Abstract
Much of the transport, tension, and movement in mitosis depends on kinesins, the ATP-powered microtubule-based motors. We report the crystal structure of a kinesin complex, the mitotic kinesin KCBP bound to its principal regulator KIC. Shown to be a Ca2+ sensor, KIC works as an allosteric trap. Extensive intermolecular interactions with KIC stabilize kinesin in its ADP-bound conformation. A critical component of the kinesin motile mechanism, called the neck mimic, switches its association from kinesin to KIC, stalling the motor. KIC denies access of the motor to its track by steric interference. Two major features of this regulation, allosteric trapping and steric blocking, are likely to be general for all kinesins.
Keywords: EF-hand, motor, calmodulin, regulation
Molecular motors, kinesins, move along microtubules and transport their cargoes by using the energy of ATP hydrolysis (1). Motor, neck, coiled-coil stalk, and tail domains comprise a molecule of kinesin. The motor domain attaches kinesin to microtubules and converts kinesin into an active enzyme that hydrolyzes ATP for each step taken along the microtubule. The tail domain anchors the specific kinesin's cargo, a vesicle, an organelle, a microtubule, or a multiprotein complex (2). In most kinesins, a flexible coiled coil connects the motor and tail domains and assembles kinesin into a dimer. The conformational changes in the motor domains of kinesins that lead to their ATP-powered directed movement along microtubules are well described both structurally and biochemically (3–5). A region of kinesin immediately before or after the motor core, which is called the neck or neck linker, is a crucial element for transmitting the conformational changes in the catalytic site into the mechanical power stroke (6, 7). During the nucleotide hydrolysis cycle, the neck linker adopts distinct positions with respect to the motor domain, being either attached to the motor core or released. In crystal structures, the N-terminal kinesins are found with the neck linker docked along the motor domain in the state reflecting their ATP-like conformation (8, 9). The neck linker is observed to be detached when the N-terminal kinesins are in the ADP-like state (10).
The regulation of kinesins is the least understood aspect of their function. Cargo-binding proteins and proteins mediating the binding of cargoes to the tail region of kinesins are presumed to activate the kinesin motor (11, 12). Pausing of the motor preventing unwanted activity depends on regulatory proteins or internal interactions within the kinesin molecule (13–18). The regulatory proteins modulating activity of the kinesins in response to different cellular signals are still being identified. The Ca2+ ion has recently been shown to regulate Kinesin-1 via the Ca2+-binding protein Miro interacting with kinesin through an adaptor protein milton (18).
For the structural studies described in this article, we have chosen the minus-end directed Kinesin-14 called KCBP (kinesin-like calmodulin-binding protein) (19). KCBP is implicated in formation of the bipolar spindles during nuclear envelope breakdown and the anaphase stage of mitosis by sliding and bundling microtubules (20, 21). During metaphase and telophase, the activity of KCBP is down-regulated to allow for greater microtubules dynamics. The intracellular motor activity of this kinesin during mitosis is presumably tuned by calcium signaling. In vitro, its microtubule-stimulated ATPase activity and affinity to microtubules is negatively regulated by calmodulin or by its recently discovered specific regulator KIC (KCBP-interacting Ca2+-binding protein) (22, 23). KIC requires 3-fold less concentration of Ca2+ (≈1 μM) than calmodulin to completely inhibit activity of KCBP. Both KCBP and KIC are also required for trichrome morphogenesis. Because the calcium signaling is thought to occur through local gradients, regulation of KCBP by KIC and calmodulin may be used differentially to produce a specific response to an appropriate calcium concentration. KCBP consists of the typical kinesin domains but, in addition, has a specialized domain for binding regulatory proteins (24). Ca2+-activated calmodulin or Ca2+-activated KIC binds to an α-helix, which is near the highly negatively charged C terminus of kinesin. Through analysis of the 3-dimensional structure of KCBP in the ATP-like conformation (25), the element preceding this helix was termed the neck mimic because of its sequence and structural resemblance to the neck linker of the N-terminal kinesins [supporting information (SI) Fig. S1]. This element is common to all C-terminal kinesins. Docking of the neck mimic along the motor core stabilizes the ATP-like conformation of the C-terminal motor (25). In the ADP-bound conformation, the interactions with the true neck, N-terminal to the motor core, stabilize the motor core of C-terminal kinesins.
