Abstract
Studying gene expression at different hematopoietic stages provides insights for understanding the genetic basis of hematopoiesis. We analyzed gene expression in human CD34+ hematopoietic cells that represent the stem-progenitor population (CD34+ cells). We collected >459,000 transcript signatures from CD34+ cells, including the de novo-generated 3′ ESTs and the existing sequences of full-length cDNAs, ESTs, and serial analysis of gene expression (SAGE) tags, and performed an extensive annotation on this large set of CD34+ transcript sequences. We determined the genes expressed in CD34+ cells, verified the known genes and identified the new genes of different functional categories involved in hematopoiesis, dissected the alternative gene expression including alternative transcription initiation, splicing, and adenylation, identified the antisense and noncoding transcripts, determined the CD34+ cell-specific gene expression signature, and developed the CD34+ cell-transcription map in the human genome. Our study provides a current view on gene expression in human CD34+ cells and reveals that early hematopoiesis is an orchestrated process with the involvement of over half of the human genes distributed in various functions. The data generated from our study provide a comprehensive and uniform resource for studying hematopoiesis and stem cell biology.
Keywords: gene expression, genome map, annotation, hemotopoiesis, stem cell
Hematopoiesis is a dynamic process. Formed in the ventral mesoderm at the embryonic stage, hematopoietic stem cells migrate progressively to yolk sac, aortic region, placenta, fetal liver, and bone marrow in the adult. During the process, the hematopoietic stem cells reproduce themselves by self-renewal and differentiate into the multipotent progenitors, the lineage-restricted progenitors, and eventually the mature cell types of erythroid cell, platelet, myeloid cell, monocyte, NK cell, T cell, and B cell in the peripheral circulation to perform specified functions (1). CD34, a cellular membrane glycoprotein, is a specific marker for the hematopoietic cells differentiated at the stem-progenitor stage in humans and other mammalian species (2, 3). The CD34+ hematopoietic stem-progenitor cells (referred to as CD34+ cells hereafter) are essential for maintaining the entire hematopoietic system and are widely used clinically to restore the hematopoietic system through bone marrow transplantation for treatment of various diseases (4). The functional importance of CD34+ cells has attracted much attention to determine the genetic basis of CD34+ cells, as exemplified by analyzing gene expression in CD34+ cells with increased scope and identifying multiple genes and pathways associated with CD34+ cell-related hematopoietic self-renewal and differentiation (5–15). However, the existing knowledge of gene expression in CD34+ cells is not comprehensive because the technologies used are limited, the data generated are fractionated in individual studies and lack consistent annotation with current genome information, and the number of genes implicated as key genes associated with CD34+ cells remains very limited.
To gain more comprehensive knowledge of the genetic basis of human CD34+ cells, we performed an integrated transcriptome analysis on human CD34+ cells. We generated a large transcript sequence dataset from human CD34+ cells. Our extensive informatics analysis of the sequence data reveals much novel information on gene expression in CD34+ cells and provides a current view for the gene expression in CD34+ cells and a comprehensive and uniform resource for studying hematopoiesis and stem cell biology.
Results
The CD34+ Transcript Sequences.
Since CD34+ cells were identified as representing the hematopoietic stem-progenitor cells, continuing efforts have identified the genes expressed in these cells with substantial progress. However, because of the limitations of the technologies, the existing data are not adequate to cover the CD34+ transcriptome. We used the following 2 approaches to maximally collect transcript sequences from CD34+ cells:
De novo CD34+ 3′ EST collection: We performed a large-scale CD34+ 3′ EST collection from normal human hematopoietic CD34+ cells, using a high-throughput generation of long sequences from serial analysis of gene expression (SAGE) tags for gene identification (GLGI) method (16, 17). By using SAGE tags as the sense primers for PCR, GLGI converts SAGE tags into 3′ ESTs. From 10,000 novel SAGE tags obtained in a previous CD34+ study (12), we generated 25,798 high-quality 3′ ESTs. This is the largest EST collection from human CD34+ cells and one of the largest EST collections from a single human cell type using the Sanger sequencing system (http://www.ncbi.nlm.nih.gov/UniGene/lbrowse2.cgi?TAXID = 9606).
