they are exposed to copper levels that WT cells can tolerate (14).
The mechanism by which copper poisons cells has been elusive. A long-standing hypothesis is that copper reacts with endogenous H2O2 to generate hydroxyl radicals in a process analogous to the Fenton reaction:
When the analogous reaction is driven by iron in vivo, the hydroxyl radicals are potent oxidants of DNA and cause both mutagenesis and lethality (15, 16). However, when copA cueO cus mutants of E. coli were overloaded with copper, growth was suppressed, but no DNA damage was detected (17). Further inspection suggested that intracellular metabolites, including glutathione, might chelate copper so that it fails to associate with DNA and/or undergo cycles of oxidation and reduction (17).
How, then, does copper “toxify” cells? The present study used copA cueO cus mutants to identify primary routes of intracellular damage. It also exploited the ability of E. coli to grow anaerobically, so that the role of oxygen in copper toxicity could be directly evaluated.
Results
Copper Is Highly Toxic Under Environmentally Relevant Conditions.
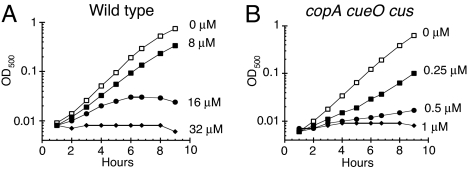
Our goal was to determine the mechanism of copper toxicity under physiological conditions. In a simple salts medium that contained glucose as the sole carbon source, WT E. coli exhibited a growth defect when Cu(II) concentrations exceeded 8 μM (Fig. 1A). Further, this sensitivity increased when systems that control copper levels were removed. A copA cueO cusCFBA mutant showed an aerobic growth defect with as little as 0.25 μM Cu(II) (Fig. 1B). This extreme sensitivity contrasts with the millimolar doses that are needed to inhibit growth in complex media (12, 14, 17).
Fig. 1.
Copper is toxic to aerobic cells at micromolar doses. E. coli cultures were grown at 37 °C in aerobic glucose medium, and CuSO4 was added. (A) W3110 (wild type) cultures were challenged with 0 μM (open squares), 8 μM (closed squares), 16 μM (closed circles), or 32 μM CuSO4 (closed diamonds). (B) LEM33 (copA cueO cusCFBA) cultures were challenged with 0 μM (open squares), 0.25 μM (closed squares), 0.5 μM (closed circles), or 1 μM CuSO4 (closed diamonds). The data are representative of 3 independent experiments.
Copper Caused a Branched-Chain Amino Acid Auxotrophy.
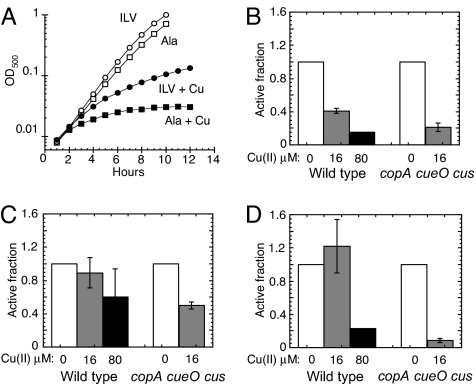
It has been hypothesized that copper produces toxicity in cells in part by generating reactive oxygen species. Indeed, we found that copper-stressed cells produced endogenous H2O2 at a rate several fold higher than did untreated cells (data not shown). Excessive levels of superoxide (O2−) and H2O2 can disrupt several amino acid biosynthetic pathways (18–20); therefore, we tested whether copper creates similar blocks. (This experiment was complicated by the fact that α-amino acids chelate copper and thereby increase the dose necessary for toxicity; to correct for this effect, alanine was added in equivalent doses to cultures from which amino acids of interest were withheld.) The copA cueO cusCFBA mutant could not grow aerobically in glucose/alanine medium to which 10 μM copper was added (Fig. 2A). The addition of branched-chain amino acids partially restored growth, indicating that this pathway is a primary target of copper toxicity. The failure to fully restore growth indicates that additional growth-limiting damage also occurred outside of this pathway.
Fig. 2.
Copper damages iron-sulfur-cluster dehydratases. (A) LEM33 (copA cueO cusCFBA) was grown at 37 °C in aerobic glucose medium with 1.5 mM alanine (Ala) (squares) or 0.5 mM each of isoleucine (I), leucine (L), and valine (V) (circles), and CuSO4 was added to 0 μM (open symbols) or 10 μM (closed symbols). The data are a representative of 3 independent experiments. (B-D) W3110 (WT) and LEM33 (copA cueO cusCFBA) were grown aerobically to an OD500 of ≈0.1, then challenged with 0 μM (open bars), 16 μM (gray bars), or 80 μM CuSO4 (black bars) for 30 min. (B and C) Cells were grown in glucose/alanine, and IPMI (B) and fumarase (C) activities were measured. (D) Cells were grown aerobically in gluconate medium supplemented with 1.5 mM alanine, and 6-phosphogluconate activity was measured. (B-D) Data are the average of 3 independent experiments, and the error bars represent SD. (B) WT cells exposed to 80 μM Cu had IPMI activities below the detection limit (<15%).
Copper Inactivated Iron-Sulfur Cluster Dehydratases Inside Aerobic Cells.