The binding of the regulator, calmodulin or KIC, disrupts interactions of KCBP with microtubules. In the absence of stimulation of its ATPase activity by microtubule binding, the motor switches off (22, 23). Our goal was to discover the regulation of this motor at high resolution and to possibly elucidate the principles applicable to the regulation of other kinesins.
Results
Crystallization of KCBP-KIC Complex.
In our crystallographic experiments, we used Arabidopsis KCBP (amino acids 876-1261), a monomer, and full-length Arabidopsis KIC. The domains and their relation to the amino acid sequence are shown in Fig. 1A. To crystallize the complex, the mutation C1131N was introduced in the region of KCBP's loop L11, which has variable conformations in crystal structures of kinesins and often makes crystal lattice contacts (26). The mutation did not affect microtubule binding of KCBP nor its regulation by KIC. The recombinant KCBP and KIC were separately expressed in Escherichia coli. The complex of 2 proteins was isolated by using Ni2+-NTA chromatography using a polyhistidine tag attached to KIC, later removed by TEV-protease, and crystallized.
Fig. 1.
Three-dimensional organization of complex between kinesin motor KCBP and regulatory Ca2+-binding protein KIC. (A) Schematic illustration of complex components. The fragments of KCBP and KIC visualized in the crystal structure are traced in orange for KCBP and in green for KIC. The domain structure of KCBP is depicted. (B) Crystal structure of the complex of kinesin KCBP and KIC solved at a resolution of 2.4 Å. The motor (light orange) and KIC (green) are shown schematically. ADP (light blue) and Ca2+ ion (black) are space-filling models. The regulatory domain of KCBP is highlighted in orange. (C and D) Two views of the structure of KIC in the complex. In C, KIC is shown in orientation allowing visualization of the part corresponding to the central helix of the Ca2+ sensors (calmodulin, troponin C). The C-terminal part of the “central” helix in KIC is less ordered than its N-terminal half. For alignment of KIC with troponin C and calmodulin, please see Table S2. In D, the EF-hands helices are indicated (A, B, C, D) as the similar elements in the structures of Ca2+ sensors. The β-sheet stabilizing the EF-hand pair is shown in cyan.
Crystal Structure of KCBP-KIC Complex.
Fig. 1B shows the crystal structure of KCBP-KIC complex refined to a resolution of 2.4 Å (see Table S1 and Methods for data collection, refinement statistics, and model building; see Fig. S2 for typical volumes of the electron density map). The model encompasses most of the sequence of the KCBP construct used for crystallization except for a few residues belonging to the termini and to some surface loops. Mg2+-ADP occupies the nucleotide pocket. A prominent 34-Å-long amphipathic helix that binds calmodulin or KIC is observed between the neck mimic and a highly negatively charged C-terminal coil of kinesin. The neck mimic, calmodulin-binding helix, and negative coil comprise the 3-unit regulatory domain of KCBP (25) (Fig. 1 A and B).
A mostly helical protein, KIC wraps around the calmodulin-binding helix of KCBP. The structural model of KIC (Fig. 1C) was built de novo and comprises most of its sequence. The model is missing the N-terminal segment preceding Asp-34 that is rich in hydrophobic residues. Residues 19–22 (ETKY) were modeled with confidence into the electron density map where the side chains of Thr-20 and Tyr-22 contribute to the sites of hydrophobic interactions.
The 2 complex components make extensive contact (1,900 Å2 are buried in the interface), with numerous specific interactions holding the 2 proteins together (identified below). Both the large size of the interface and the specificity of the interprotein interactions suggest that the observed conformation of the complex is physiologically relevant and is not influenced by crystal packing forces.
KIC Is a Previously Uncharacterized Ca2+ Sensor.