Existing CD34+ mRNA sequences collected: We performed database and literature mining to identify publicly available mRNA sequences originating from human CD34+ cells. These include full-length cDNA sequences (8), 5′ and 3′ ESTs (7, 8, 16), 21-bp-longSAGE tags (15), and 14-bp SAGE tags (10, 13). To ensure that the information generated from the study represents the normal CD34+ transcriptome, we used only the sequences generated from normal primary CD34+ cells.
A total of 459,482 CD34+ transcript signatures were identified through these processes (Table 1, Dataset S1). They represent the achievement of CD34+ transcript identification in the past decade using the Sanger sequencing system and provide a solid base for comprehensive CD34+ transcriptome annotation.
Table 1.
The sequence sources and genes identified in CD34+ cells
| Sequence type | Number (%) | Matched (%) | Reference |
|---|---|---|---|
| Sources of CD34+ transcript sequences | |||
| Full-length cDNA | 298 (0.1) | 8 | |
| ESTs | |||
| 3′ EST | 25,798 | Current study | |
| 3′ EST | 1,591 | dbEST | |
| 3′ EST | 214 | 10 | |
| 3′ EST | 177 | 6 | |
| 5′ EST | 13,493 | 8 | |
| Oligo-capped 5′ EST | 1,295 | 14 | |
| EST | 329 | dbEST | |
| Subtotal | 42,897 (9) | ||
| SAGE tags | |||
| 14-bp SAGE tag | 99,954 | 10 | |
| 14-bp SAGE tag | 117,939 | 13 | |
| 21-bp longSAGE tag | 198,394 | 15 | |
| Subtotal | 416,287 (91) | ||
| Total signature | 459,482 (100) | ||
| Human genes matched by CD34+ transcript sequences | |||
| Sequence type | Matched genes | ||
|---|---|---|---|
| Full-length sequence | 298 | 293 (98) | 265 |
| EST | 42,897 | 10,273 (24) | 4,989 |
| SAGE tag | 416,287 | 210,496 (51) | 11,629 |
| Total | 459,482 | 221,017 (48) | 12,759* |
*The number refers to the nonredundant genes.
The Genes Expressed in CD34+ Cells.
We compared the CD34+ transcript sequences with 22,828 human genes, including the 18,013 human genes represented by 26,829 RefSeq mRNA sequences and the 14,129 human genes represented by the SAGEmap reference database. A total of 12,759 (56%) human genes were matched by 221,017 CD34+ transcript sequences (Table 1, Dataset S2), indicating that over half of the human genes are expressed in CD34+ cells.
The Functional Categories for the Genes Expressed in CD34+ Cells.
Using the Gene Ontology reference database (18), we performed a global functional classification for the genes expressed in CD34+ cells (Dataset S3). Under the “Cellular component” category, the genes most commonly expressed were those involved in “cell,” “cell part,” “organelle,” and “organelle part”; under the “Biological process” category, the genes most commonly expressed were those involved in “cellular process,” “metabolic process,” “biological regulation,” and “gene expression”; under the “Molecular function” category, the most commonly expressed genes were those involved in “binding,” “catalytic activity,” “molecular transducer activity,” and “transcription regulator activity.” The broad functional categories imply that CD34+ cells execute many basic biological activities.
We further characterized several groups of functionally important genes relating to hematopoiesis.
Genes Involved in Stem Cell Self-Renewal.