Reactive oxygen species block branched-chain biosynthesis because they directly damage the iron-sulfur clusters of 2 dehydratases: dihydroxy-acid dehydratase in the common branched-chain pathway and isopropylmalate isomerase (IPMI) in the leucine-specific branch (18, 21–23). In conformity with the growth data, we found that the IPMI activity of WT cells did not decrease when they were exposed to 10 μM Cu(II) for 8 h, whereas a similar treatment to copA cueO cusCFBA mutant cells reduced the activity to less than 15% (the detection limit) of untreated WT controls.
In principle, protracted exposure to copper might diminish enzyme activity either by damaging extant enzymes or by blocking new enzyme synthesis or activation. A higher dose of 16 μM copper diminished the total IPMI activity of a culture of WT cells by 60% within 30 min, suggesting that the former explanation pertains. Similarly, activity decreased in copA cueO cusCFBA mutant cells by 80% (Fig. 2B).
Furthermore, the activities of fumarase A and 6-phosphogluconate dehydratase also declined when cells were exposed to copper (Fig. 2 C and D). Like IPMI and dihydroxy-acid dehydratase, these enzymes are dehydratases that employ iron-sulfur clusters, and so it appears that all members of this enzyme class are vulnerable to copper stress. Whereas growth on glucose does not require large substrate fluxes through these enzymes, other growth conditions do: the copA cueO cusCFBA mutant failed to grow aerobically on succinate medium even when it was supplemented with branched-chain amino acids [supporting information (SI) Fig. S1A], a phenotype consistent with the inactivation of fumarase A. Indeed, overexpression of fumarase A restored growth.
The Mechanism of Copper Toxicity Is Independent of Oxygen.
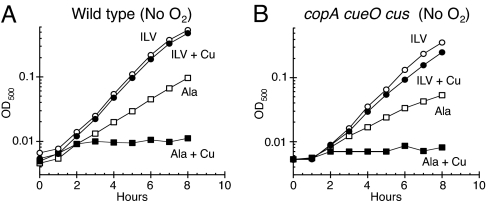
Other investigators have noted that copper can poison bacterial cells in anaerobic medium (12, 17, 24), and we observed, in fact, that both WT cells and the copA cueO cusCFBA mutant were much more sensitive to growth inhibition by copper when oxygen was absent (Fig. S2). The anaerobic growth of the WT and mutant strains was effectively blocked by 1 μM and 125 nM copper, respectively. At the very least, this means that copper has an acute mechanism of toxicity that does not involve reactive oxygen species. Surprisingly, experiments suggested that the primary target of anaerobic toxicity was no different from that in aerobic cells. Growth resumed when branched-chain amino acids were added to anaerobic medium (Fig. 3). This restoration of growth was not caused by chelation of copper, because the D-enantiomers of the amino acids failed to restore growth (data not shown). Further, an overexpression plasmid carrying leuCD fully suppressed the growth defect, indicating that it arose from the lack of IPMI activity (Fig. S1B).
Fig. 3.
Copper inhibits branched-chain biosynthesis even in the absence of oxygen. (A and B) E. coli cultures were grown anaerobically in glucose medium supplemented with either 1.5 mM alanine (ala) (squares) or 0.5 mM each of isoleucine (I), leucine (L), and valine (V) (circles), and they were then challenged with 0 μM (open symbols) or 2 μM CuSO4 (closed symbols). (A) W3110 (WT). (B) LEM33 (copA cueO cusCFBA). The data are representative of 3 independent experiments.
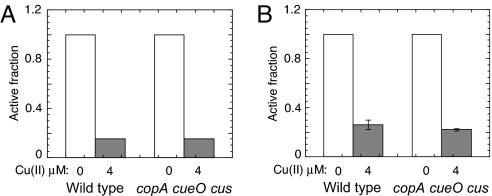
Indeed, during anaerobic exposure to 4 μM copper, the IPMI activities of both WT cells and copA cueO cusCFBA mutant cells decreased to less than 15% of the activity of untreated cells (Fig. 4A). Copper also damaged fumarase B, the anaerobic isozyme of fumarase A (Fig. 4B). These results indicated that the poisoning of these enzymes by copper occurred through a non-oxidative mechanism. It is likely that the same was true in aerobic media.
Fig. 4.
Copper damages iron-sulfur cluster dehydratases in the absence of oxygen. (A and B) W3110 (WT) and LEM33 (copA cueO cusCFBA) cultures were grown anaerobically in glucose/alanine medium to an OD500 of ≈0.1, and then challenged with 0 μM (open bars) or 4 μM CuSO4 (gray bars) for 30 min. (A) IPMI activity: copper-treated cells had IPMI activities below the detection limit (<15%). (B) Fumarase activity: the data are the average of 3 independent experiments, and the error bars represent SD.
CueR is the regulatory protein that senses cytoplasmic copper and activates synthesis of the efflux pump CopA and the copper oxidase CueO. When WT cells were exposed to 2 μM copper—half the level that inactivated IPMI and fumarase A in the preceding experiment—transcription of a copA-lacZ fusion was activated 5-fold (Fig. S3A). Strikingly, transcription of the fusion was fully activated in a copA cueO cus mutant even without copper supplements, indicating that the efflux system was needed to avoid stress from the trace copper that contaminates standard media.
Copper-Damaged Fumarase A Was Repaired in Vivo.