Sequence analysis of the recently discovered KIC defined a group of Ca2+ ion-binding proteins featuring a single EF-hand motif (23). Surprisingly, our structure revealed 2 EF-hands in KIC, one with a canonical Ca2+-binding loop (Fig. S2B) (27) (helices C and D in Fig. 1D) and another of nearly identical conformation (helices A and B in Fig. 1D). However, because of its amino acid composition, the loop of this EF-hand will not bind metal ions (Table S2). Both the Ca2+-independent EF-hand and the classical EF-hand loaded with the Ca2+ ion are found in the conformation usually described as “open” and observed for EF-hand pairs of Ca2+-binding proteins [fast skeletal troponin C (28), calmodulin (29)] loaded with Ca2+ ions (Fig. S3). The familiar short antiparallel β-sheet (27) (Fig. 1D) stabilizing EF-hands pairing is formed. The hydrophobic interactions between the helices of the EF-hands further support the open conformation of the EF-hands in KIC. An extensive surface-exposed hydrophobic pocket consistent with the open conformation of the EF-hands embraces the calmodulin-binding helix of KCBP. Remarkably, the 3-dimensional organization of KIC reminds one of troponin C without its C-terminal domain (Fig. S3 B and E). Even the segment corresponding to the troponin C central helix is present in KIC (Fig. 1C). The superposed positions of α-carbon atoms of KIC (amino acids 35–107) and calmodulin (amino acids 5–76) (PDB ID code 1mxe) (30), or KIC (amino acids 34–111) and troponin C (amino acids 13–89) (PDB ID code 1ytz) (28) show RMS deviations of 1.73 and 2.03 Å, respectively. Thus, KIC, troponin C, and calmodulin are structural relatives and belong to the same group of EF-hand proteins, namely the subfamily of Ca2+ sensors (31).
Specific Interactions Between KCBP and KIC Define 2 Extensive Interaction Sites.
There are 2 major sites of interactions between KCBP and KIC. One was anticipated from the secondary structure predictions of KCBP and biochemical experiments; this is the classical target-recognition mechanism used by Ca2+-binding proteins. The amphipathic calmodulin-binding helix of KCBP is localized in the hydrophobic pocket of KIC. In the heart of the hydrophobic pocket, Trp-1233 of calmodulin-binding helix of KCBP is found (Fig. 2). The large hydrophobic side chain of this residue coordinates ≈9 residues of KIC, whereas the side chains of other hydrophobic residues of KCBP (Leu-1226, Leu-1229, Val-1230, Tyr-1232) make further complementary hydrophobic contacts. These interactions define the position and orientation of the calmodulin-binding helix inside KIC.
Fig. 2.
Interactions of calmodulin-binding helix of KCBP with the hydrophobic pocket of KIC. KCBP is in orange; KIC is in green. The selected interacting residues are shown as stick models and specified. The calmodulin-binding helix is shown as a ribbon. The secondary structure elements of KIC are shown schematically.
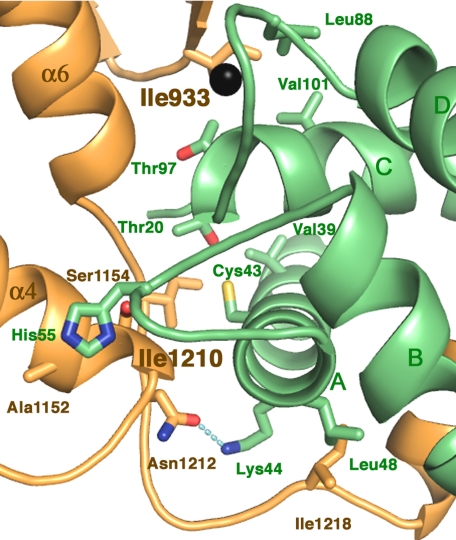
The calmodulin-binding helix, placed within the pocket in KIC is only part of the extensive interface between the motor and its regulator. The 2 proteins interact through a large surface area with less than half (900 Å2) because of interaction via the binding helix. The further contacts are forged between residues of the KCBP neck mimic as well as the part of the kinesin core called the β-domain (32) (indicated in Fig. 1B) with the surface of KIC including the loops of the EF-hands (Fig. 3). These contacts with the surface of KIC, opposite from the traditional target-binding site in the Ca2+-sensors, contribute another 1,000 Å2 into the complex interface and comprise the second interaction site.