A group of genes has been clearly demonstrated to control self-renewal of hematopoietic stem cells (5, 19). These genes include growth factors, chromatin association factors, homeobox genes, transcription factors, and cell cycle regulators. Searching the CD34+ gene list shows that 59 such genes were detected (Dataset S4A). MYB is an important transcription factor for hematopoiesis. It was detected by 12 ESTs, 6 SAGE tags (33 copies), and 1 longSAGE tag (13 copies), indicating the presence of multiple transcript isoforms from this gene in CD34+ cells. MLL, a Drosophila trithorax homolog, methylates histone H3K4 and regulates expression of many developmental genes including HOX genes. It is frequently involved in acute leukemia through chromosomal translocation. Five MLL isoforms were detected in CD34+ cells, of which 4 were detected by both EST and SAGE tags. HOX genes are the master regulators of cellular differentiation. Of the 39 human HOX genes, 14 were detected in CD34+ cells. Of those 14 genes, HOXA9, HOXA10, and HOXB4 are known regulators of hematopoiesis, and the remaining 11 were newly detected.
Transcription Factor Genes.
Transcription factors are vital for gene expression regulation. Comparing the 1,023 human transcription factor genes grouped in 220 transcription factor families showed that 574 (56%) transcription factor genes were expressed in CD34+ cells (Dataser S4B). These 574 genes were distributed in 197 (90%) transcription factor families (Table 2, Dataset S4C). zf-C2H2, a zinc finger protein family, contains the largest number of genes of all gene families; of the 574 genes detected, 327 belong to this family.
Table 2.
Trancription factor, signal, and kinase genes in CD34+ cells
| Items | Total genes | Detected* (%) |
|---|---|---|
| Examples of transcription factor gene families | ||
| zf-C2H2 | 549 | 327 (60) |
| KRAB | 268 | 162 (60) |
| Homeobox | 204 | 68 (33) |
| HLH | 104 | 46 (44) |
| BTB | 53 | 35 (66) |
| SCAN | 50 | 34 (68) |
| Hormone receptor | 48 | 29 (60) |
| zf-C4 | 48 | 29 (60) |
| bZIP 1 | 33 | 26 (79) |
| ETS | 27 | 18 (67) |
| Fork head | 47 | 18 (38) |
| PAS | 18 | 15 (83) |
| TIG | 14 | 13 (93) |
| PHD | 14 | 12 (86) |
| GATA | 14 | 11 (79) |
| MYB DNA-binding | 17 | 11 (65) |
| Signal pathway genes detected in CD34+ cells | ||
| Calcium signaling pathway | 176 | 44 (25) |
| ERB B signaling pathway | 87 | 29 (33) |
| Hedgehog signaling pathway | 57 | 12 (21) |
| JAK-STAT signaling pathway | 155 | 35 (23) |
| MAPK signaling pathway | 265 | 66 (25) |
| mTOR signaling pathway | 51 | 14 (27) |
| Notch signaling pathway | 46 | 12 (26) |
| Phosphatidylinositol signaling system | 79 | 63 (80) |
| TGF-β signaling pathway | 89 | 27 (30) |
| VEGF signaling pathway | 73 | 20 (27) |
| WNT signaling pathway | 148 | 50 (34) |
| Kinase genes detected in CD34+ cells | ||
| AGC | 69 | 6 (9) |
| Atypical | 39 | 9 (23) |
| Calcium/calmodulin regulated kinases | 113 | 10 (9) |
| Casein kinase 1 | 17 | 5 (29) |
| CMGC | 76 | 9 (12) |
| Receptor guanylate cyclase | 8 | 0 (0) |
| STE | 53 | 10 (19) |
| Tyrosine kinase-like | 49 | 11 (22) |
| Tyrosine kinases | 95 | 18 (19) |
| Other | 101 | 15 (15) |
| Total | 620 (100) | 94 (15) |
*The same genes can be classified in different families.
Signal Transduction Genes.
We searched the CD34+ gene list to identify the genes involved in signal transduction pathways and identified the genes involved in at least 10 pathways (Table 2, Dataset S5A). Notch, Wnt, and TGFB pathways are well known to regulate hematopoietic self-renewal and differentiation (5). Multiple genes involved in these pathways were detected (Dataset S5B). An example is SMAD3, a gene involved in the WNT pathway. It was detected by full-length cDNA, EST, and SAGE tags. More genes involved in other signal transduction pathways were also detected. For example, of the 79 known genes in the phosphatidylinositol signaling system, 63 were detected in CD34+ cells.