E. coli can repair damaged clusters (25), and so we tested whether the activity of these enzymes would be restored in vivo after toxic doses of copper were removed (see SI Methods). These experiments were conducted in aerobic medium, to facilitate the export of residual copper by the pmf-driven Cus system. Chloramphenicol was included to ensure that any regained activity was not caused by new protein synthesis. We observed that the activity that had been lost during copper treatment gradually reappeared after copper was removed, with half of the activity regained within 30 min (Fig. S4). The fact that the enzyme damage was reversible is consistent with the notion that inactivation was caused by damage to the cluster itself, rather than to polypeptide.
Cuprous Copper Directly Damages Fumarase A in Vitro.
Our previous EPR analyses showed that intracellular copper in overloaded E. coli is in the reduced, Cu(I), valence (17). Therefore, we tested the ability of Cu(I) to directly damage a purified iron-sulfur cluster dehydratase in an anaerobic reaction system. Because of the poor solubility of Cu(I) salts, we generated Cu(I) by co-incubation of Cu(II) with ascorbic acid before its addition to the target enzyme.
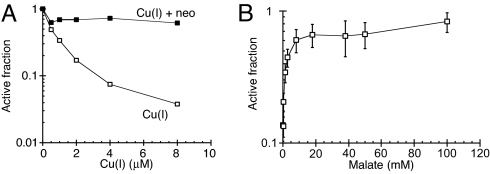
Low micromolar concentrations of Cu(I), but not ascorbate alone, rapidly inactivated purified fumarase A (Fig. 5A). The Cu(I)-specific chelator neocuproine completely protected fumarase activity. To emphatically exclude the possibility that reactive oxygen species might be generated through leaked oxygen, superoxide dismutase (SOD) and catalase were added to the reaction. As expected, these enzymes, together or separately, did not protect fumarase at all (data not shown). We conclude that the enzyme is directly damaged by Cu(I). Separate experiments showed that Cu(II) is also capable of damaging fumarase, but Cu(II) does so at a rate significantly slower than Cu(I) (data not shown).
Fig. 5.
Fumarase A is damaged by Cu(I) and protected by substrate. (A) Purified fumarase A (0.1 μM) was anaerobically challenged in 50 mM Tris-HCl, pH 7.6, with Cu(I) for 3 min at 27 °C either without a chelator (open squares) or in the presence of 100 μM neocuproine (neo) (closed squares). The data are a representative of 3 independent experiments. (B) Fumarase A was exposed to 3 μM Cu(I) for 3 min at 27 °C in the presence of the indicated concentrations of malate, and final activity was determined. The data are normalized to the starting activity. The data are the average of 3 independent experiments, and the error bars represent the SD.
Copper Damaged the Iron-Sulfur Cluster of Fumarase A.
Saturating concentrations of malate, a substrate of fumarase A, fully protected the enzyme activity from copper (Fig. 5B). This effect was probably mediated by its shielding the enzyme active site, as the protective effect tracked closely to the dose needed to form an ES complex.
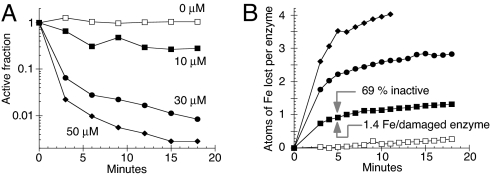
The ferrous-iron chelator dipyridyl was used to monitor any loss of iron from the enzyme. The addition of 10 μM Cu(I) caused a rapid release of more than one atom of Fe(II) per inactivated enzyme (Fig. 6B), with the loss of a second iron atom following quickly after the first (e.g., arrows in Fig. 6B). In contrast, oxidants displace a single iron atom, and the residual [3Fe-4S] cluster is stable to further exposure (18, 26). Higher concentrations of copper lead to total disintegration of the cluster.
Fig. 6.
Cu(I) damages the iron-sulfur cluster of fumarase A. (A and B) Fumarase A (4 μM) was challenged anaerobically at 27 °C with 0 μM (open squares), 10 μM (closed squares), 30 μM (closed circles), or 50 μM (closed diamonds) Cu(I) for 3 min. (A) Fumarase activity after challenge with Cu(I). (B) Time course of iron release from fumarase during copper challenge in A. The data are representative of 3 independent experiments.
Two aspects of the kinetics were unexpected. First, the concentration of copper that was needed to inactivate fumarase A was higher in experiments that used elevated concentrations of fumarase A, even though copper was substantially super-stoichiometric to enzyme. Second, the rate of enzyme inactivation quickly declined during time courses. We speculated that both phenomena reflect the adventitious binding of copper to the protein surface, causing the copper to become progressively immobilized and unable to generate further damage.
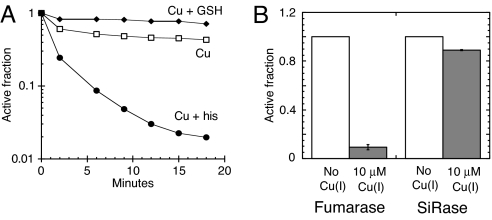
We tested this notion by adding 100 μM histidine as a moderate-affinity solubilizing agent that might lessen the tendency of copper to adhere to the surface of the protein. The histidine significantly enhanced the rate of enzyme inactivation, and it extended the period during which copper was active against the enzyme (Fig. 7A). Presumably histidine and analogous metabolites mobilize copper in vivo, too, and enhance its ability to selectively access higher-affinity binding sites. Conversely, glutathione is a small molecule that binds copper very tightly, and its addition (100 μM) to the reaction system completely blocked damage to the enzyme (Fig. 7A).