Fig. 3.
Interactions formed between the residues of the neck mimic and KIC and between the EF-hands loops of KIC and KCBP. KCBP is in orange; KIC is in green. Ca2+ is shown as a black sphere. The secondary structure elements of both proteins are shown schematically and labeled as in Fig. 1. Principal residues interacting at the interface are shown as stick models and labeled. For better figure presentation, we chose not to show Gln96 of KIC.
In the second interaction site, there are 2 key clusters of hydrophobic side chains. The side chain of KCBP's Ile-1210 forms hydrophobic interactions with 3 side chains of KIC (Cys-43, Val-39, and Thr-20). Another site of strong hydrophobic interactions is formed around Ile-933 of KCBP, a residue conserved in all KCBPs. This residue interacts with 4 hydrophobic residues of KIC including 2 located in the Ca2+-binding loop region, Leu-88 and Thr-97. The side chain of His-55 located in the Ca2+-independent EF-hand loop interacts with KCBP residues Ala-1152 and Ser-1154 at the C-terminal part of its helix α4; the latter is part of the switch II cluster of kinesins. Hydrogen bonds also contribute to this interaction site; one links the main chain of Ile-1210 and the side chain of Gln 96 of KIC, and another is formed with the side chains of Asn 1212 of KCBP and Lys-44 of KIC. The described interactions found in the second site of the KCBP-KIC interface define the position and orientation of KIC with respect to KCBP in the complex.
Conformational State of Kinesin.
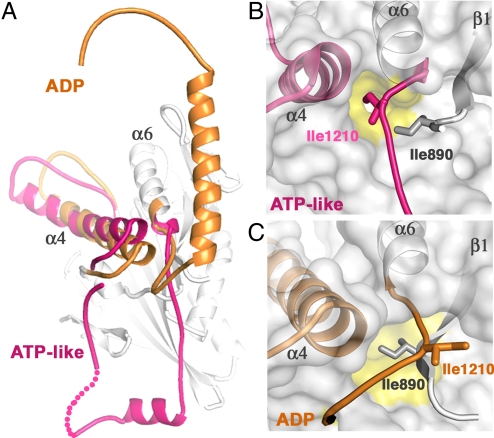
The observed conformation of kinesin in the complex is clearly different from those determined previously without KIC (Fig. 4A) (25, 26). Specifically, the regulatory domain of KCBP is dislodged from the motor surface and is configured differently. The neck mimic is unzipped from its docked position along the motor core and now interacts with KIC (Fig. 3). The calmodulin-binding helix grows longer by 4 helical turns within the KIC hydrophobic pocket. ADP occupies the nucleotide-binding pocket in all KCBP structures, but the positions of the switch II cluster helices (33) differ, representing the ADP-bound conformation of the motor in the complex and the ATP-like conformation in free KCBP (Fig. 4A). In the absence of microtubules, these conformational states of kinesins are of nearly equal free energy (9). Changes in buffer conditions will favor one state over the other, resulting in different conformations observed in the crystal structures.
Fig. 4.
The conformational change in KCBP accompanying ATP hydrolysis. (A) The structure of KCBP-KIC complex is superposed with the structure of KCBP alone (PDB ID code 3cob, chain A). Only the switch II helix α4 and the regulatory elements (in pink) of the solo KCBP structure are shown. The analogous elements of KCBP in the structure of the complex are highlighted in orange. For our previous crystallographic studies of KCBP (PDB ID codes 1sdm, 3cob, 3cnz), we used KCBP from potato (amino acids 884-1252) that is 80% identical to Arabidopsis KCBP. (B) The hydrophobic pocket (marked in yellow) on the kinesin surface (gray) is occupied by Ile-1210 in the ATP-like conformation. (C) The shifted hydrophobic pocket (marked in yellow) on the kinesin surface (gray) is occupied by Ile-890 of the β1 strand as found in the ADP state. Ile-1210 is expelled from the hydrophobic pocket on the kinesin surface and interacts with KIC (Fig. 3). Hydrophobic residues are conserved at positions 890 and 1210 in all kinesins. Helices α4 and α6 and the β1 strand of KCBP (indicated) are shown schematically and are visible through the translucent surface. The neck mimic is shown as a coil and is colored according to the conformational state.