Kinase Genes.
Protein kinases play essential roles in regulating a wide range of biological activities. Of the 620 known human kinase genes (20), 94 were detected in CD34+ cells (Table 2, Dataset S5 C–E), including 19% of tyrosine kinase genes. FLT3 is a receptor tyrosine kinase that regulates self-renewal of hematopoietic stem cells, and it is frequently mutated in acute myeloid leukemia. Studies in mouse and human cells have not determined the specific hematopoietic stages for FLT3 expression (21). The detection of FLT3 transcripts by both EST and longSAGE in CD34+ cells indicates that FLT3 is expressed at the stem-progenitor stages. Interestingly, none of the 8 kinase genes belonging to the receptor guanylate cyclase were detected in CD34+ cells. It remains to be determined if this type of kinase plays no role in early hematopoiesis.
microRNA Genes.
Evidence shows that microRNAs are involved in regulating hematopoiesis (22, 23). The primary microRNA transcripts are processed by 5′ capping and 3′ polyadenylation into the precursors before being further processed into mature microRNA (24). Matching CD34+ ESTs and SAGE tags to known human microRNA precursor sequences identifies 45 microRNAs expressed in CD34+ cells, most of which are not known to relate to hematopoiesis (Dataset S6 A–C). Several microRNA precursors are present at high levels, such as hsa-mir-566 (45 EST copies and 56 SAGE copies), hsa-mir-619 (53 EST copies and 187 SAGE copies), and hsa-mir-1273 (195 EST copies). Their high abundance suggests their functional importance in regulating early hematopoiesis.
Alternative Transcription.
The coding sequences of known genes in the genomic DNA have defined structures. However, the transcripts expressed from the genomic coding sequences can be substantially different because of transcriptional regulation. The resulting transcript isoforms substantially increase genomic complexity and can result in altered biological activities. We addressed this issue by analyzing differential transcriptional initiation, alternative splicing and adenylation, and antisense and noncoding transcription.
Alternative transcriptional initiation.
A set of 1,090 5′ ESTs was generated from an oligo-capping CD34+ cDNA library (CD34C, ref. 16). Our evaluation of those sequences with human 5′ cap-analysis gene expression (CAGE) tags shows that the 5′ ends of 83% of the sequences map to 5′ CAGE tags, confirming that the 5′ ESTs from this CD34+ library provide high-quality bona fide 5′ end information. A total of 503 promoters for 495 genes were identified by the 1,090 5′ ESTs, of which 157 belong to the genes with a single promoter and 346 belong to the genes with multiple promoters. Of the 503 promoters, 333 are TATA− CpG+, 52 are TATA+ CpG+, 27 are TATA+ CpG−, and 91 are TATA− CpG− (Table 3, Dataset S7A). The distribution pattern is consistent with that for most human genes (25). IL2RA (interleukin-2 receptor alpha subunit) is a gene important for interleukin 2-regulated T cell proliferation. A promoter of this gene identified by a 5′ EST (DA419380) is TATA− CpG−. The wide use of multipromoter genes with atypical promoter structure suggests that alternative transcriptional initiation is commonly used by the genes expressed in CD34+ cells.
Table 3.