Fig. 7.
The ability of Cu(I) to damage iron-sulfur clusters is modulated by copper-binding metabolites and requires that clusters be solvent-exposed. (A) Fumarase A (4 μM) was anaerobically challenged with 10 μM Cu(I) at 27 °C without supplementation (open squares) or in the presence of 100 μM histidine (his) (closed circles) or 100 μM reduced glutathione (GSH) (closed diamonds). The data are a representative of 3 independent experiments. (B) Fumarase A (0.1 μM) was mixed with sulfite reductase (SiRase), and the enzyme mixture was then challenged anaerobically with 0 μM (open bars) or 10 μM Cu(I) (gray bars) for 3 min at 27 °C. The data are the average of 3 independent experiments, and the error bars represent SD.
When a copper-binding protein, mammalian CuZnSOD, was heterologously expressed in WT anaerobic E. coli, 40 U/mg SOD activity was recovered in cell extracts, representing the activation of 100 μM active CuZnSOD monomers in the cytoplasm. At the same time, the presence of the CuZnSOD protein shielded fumarase from inactivation when these cells were exposed to copper (Fig. S3B), presumably by immobilizing the copper (the experiment was performed anaerobically to ensure that the catalytic function of CuZnSOD could not play a role in the protection of fumarase). These results support the implications of the histidine and glutathione experiments: inside the cell, metabolites allow copper to find tight binding sites, and sequestration can restrict the tendency of copper to damage dehydratases.
What Is the Structure of the Damaged Cluster?
Copper-damaged fumarase could be substantially reactivated in vitro. The enzyme was incubated with Cu(I) until 5% of the initial activity remained; neocuproine was then added to chelate the copper and block further damage. The subsequent addition of ferrous iron and DTT reactivated fumarase to 50% of its initial activity, suggesting that a significant fraction of the damaged enzyme molecules contained [3Fe-4S] clusters that retained their inorganic sulfur atoms. The remaining enzymes that could not be reactivated likely had clusters that had degraded further, which would have required the provision of a sulfur species for repair. Unfortunately, the addition of a moderate level of sulfide was inhibitory, causing less recovery of activity than did iron/DTT alone, and higher concentrations led to precipitation. As these treatments have been successful for us in other circumstances (26), we suspect that residual copper interfered with the process.
The stoichiometry of iron release suggested that a population of copper-treated fumarase molecules would likely include clusters at different stages of disintegration. EPR spectroscopy has been used to identify [3Fe-4S]+ (18, 27), [2Fe-2S]+ (28), and [Cu-3Fe-4S]+ clusters (29, 30), and we attempted to find evidence for any of these species (see SI Methods). Enzyme was exposed to sufficient copper to inactivate 85% of the population. This treatment did not generate any significant [3Fe-4S]+ signal, in contrast to a large signal obtained with H2O2 (Fig. S5); this result was expected, as a non-oxidative event that released ferrous iron would be expected to create the EPR-silent [3Fe-4S]0 species. No [2Fe-2S]+ signal was detected, even after the addition of dithionite to reduce any [2Fe-2S]2+ clusters to the EPR-visible [2Fe-2S]+ form, consistent with the limited extent of cluster destruction. We had thought that we might detect a [Cu-3Fe-4S] cluster species similar to those that have been artificially generated in ferredoxins (29, 30). However, no such signal was observed, suggesting that the presumptive displacement of iron atoms by copper does not produce a stable intermediate form that contains copper. Thus, the current data support the idea that copper treatment first converts the [4Fe-4S]2+ native enzyme to a [3Fe-4S]0 form, and that subsequent events further degrade the enzyme through undefined intermediates ultimately to an apoenzyme.
Enzymes with Buried Iron-Sulfur Clusters Were Resistant to Copper.
The vulnerability of dehydratases to chemical agents reflects the accessibility of their clusters to solutes. This principle was reaffirmed by the protective effect that bound substrate had upon the fumarase cluster (Fig. 5B). It follows that enzymes whose clusters are largely occluded by polypeptide are unlikely to be targets of dissolved copper. Indeed, metabolic pathways that include non-dehydratase cluster enzymes—such as sulfate reduction (sulfite reductase), nicotinamide biosynthesis (quinolinate synthase), and ammonia assimilation (glutamate synthetase)—remained functional when cells were exposed to copper, as evidenced by growth in glucose medium supplemented with branched-chain amino acids. Further, when purified fumarase A was co-incubated with sulfite reductase or with everted membrane vesicles containing succinate dehydrogenase, copper doses that eliminated fumarase activity had virtually no effect on these other enzymes, which rely upon buried iron-sulfur clusters (Fig. 7B and Fig. S6). Thus, the toxicity of copper is focused upon cell processes that rely upon proteins with solvent-exposed clusters.
Glutathione and the Suf Iron-Sulfur Cluster Assembly System Contributed to Copper Resistance.
As demonstrated earlier, glutathione protected purified fumarase from damage by copper in vitro. To test whether glutathione might also play this role in vivo, we deleted the gene (gshA) that catalyzes the first step in glutathione biosynthesis. The resultant ΔgshA mutant was indeed more sensitive to growth inhibition by copper than were WT cells (Fig. S7A).