The ADP-bound conformation of KCBP differs significantly from its ATP-like conformation not only by positions of the nucleotide-sensing elements (switch I and switch II) but also by positions of the whole regulatory domain including the neck mimic.
KIC Recognizes ADP-Bound Conformation of KCBP.
KIC binds tightly to residues of KCBP that are exposed in the ADP-bound state but would not be available in the ATP state. Upon hydrolysis of ATP, helix α4 of switch II moves toward the tip of the motor core (Fig. 4A). Influenced by the movement of helix α4, the hydrophobic pocket on the surface of kinesin shifts and exchanges Ile-1210 at the base of the neck mimic (Ile-1210 is conserved in kinesins and positioned like Ile-325 in Kinesin-1) to Ile-890 of the motor β1 strand (Fig. 4 B and C). The liberated Ile-1210 dips into a hydrophobic cavity provided by KIC (Fig. 3). The released neck mimic is fixed to the surface of KIC by several complementary contacts including electrostatic and hydrophobic side-chain interactions (Fig. 3). Additionally, because helix α4 is moved, His-55 of KIC is accommodated between Ala-1152 and Ser-1154 at the C-terminal part of the helix α4. Thus, binding of KIC stabilizes the ADP-bound conformation of the motor.
Modeling of Complex on Microtubules.
We modeled the KCBP-KIC structure on microtubules (Fig. S4) to investigate how binding of KIC affects the relationships of this kinesin with microtubules. The model was built by superposition of the KCBP structure in a complex with KIC and the structure of KAR3, a Kinesin-14 motor, bound to microtubules in ADP state (34). KIC clashes with tubulin, and this fact is sufficient to promote the dissociation of KCBP from microtubules. In addition, the calmodulin-binding helix of KCBP is oriented by KIC so that the negative coil segment after the helix is pointed toward the negatively charged C terminus of tubulin. The repelling electrostatic interactions between this C-terminal region of KCBP and microtubules surface might contribute to the destabilization of the KCBP–microtubules complex in the presence of KIC. However, the effect of the charge repulsion as a part of regulatory mechanism of KCBP seems not to be crucial, because the deletion of the negative coil does not affect the regulation of KCBP by KIC.
Discussion
Many biochemical and functional studies of this complex were recently published (23). We determined and analyzed the crystal structure of kinesin motor KCBP in complex with its physiological regulator KIC, a previously uncharacterized EF-hand Ca2+ sensor. The recognition site formed between KCBP and its regulator extends beyond the motor's special binding domain to engage complementary surfaces of 2 proteins. Our results show that the interface supporting interactions between the motor and its regulator is tuned in response to specific signaling, first of all, by binding of the Ca2+ ion and subsequent conformational changes modifying the entire surface of the Ca2+ sensor. Upon binding of a single Ca2+ ion, the surface of the EF-hand Ca2+ sensor undergoes significant changes that have 2 consequences, the first being the opening of the hydrophobic pocket for binding the specialized helical motif of its target. The second, previously unappreciated, consequence is that the surface of the EF-hands loops, transformed by Ca2+ ion binding, is an equal player in the target-recognition mechanism. This surface is designed to fit the particular conformation of its motor target and makes the regulation highly specific.
The conformation of kinesin in the complex, producing the matching surface for interaction with KIC, is ADP bound. Consistent with the proposed role of imitating the properties of the N-terminal kinesin neck (7, 25), a critical component of motility, the neck mimic stabilizes the ATP conformation of KCBP by being zipped along the motor. This element was predicted to become free when the motor conformation changes to the ADP-bound. The dislocation of the regulatory domain in the ADP state of KCBP is the direct result of the conformational transformation of the motor reflecting its nucleotide state. Indeed, in the ADP state of KCBP, the neck mimic is unbound from the motor core. Therefore, the structure is important evidence of the nucleotide dependent transformations of the neck mimic, supporting the assigned role of this element in the kinesin catalytic cycle.