Alternative gene expression in CD34+ cells
| Type | Mapped sequences (%) | Mapped by | Upstream (%) | 3′ end (%) | Mapped antisense (%) | Mapped noncoding (%) |
|---|---|---|---|---|---|---|
| A. Alternative initiation | ||||||
| 5′ EST | 1,090 | |||||
| Mapped promoter | 503 (100) | |||||
| Single promoter gene | 157 (31) | |||||
| Multiple promoter gene | 346 (69) | |||||
| Promoter structure | ||||||
| TATA−, CpG island+ | 333 | |||||
| TATA+, CpG island− | 27 | |||||
| TATA−, CpG island− | 91 | |||||
| TATA+, CpG island+ | 52 | |||||
| B. Alternative splicing and adenylation | ||||||
| 3′ EST | 2,786 (100) | 894 (32) | 1,892 (68) | |||
| SAGE tag | 11,136 (100) | 8,118 (73) | 3,018 (27) | |||
| longSAGE tag | 7,512 (100) | 3,723 (50) | 3,788 (50) | |||
| C. Antisense transcription | ||||||
| Known antisense transcripts | 7,356 (100) | |||||
| EST | 697 | 441 | ||||
| SAGE tag | 1,346 | 1,478 | ||||
| longSAGE tag | 993 | 993 | ||||
| Total | 1,864 (25)* | |||||
| D. Noncoding transcripts | ||||||
| Known noncoding transcripts | 2,354 (100) | |||||
| EST | 958 | 660 | ||||
| SAGE tag | 345 | 405 | ||||
| longSAGE tag | 112 | 144 | ||||
| Total | 923 (39)* |
*The numbers refer to nonredundant sequences.
Alternative Splicing and Adenylation.
Alternative splicing and adenylation are 2 mechanisms of posttranscriptional regulation (26). A 3′ EST is located at the 3′ end of the detected transcript. Their 3′ end location in the mapped RefSeq can be used to determine the alternatively spliced and/or adenylated transcripts; a SAGE tag is located at the 3′ end after the last CATG of the detected transcript. A SAGE tag matching to the upstream CATG position of the RefSeq implies that the SAGE tag is derived from an alternatively spliced or adenylated transcript. Mapping the poly(A) signal + 3′ ESTs to RefSeq shows that 28% of the 3′ ESTs matched upstream of the 3′ ends (Table 3, Dataset S7B). FTH1 (NM_002032) represents an example. Of the 7 ESTs mapped to its 1,245-base full-length sequence, all matched 297 bases upstream of the 3′ end. SAGE tag mapping shows that 50–73% of the SAGE tags matched upstream locations (Table 3, Dataset S7 C and D). For example, of the 5 SAGE tags matching to the FOXK1 sequence (NM_001037165), 4 matched upstream CATG sites and 1 matched the 3′-end CATG site (Dataset S7C).
Antisense and Noncoding Transcripts.
Antisense transcription is a mechanism for gene expression regulation. Up to 70% of human genes have been shown to express antisense transcripts (27). Using the 7,356 well-annotated antisense sequences as the reference, 25% are mapped by CD34+ ESTs and SAGE tags (Table 3, Dataset S8A). AA687703, an antisense transcript to CSDE1, was mapped by an EST, a SAGE tag, and a longSAGE tag. Noncoding transcripts are involved in gene expression regulation, RNA editing, genomic imprinting and epigenetic activities (32), and stem cell differentiation (28). Of the 2,354 known noncoding transcript sequences, 39% are mapped by CD34+ ESTs and SAGE tags (Table 3, Dataset S8B). For example, a noncoding xi transcript AF001545 was detected by a SAGE tag and a longSAGE tag.
CD34+ Cell-Specific Gene Expression Signatures.
Reflecting the dynamic process of hematopoietic differentiation, the spectrum of the transcriptome changes at different stages. We compared gene expression between CD34+ cells, upstream embryonic stem cells, and downstream mature hematopoietic cell types. SAGE tags provide both high coverage and quantitative information and were used for the comparison. The results show that CD34+ cells do have specific expression signatures, as reflected by the differences of thousands of SAGE tags between CD34+ cells and other cell types (Table 4, Dataset S9). Detailed comparison identified a set of core genes that distinguishes CD34+ cells from multiple cell types, including 220 genes highly or only detected in CD34+ cells and 98 genes highly or only detected in other cell types (Table 5, Dataset S9B). The functional categories of these genes cover a wide range. These genes provide new candidate genes associated with early hematopoiesis and new candidate gene markers specific for CD34+ cells. HLA-DRA, an MHC class II gene, plays a critical role in antigen presentation in the immune reaction. However, the highest expression level of this gene is in CD34+ cells but not in antigen presenting cell types. XIST encodes noncoding transcripts that silence one of the 2 X chromosomes through X chromosome imprinting. XIST is considered to be active only at the early embryonic stages. The high-level expression of multiple XIST transcript isoforms in CD34+ cells as reflected by 3 different SAGE tags at 32, 13, and 10 copies (Dataset S9B) supports the notion that XIST is reactivated in early hematopoiesis (29).