The iron-sulfur cluster assembly system (Isc) is responsible for de novo synthesis and transfer of iron-sulfur clusters. The exact mechanism of Isc function is not yet clear; however, the assembly and/or transfer of an iron-sulfur cluster could plausibly involve its exposure to solvent and, as a consequence, vulnerability to small solutes like copper. In fact, the Isc system is inhibited by H2O2 and O2− (S. Jang, unpublished observations). In E. coli, when Isc function is inhibited, the cell induces a second iron-sulfur cluster assembly system, the Suf system. The Suf system, unlike the Isc system, is resistant to H2O2 and O2−, perhaps because it builds and transfers clusters in a more cloistered environment (S. Jang, unpublished observations). Therefore, we wondered whether copper might similarly inhibit the Isc system and create a requirement for the Suf system during copper stress. To test this idea, cells were grown anaerobically on medium containing glycerol and nitrate to maximize the need for iron-sulfur cluster enzymes. The suf mutant was more sensitive to copper than was the WT strain (Fig. S7B), suggesting both the failure of the Isc system and the resistance of the Suf system.
Discussion
Life evolved in an anaerobic world that was rich in iron. Evolution exploited the versatile surface chemistry of this metal by integrating it into iron-sulfur clusters that serve to bind substrates into enzyme active sites. This arrangement succeeded for 2 billion years, until the emergence of photosystem 2 caused the gradual oxygenation of the environment. As molecular oxygen accumulated, it oxidized dissolved sulfur compounds, which until that time had sequestered soft metals into mineral precipitates. The subsequent solubilization of these metals evidently posed a new problem for extant organisms. The present study reveals that one of these metals, copper, rapidly inactivates the catalytic clusters of dehydratases. This enzyme family has representatives in central catabolic and biosynthetic pathways, which therefore become dysfunctional in copper-exposed cells.
These enzymes are uniquely vulnerable to chemical damage because their clusters are substantially exposed to solvent (31). Previous studies show that O2− and H2O2 are small enough to invade the active site, where they coordinate and oxidize the iron-sulfur cluster to an unstable valence (18, 21, 32). In this work, we found that these clusters are also the primary targets of copper, which evidently displaces their iron atoms when it coordinates their thiolate or inorganic sulfur ligands. The extreme avidity with which it does so means that cellular environments that contain such enzymes cannot tolerate even modest levels of active copper.
Mechanisms of Protection: Compartmentalization or Chelation?
After the biosphere became aerobic, rather than discard and replace their many iron-sulfur-based enzymes in piecemeal fashion, ancient organisms developed comprehensive strategies that shield them from the toxic agents that might damage them. Peroxidases and SODs reduce reactive oxygen species to tolerable levels by converting them to innocuous products. Copper (and other soft metals) cannot be chemically degraded, and so E. coli employs efflux systems that drive copper out of the cytosol (7, 13). Mutants that lacked these enzymes were damaged by nanomolar levels of copper, indicating that such defenses must be requisite for organisms to live in many natural habitats.
The redox activity of copper also induced microbes to integrate it into novel enzymes that facilitate aerobic metabolism, including the copper-zinc SOD, cytochrome bo oxidase, and amine oxidase. However, this advance posed the problem of ensuring that copper was available as a cofactor for these proteins but somehow still blocked from damaging hypersensitive iron-sulfur enzymes. E. coli appears to have solved this conundrum by compartmentalizing its copper-dependent enzymes in the periplasm or the outer aspect of the cytoplasmic membrane, whereas its unstable dehydratases are exclusively inside the cytoplasm. The periplasm accumulates significant copper (17); it contains the Cus copper export system, but the synthesis of the exporter is not triggered until copper levels substantially exceed what is needed to charge these enzymes. In contrast, CopA, which pumps copper out of the cytoplasm, is activated by very low environment copper levels (12) and may be designed to drive the amount of cytoplasmic copper as low as possible. Interestingly, the periplasmic Cus system probably assists the CopA system: by limiting the copper concentration in the periplasm to a reasonable level, it can slow the incidental leakage of copper into the cytoplasm.
Eukaryotes appear to shield dehydratases from copper by chelation rather than compartmentalization. In their cytosol, enzymes that require copper, such as SOD, commingle with enzymes of the dehydratase class, including IPMI. To circumvent possible problems, chaperone systems deliver copper directly to SOD and cytochrome oxidase (33–37). Calculations demonstrate that the level of fully aquated copper inside these cells must be vanishingly low (38), although an unknown amount of un-incorporated copper might still be bound to metabolites and to the surfaces of bio-molecules. It is possible that glutathione plays an accessory role in sequestering loose copper, as it does in E. coli.
Secondary Effects of Cluster Damage.
Workers in eukaryotic systems have reported that copper stress elicits markers of oxidative damage, including DNA damage (39–42). Speculation as to the basis of this phenomenon has centered on the possibility that copper generates hydroxyl radicals by participating in Fenton-like chemistry, after the fashion of iron. However, the discovery that copper destroys iron-sulfur clusters points to another possibility: that the released iron atoms overload the cell and proportionately accelerate iron-based Fenton chemistry. Superoxide stress facilitates DNA damage through a similar mechanism (43, 44). Further, because the eukaryotic iron-responsive protein belongs to the dehydratase class of cluster enzymes (45), it follows that copper may adventitiously convert it into the apo-protein form and thereby stimulate excessive iron import. Consistent with these ideas, workers have shown that iron chelators can suppress some aspects of copper toxicity (40).