As a result of the changed motor conformation, many residues become exposed and available for interactions with KIC. These mutually stabilizing, complementary interactions between the motor and its regulator form the extensive specific interface and unambiguously define the relative orientation of the complex components. In the observed conformation of the complex, the neck mimic is found sequestered and, therefore, disengaged from further participation in the conformational events associated with nucleotide hydrolysis and related mechanical cycles of KCBP.
In this work, the previously proposed mechanism of KCBP regulation (25) is elaborated (Fig. S5). The essential strategy for this regulation is that binding of Ca2+-activated KIC stabilizes the motor in the ADP state or in a similar conformation in the absence of nucleotide. KIC disallows ATP hydrolysis by KCBP by fixing the position of the neck mimic on its surface. The immobilized neck mimic is no longer available for interactions with the motor core that are required for achieving a stable conformation when ATP binds. The motor is arrested in a state characterized by weak affinity for microtubules. The interactions of KCBP with microtubules are destabilized further by steric hindrance between KIC and tubulin and electrostatic repulsion between the negatively charged clusters in the C-terminal regions of KCBP and tubulin.
The native KCBP may form a dimer like some other Kinesins-14 (6, 22). Binding of KIC takes place, likely within the proximity of the second motor head, and so may influence its orientation and, therefore, the behavior of the dimer. However, the relevance of this suggestion needs further experimental justification and cannot be warranted based on available data.
The mechanism suggested by findings presented in this article advance our knowledge of kinesin regulation. Our work shows that one strategy to stop a kinesin motor is to trigger its association with a regulatory protein that interferes with the catalytic or mechanical cycles of kinesin by blocking nucleotide-responsive elements of the motor. Given the structural similarity and the analogous roles in nucleotide-induced conformational transformations, we hypothesize that the neck linkers in plus-end kinesins and the neck mimics in minus-end kinesins could be targeted by regulation in an analogous way. Sequestering the neck linker or its mimic would interrupt the nucleotide cycle even in the presence of ATP and microtubules and cause the motor to pause. In addition, similar to the regulation of KCBP by KIC, steric hindrance of microtubule binding is likely to be exploited in the regulation of other kinesins.
Methods
Protein Expression and Complex Isolation.
Arabidopsis KIC was subcloned into a modified pRSFduet plasmid (Novagen) adapted for Gateway cloning technology (Invitrogen). The plasmid encoded the N-terminal His6 tag separated from the expression gene by a linker with the TEV-protease cleavage site. Arabidopsis KCBP (amino acids 876-1261) was cloned into pET28b vector (Novagen) by using NcoI-EcoRI sites carrying no tag. Mutation C1131N was introduced into cDNA of KCBP according to a protocol and by using the reagents of the QuikChange site-directed mutagenesis kit (Stratagene). For protein expression, these constructs were transformed into E. coli-competent cells BL21 (DE3) separately. The protein production was induced by adding 0.1 mM IPTG to the cell cultures. After 3–16 h of expression at 25 °C, the cells were harvested. The cell pellets containing the recombinant KCBP and KIC were combined and subjected to lysis by sonication in the buffer containing 50 mM Tris (pH7.5), 50 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 0.1 mM ATP, 1 mM TCEP, and protease inhibitors mixture. KCBP was provided in excess to ensure that all KIC is bound in a complex. The soluble fraction of the lysate was loaded on the Ni-NTA beads (Amersham). The bound KCBP-KIC complex was eluted in the presence of 100 mM imidazole. The complex was treated with TEV-protease and dialyzed overnight against the original buffer. Then, the sample was passed through the Ni-NTA beads again. The unbound fraction containing the tag-free KCBP-KIC complex was collected. Before crystallization, the complex sample was concentrated up to 15 mg/mL.
Crystallization and Data Collection.
Crystals were grown by using the vapor-diffusion method, in sitting drops under the following conditions: 30% PEG 400, 100 mM Tris (pH 8.5), 200 mM MgCl2, at +4 °C. Before data collection, the crystals were frozen in liquid nitrogen. Data collection was done at the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA) Beamline 8.3.1 (λ = 1.1 Å) by using a single crystal. Data were integrated by using DENZO and scaled with SCALEPACK. The crystal of the complex was of the hexagonal space group P6522 with cell dimensions a = b = 118.8 Å and c = 142.1 Å. The asymmetric unit contained 1 molecule of the complex with molar ratio of KCBP/KIC of 1:1 (57 kDa). The solvent fraction of the crystal is 49.7%, and the Matthews coefficient is 2.5 A3/Da.