It is notable that copper may also inhibit the Isc iron-sulfur-cluster assembly system. Many bacteria express the Suf system as a more-robust back-up, as in the case of the E. coli studied here, but eukaryotes rely exclusively on Isc and therefore may suffer a more expansive set of disabled pathways. Interestingly, Ranquet et al. recently reported that cobalt disrupts Isc function (46). We found that 100 μM cobalt did not directly inactivate fumarase A in vitro (data not shown), which suggests that the mechanism by which metals impede Isc may not directly mimic the mechanism by which they displace iron from dehydratases.
Materials and Methods
Reagents.
Djenkolic acid and D-leucine were obtained from Aldrich; glycerol, D-glucose, potassium cyanide, and potassium nitrate from Fisher Scientific; sodium dithionite from Fluka; Amplex red from Invitrogen; and neocuproine hydrochloride from MP Biomedicals. L-amino acids, D-isoleucine, D-valine, L-ascorbic acid, bovine liver catalase, bovine erythrocyte SOD, bovine milk xanthine oxidase, horse heart cytochrome C, rabbit muscle lactate dehydrogenase, chloramphenicol, copper sulfate, ferrous ammonium sulfate, 2,2′-dipyridyl, disodium L(-) malic acid, D,L-DTT, flavin mononucleotide, hydrogen peroxide, isopropyl β-D-thiogalactopyranoside, β-lactose, NADH, NADPH, potassium ferricyanide, potassium ferrocyanide, potassium D-gluconate, glutathione (reduced), sodium sulfide, succinic acid, tri(cyclohexylammonium) 6-phosphogluconic acid, plumbagin, thiamine, and xanthine were obtained from Sigma.
Fumarase A was purified and quantified (18, 27) from anaerobic cultures of an overexpressing strain (SJ50) that lacks catalase and peroxidase, to ensure that hydrogen peroxide could be used to inactivate the enzyme in subsequent experiments. All purification steps (27) were conducted anaerobically at chamber temperature (27 °C), and all buffers were anaerobic. Sulfite reductase was purified aerobically from an overproducing strain (PD401) grown with djenkolic acid as the sole sulfur source (47).
Bacterial Growth.
Strains are listed in Table S1 in the SI Methods. Luria broth and base M9 minimal salts were of standard composition (48). Where indicated, M9 medium was supplemented with 0.2% glucose, 0.4% gluconate, 0.2% lactose, 40 mM succinate, 40 mM glycerol/40 mM nitrate, 1.5 mM alanine, 0.5 mM L-isoleucine/L-leucine/L-valine, or 0.5 mM of the standard amino acids. Media were supplemented with 100 μg/mL ampicillin, 20 μg/mL chloramphenicol, 30 μg/mL kanamycin, or 12 μg/mL tetracycline when antibiotic selection was needed. Anaerobic cultures were grown in a Coy chamber (Coy Laboratory Products) using pre-equilibrated growth media under 85% N2, 10% H2, and 5% CO2.
To ensure that all studies were conducted upon exponentially growing cells, overnight cultures were diluted to OD500 0.005, and cells were grown at 37 °C to an OD500 of ≈0.1. They were then subcultured again to OD500 0.005 and grown at 37 °C to an OD500 of ≈0.1 before analyses. Measurements were made at 500 nm to minimize interference by dissolved copper.
Enzyme activities from copper-treated cells were typically measured after 30 min of incubation with copper at 37 °C.
Enzyme Assays.
To prepare extracts, cells were washed twice and resuspended in anaerobic buffer. Lysis and the removal of cell debris by centrifugation were also performed in the anaerobic chamber. All enzyme reactions were monitored at room temperature (25 °C). IPMI (49), fumarase (50), β-galactosidase (48), SOD (51), 6-phosphogluconate dehydratase (52), and sulfite reductase (47) activities were measured according to the cited methods. Protein concentrations were determined using the Coomassie protein assay reagent (Pierce) with BSA as the standard.
In Vitro Studies.
Stock solutions of Cu(I) were prepared by mixing Cu(II) with 350 μM ascorbic acid in anaerobic 50 mM Tris-HCl, pH 7.6. The reduction of Cu(II) to Cu(I) was confirmed by the complete loss of the Cu(II) absorption peak at 630 nm.
Purified fumarase was inactivated by addition of copper or H2O2 at 27 °C (i.e., ambient chamber temperature) to enzyme dissolved in anaerobic buffer (50 mM Tris-HCl/10 mM MgCl2, pH 8.0). Where indicated, 100 μM neocuproine, 100 μM reduced glutathione, 100 μM histidine, varied L-malic acid, 160 U of SOD, or 28,000 U of catalase was present.
In some experiments, damaged fumarase was reactivated after copper treatment. Neocuproine (100 μM) was added after the damage period to sequester copper, and 10 μM Na2S/500 μM D,L-DTT/100 μM Fe(NH4)2(SO4)2 were then added.
The loss of iron from the cluster of fumarase was monitored by trapping with dipyridyl (εmM, 9.2 mM−1 cm−1 at 522 nm for the ferrous complex). The reaction contained 4 μM fumarase A/1 mM dipyridyl/50 mM Tris-HCl/10 mM MgSO4, pH 8. Fe(NH4)2(SO4)2 was used to create a standard curve.
Supplementary Material
Acknowledgments.
We thank Mark Nilges (Illinois EPR Research Center, Urbana, IL) for assistance with EPR experiments; Rudiger Laufhutte (University of Illinois Microanalysis Laboratory) for ICP measurements; and Chris Rensing (Universty of Arizona, Tucson, AZ), Jeff Gardner (University of Illinois), John Guest (University of Sheffield, Sheffield, UK), Nicholas Kredich (Duke University, Durham, NC), Tom O'Halloran (Northwestern University, Evanston, IL), and Jim Slauch (University of Illinois) for strains and plasmids. This study was supported by National Institutes of Health Grant GM049640.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812808106/DCSupplemental.
References
- 1.Muller T, Feichtinger H, Berger H, Muller W. Endemic Tyrolean infantile cirrhosis: an ecogenetic disorder. Lancet. 1996;347:877–880. doi: 10.1016/s0140-6736(96)91351-3. [DOI] [PubMed] [Google Scholar]
- 2.Tanner MS, et al. Increased hepatic copper concentration in Indian childhood cirrhosis. Lancet. 1979;8128:1203–1205. doi: 10.1016/s0140-6736(79)91893-2. [DOI] [PubMed] [Google Scholar]
- 3.Bull PC, Thomas GR, Rommens JM, Forbes JR, Cox DW. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat Genet. 1993;5:327–337. doi: 10.1038/ng1293-327. [DOI] [PubMed] [Google Scholar]
- 4.Shah AB, et al. Identification and analysis of mutations in the Wilson disease gene (ATP7B): population frequencies, genotype-phenotype correlation, and functional analyses. Am J Hum Genet. 1997;61:317–328. doi: 10.1086/514864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanzi RE, et al. The Wilson disease gene is a copper transporting ATPase with homology to the Menkes disease gene. Nat Genet. 1993;5:344–350. doi: 10.1038/ng1293-344. [DOI] [PubMed] [Google Scholar]
- 6.Thomas GR, Forbes JR, Roberts EA, Walshe JM, Cox DW. The Wilson disease gene: spectrum of mutations and their consequences. Nat Genet. 1995;9:210–217. doi: 10.1038/ng0295-210. [DOI] [PubMed] [Google Scholar]
- 7.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci USA. 2000;97:652–656. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grass G, Rensing C. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem Biophys Res Commun. 2001;286:902–908. doi: 10.1006/bbrc.2001.5474. [DOI] [PubMed] [Google Scholar]
- 9.Kim C, Lorenz WW, Hoopes JT, Dean JFD. Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene. J Bacteriol. 2001;183:4866–4875. doi: 10.1128/JB.183.16.4866-4875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoyanov JV, Hobman JL, Brown NL. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol Microbiol. 2001;39:502–511. doi: 10.1046/j.1365-2958.2001.02264.x. [DOI] [PubMed] [Google Scholar]
- 11.Outten FW, Outten CE, Hale J, O'Halloran TV. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, CueR. J Biol Chem. 2000;275:31024–31029. doi: 10.1074/jbc.M006508200. [DOI] [PubMed] [Google Scholar]
- 12.Outten FW, Huffman DL, Hale JA, O'Halloran TV. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem. 2001;276:30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 13.Munson GP, Lam DL, Outten FW, O'Halloran TV. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol. 2000;182:5864–5871. doi: 10.1128/jb.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grass G, Rensing C. Genes involved in copper homeostasis in Escherichia coli. J Bacteriol. 2001;183:2145–2147. doi: 10.1128/JB.183.6.2145-2147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 16.Mello Filho AC, Hoffmann ME, Meneghini R. Cell killing and DNA damage by hydrogen peroxide are mediated by intracellular iron. Biochem J. 1984;218:273–275. doi: 10.1042/bj2180273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imlay JA, Fridovich I. Suppression of oxidative envelope damage by pseudoreversion of a superoxide dismutase-deficient mutant of Escherichia coli. J Bacteriol. 1992;174:953–961. doi: 10.1128/jb.174.3.953-961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo CF, Mashino T, Fridovich I. α, β-Dihydroxyisovalerate dehydratase. A superoxide-sensitive enzyme. J Biol Chem. 1987;262:4724–4727. [PubMed] [Google Scholar]
- 22.Smyk-Randall E, Brown OR, Wilke A, Eisenstark A, Flint DH. Near ultraviolet light inactivation of dihydroxyacid dehydratase in Escherichia coli. Free Radic Biol Med. 1993;14:609–613. doi: 10.1016/0891-5849(93)90142-h. [DOI] [PubMed] [Google Scholar]
- 23.Brown OR, Smyk-Randall E, Draczynska-Lusiak B, Fee JA. Dihydroxy-acid dehydratase, a [4Fe-4S] cluster-containing enzyme in Escherichia coli: effects of intracellular superoxide dismutase on its inactivation by oxidant stress. Arch Biochem Biophys. 1995;319:10–22. doi: 10.1006/abbi.1995.1262. [DOI] [PubMed] [Google Scholar]
- 24.Beswick PH, et al. Copper toxicity: evidence for the conversion of cupric to cuprous copper in vivo under anaerobic conditions. Chem Biol Interact. 1976;14:347–356. doi: 10.1016/0009-2797(76)90113-7. [DOI] [PubMed] [Google Scholar]