Molecular Replacement and Model Building.
The structure of the complex was determined by the molecular-replacement method using the CNS algorithms with a starting model derived from the atomic coordinates for residues 884-1252 of potato KCBP (PDB ID code 1SDM). Electron-density maps based on coefficients 2Fo-Fc were calculated from the phases of the initial model. Subsequent rounds of model building and refinement were performed by using the programs Coot (CCP4 Package) and CNS, respectively. The electron density corresponding to the regulatory protein (KIC) was visualized at early stages of model building. The crystallographic model is refined to 2.4 Å, with R/Rfree values of 22.3/22.8, and contains the polypeptide chains of KCBP (359 aa) and KIC (96 aa), 160 water molecules, 1 Mg2+–ADP complex, 1 Mg2+ ion bound to kinesin, and 1 Ca2+ ion bound to KIC. The model does not include the N-terminal amino acid residues of KCBP (amino acids 876–879 and 881–885), although Tyr-880 was modeled into electron density. There was no visible electron density for loop regions of KCBP (amino acids 1030–1034 and amino acids 1075–1083), and the C-terminal region (amino acids 1256–1261). Only 4 aa (amino acids 19–22) could be modeled for the N-terminal region of KIC. The model continues for amino acids 34–125 of KIC. The following residues (amino acids 126–131) remained invisible. See also Table S1 for more details on data collection and refinement statistics.
Supplementary Material
Acknowledgments.
We thank Dr. K. Hirose for providing the EM models of KAR3 bound on microtubules. This work was supported by National Institutes of Health Grant P01 AR42895 (to Roger Cooke) and National Science Foundation Grant MCB-0079938 (to A.S.N.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structure of KCBP-KIC complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3H4S).
This article contains supporting information online at www.pnas.org/cgi/content/full/0811131106/DCSupplemental.
References
- 1.Schliwa M, Woehlke G. Molecular motors. Nature. 2003;422:759–765. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- 2.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 3.Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- 5.Vale RD, Milligan RA. The way things move: Looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 6.Sablin EP, et al. Direction determination in the minus-end-directed kinesin motor ncd. Nature. 1998;395:813–816. doi: 10.1038/27463. [DOI] [PubMed] [Google Scholar]
- 7.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 8.Kozielski F, et al. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 9.Sindelar CV, et al. Two conformations in the human kinesin power stroke defined by X-ray crystallography and EPR spectroscopy. Nat Struct Biol. 2002;9:844–848. doi: 10.1038/nsb852. [DOI] [PubMed] [Google Scholar]
- 10.Turner J, et al. Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J Biol Chem. 2001;276:25496–25502. doi: 10.1074/jbc.M100395200. [DOI] [PubMed] [Google Scholar]
- 11.Verhey KJ, et al. Light chain-dependent regulation of kinesin's interaction with microtubules. J Cell Biol. 1998;143:1053–1066. doi: 10.1083/jcb.143.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blasius TL, Cai D, Jih GT, Toret CP, Verhey KJ. Two binding partners cooperate to activate the molecular motor Kinesin-1. J Cell Biol. 2007;176:11–17. doi: 10.1083/jcb.200605099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hackney DD, Stock MF. Kinesin's IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat Cell Biol. 2000;2:257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 14.Lee JR, et al. An intramolecular interaction between the FHA domain and a coiled coil negatively regulates the kinesin motor KIF1A. EMBO J. 2004;23:1506–1515. doi: 10.1038/sj.emboj.7600164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bathe F, et al. The complex interplay between the neck and hinge domains in kinesin-1 dimerization and motor activity. Mol Biol Cell. 2005;16:3529–3537. doi: 10.1091/mbc.E04-11-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espeut J, et al. Phosphorylation relieves autoinhibition of the kinetochore motor Cenp-E. Mol Cell. 2008;29:637–643. doi: 10.1016/j.molcel.