- 25.Djaman O, Outten FW, Imlay JA. Repair of oxidized iron-sulfur clusters in Escherichia coli. J Biol Chem. 2004;279:44590–44599. doi: 10.1074/jbc.M406487200. [DOI] [PubMed] [Google Scholar]
- 26.Varghese S, Tang Y, Imlay JA. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J Bacteriol. 2003;185:221–230. doi: 10.1128/JB.185.1.221-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flint DH, Emptage MH, Guest JR. Fumarase A from Escherichia coli: purification and characterization as an iron-sulfur cluster containing enzyme. Biochemistry. 1992;31:10331–10337. doi: 10.1021/bi00157a022. [DOI] [PubMed] [Google Scholar]
- 28.Dailey HA, Finnegan MG, Johnson MK. Human ferrochelatase is an iron-sulfur protein. Biochemistry. 1994;33:403–407. doi: 10.1021/bi00168a003. [DOI] [PubMed] [Google Scholar]
- 29.Butt JN, Niles J, Armstrong FA, Breton J, Thomson AJ. Formation and properties of a stable ‘high-potential’ copper-iron-sulphur cluster in a ferredoxin. Nat Struct Biol. 1994;1:427–433. doi: 10.1038/nsb0794-427. [DOI] [PubMed] [Google Scholar]
- 30.Staples CR, et al. Electronic, magnetic, and redox properties of [MFe3S4] clusters (M = Cd, Cu, Cr) in Pyrococcus furiosus ferredoxin. Inorg Chem. 1997;36:5740–5749. doi: 10.1021/ic970200k. [DOI] [PubMed] [Google Scholar]
- 31.Lauble H, Kennedy MC, Beinert H, Stout CD. Crystal structures of aconitase with isocitrate and nitroisocitrate bound. Biochemistry. 1992;31:2735–2748. doi: 10.1021/bi00125a014. [DOI] [PubMed] [Google Scholar]
- 32.Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 33.Glerum DM, Shtanko A, Tzagoloff A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J Biol Chem. 1996;271:14504–14509. doi: 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
- 34.Glerum DM, Shtanko A, Tzagoloff A. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J Biol Chem. 1996;271:20531–20535. doi: 10.1074/jbc.271.34.20531. [DOI] [PubMed] [Google Scholar]
- 35.Cobine PA, Pierrel F, Winge DR. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim Biophys Acta. 2006;1763:759–772. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Culotta VC, Yang M, O'Halloran TV. Activation of superoxide dismutases: Putting the metal to the pedal. Biochim Biophys Acta. 2006;1763:747–758. doi: 10.1016/j.bbamcr.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Culotta VC, et al. The copper chaperone for superoxide dismutase. J Biol Chem. 1997;272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 38.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 39.Arciello M, Rotilio G, Rossi L. Copper-dependent toxicity in SH-SY5Y neuroblastoma cells involves mitochondrial damage. Biochem Biophys Res Commun. 2005;327:454–459. doi: 10.1016/j.bbrc.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 40.Krumschnabel G, Manzl C, Berger C, Hofer B. Oxidative stress, mitochondrial permeability transition, and cell death in Cu-exposed trout hepatocytes. Toxicol Appl Pharmacol. 2005;209:62–73. doi: 10.1016/j.taap.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 41.Rau MA, Whitaker J, Freedman JH, Di Giulio RT. Differential susceptibility of fish and rat liver cells to oxidative stress and cytotoxicity upon exposure to prooxidants. Comp Biochem Physiol C Toxicol Pharmacol. 2004;137:335–342. doi: 10.1016/j.cca.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 42.Saleha Banu B, Ishaq M, Danadevi K, Padmavathi P, Ahuja YR. DNA damage in leukocytes of mice treated with copper sulfate. Food Chem Toxicol. 2004;42:1931–1936. doi: 10.1016/j.fct.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Liochev SI, Fridovich I. The role of O2.− in the production of HO.: in vitro and in vivo. Free Radic Biol Med. 1994;16:29–33. doi: 10.1016/0891-5849(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 44.Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaptain S, et al. A regulated RNA binding protein also possesses aconitase activity. Proc Natl Acad Sci USA. 1991;88:10109–10113. doi: 10.1073/pnas.88.22.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ranquet C, Ollagnier-de-Choudens S, Loiseau L, Barras F, Fontecave M. Cobalt stress in Escherichia coli. The effect on the iron-sulfur proteins. J Biol Chem. 2007;282:30442–30451. doi: 10.1074/jbc.M702519200. [DOI] [PubMed] [Google Scholar]
- 47.Messner KR, Imlay JA. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem. 1999;274:10119–10128. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- 48.Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1972. [Google Scholar]
- 49.Kohlhaw GB. Isopropylmalate dehydratase from yeast. Methods Enzymol. 1988;166:423–429. doi: 10.1016/s0076-6879(88)66055-1. [DOI] [PubMed] [Google Scholar]
- 50.Massey V. Fumarase. Methods Enzymol. 1955;1:729–735. [Google Scholar]
- 51.McCord JM, Fridovich I. Superoxide dismutase: an enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 52.Gardner PR, Fridovich I. Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J Biol Chem. 1991;266:1478–1483. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.