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Dietrich KA, et al. The kinesin-1 motor protein is regulated by a direct interaction of its head and tail. Proc Natl Acad Sci USA. 2008;105:8938–8943. doi: 10.1073/pnas.0803575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Schwarz TL. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009;136:163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy AS, Safadi F, Narasimhulu SB, Golovkin M, Hu X. A novel plant calmodulin-binding protein with a kinesin heavy chain motor domain. J Biol Chem. 1996;271:7052–7060. doi: 10.1074/jbc.271.12.7052. [DOI] [PubMed] [Google Scholar]
- 20.Vos JW, Safadi F, Reddy ASN, Hepler PK. The kinesin-like calmodulin binding protein is differentially involved in cell division. Plant Cell. 2000;12:979–990. doi: 10.1105/tpc.12.6.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao YL, Deavours BE, Phelps KK, Walker RA, Reddy AS. Bundling of microtubules by motor and tail domains of a kinesin-like calmodulin-binding protein from Arabidopsis: Regulation by Ca(2+)/calmodulin. Biochem Biophys Res Commun. 2000;267:201–207. doi: 10.1006/bbrc.1999.1896. [DOI] [PubMed] [Google Scholar]
- 22.Deavours BE, Reddy AS, Walker RA. Ca2+/calmodulin regulation of the Arabidopsis kinesin-like calmodulin-binding protein. Cell Motil Cytoskeleton. 1998;40:408–416. doi: 10.1002/(SICI)1097-0169(1998)40:4<408::AID-CM8>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 23.Reddy VS, Day IS, Thomas T, Reddy AS. KIC, a novel Ca2+ binding protein with one EF-hand motif, interacts with a microtubule motor protein and regulates trichome morphogenesis. Plant Cell. 2004;16:185–200. doi: 10.1105/tpc.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narasimhulu SB, Reddy AS. Characterization of microtubule binding domains in the Arabidopsis kinesin-like calmodulin binding protein. Plant Cell. 1998;10:957–965. doi: 10.1105/tpc.10.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vinogradova MV, Reddy VS, Reddy AS, Sablin EP, Fletterick RJ. Crystal structure of kinesin regulated by Ca(2+)-calmodulin. J Biol Chem. 2004;279:23504–23509. doi: 10.1074/jbc.M400741200. [DOI] [PubMed] [Google Scholar]
- 26.Vinogradova MV, Malanina GG, Reddy VS, Reddy AS, Fletterick RJ. Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein. J Struct Biol. 2008;163:76–83. doi: 10.1016/j.jsb.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix–loop–helix EF-hand motifs. Biochem J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 28.Vinogradova MV, et al. Ca(2+)-regulated structural changes in troponin. Proc Natl Acad Sci USA. 2005;102:5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ménétrey J, et al. The post-rigor structure of myosin VI and implications for the recovery stroke. EMBO J. 2008;27:244–252. doi: 10.1038/sj.emboj.7601937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clapperton J, et al. Structure of the complex of calmodulin with the target sequence of calmodulin-dependent protein kinase I: Studies of the kinase activation mechanism. Biochemistry. 2002;41:14669–14679. doi: 10.1021/bi026660t. [DOI] [PubMed] [Google Scholar]
- 31.Kawasaki H, Nakayama S, Kretsinger RH. Classification and evolution of EF-hand proteins. BioMetals. 1998;11:277–295. doi: 10.1023/a:1009282307967. [DOI] [PubMed] [Google Scholar]
- 32.Sablin EP, Fletterick RJ. Coordination between motor domains in processive kinesins. J Biol Chem. 2004;279:15707–15710. doi: 10.1074/jbc.R300036200. [DOI] [PubMed] [Google Scholar]
- 33.Kikkawa M, et al. Switch-based mechanism of kinesin motors. Nature. 2001;411:439–445. doi: 10.1038/35078000. [DOI] [PubMed] [Google Scholar]
- 34.Hirose K, Akimaru E, Akiba T, Endow SA, Amos LA. Large conformational changes in a kinesin motor catalyzed by interaction with microtubules. Mol Cell. 2006;23:913–923. doi: 10.1016/j.molcel.2006.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.