Table 4.
CD34-specific gene expression signature: Differences between CD34+ cells and multiple cell types*
| Comparison to | Expression status in CD34+ cells |
Total | |||
|---|---|---|---|---|---|
| Present | High | Absent | Low | ||
| ES cells | 1,077 | 436 | 1,257 | 1,063 | 3,833 |
| Erythroids | 327 | 198 | 387 | 306 | 1,218 |
| Monocyte | 767 | 269 | 522 | 568 | 2,126 |
| Immature dendritic cells | 486 | 299 | 897 | 677 | 2,359 |
| Mature dendritic cells | 229 | 177 | 484 | 391 | 1,281 |
| CD4 T cells | 396 | 254 | 714 | 397 | 1,761 |
| CD8 T cells | 394 | 216 | 682 | 481 | 1,773 |
| NK cells | 276 | 178 | 482 | 491 | 1,427 |
| B cells | 393 | 255 | 712 | 543 | 1,903 |
| Myeloid cells | 507 | 535 | 711 | 565 | 2,318 |
| Total | 4,852 | 2,817 | 6,848 | 5,482 | 19,999 |
*Each tag is determined under P < 0.05 and fold change ≥ 3 between CD34 and given cell type. ES, embryonic stem cells; ER, erythroid cells; MC, monocytes; ID, inmature dendritic cells; MD, mature dendritic cells; CD4 T, CD4+ T cells; CD8 T, CD8+ T cells; B, B cells; Mye, myeloid cells.
Table 5.
CD34-specific gene expression signature: Examples of the signature genes between CD34 cells and multiple cell types
| Gene | SAGE tag | SAGE tag copy* |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD34 | ES | ER | MC | ID | MD | CD4 T | CD8 T | NK | B | Mye | ||
| High in CD34+ cells | ||||||||||||
| ANXA11 | TGGCGTACGG | 97 | 1 | — | 4 | — | 3 | 6 | — | 3 | 4 | 9 |
| ATCAY | GGGACCACCG | 26 | — | — | — | — | — | — | — | — | — | — |
| EEF1A1 | TTTTTGATAA | 156 | 8 | — | 2 | 1 | 1 | 2 | 2 | — | 1 | 34 |
| HLA-DRA | ATTCCTGAGC | 17 | — | 2 | — | 4 | — | — | — | 1 | — | — |
| HMGB1 | TCTGCTAAAG | 58 | 19 | 3 | 2 | 5 | — | 2 | 1 | — | — | — |
| HSPD1 | CTCTTAAAAG | 32 | — | 1 | — | — | — | — | — | — | — | 2 |
| KIF5C | GAGCGGCGCT | 114 | — | — | — | — | — | — | — | — | — | — |
| MALAT1 | CCAGAGAACT | 165 | — | 29 | 11 | 13 | 4 | 7 | 2 | 2 | 1 | — |
| MPO | GCTCCCCTTT | 126 | — | 10 | — | — | — | — | — | — | — | 37 |
| PRDX1 | ACCCGCCGGG | 918 | 6 | 6 | 10 | 24 | 8 | 3 | 1 | 4 | 1 | 103 |
| SET | GAGTAGAGAA | 17 | — | — | 2 | 5 | — | 3 | — | — | 3 | — |
| SP1 | ATGATCTGCC | 13 | 1 | 1 | — | 3 | — | — | — | — | — | — |
| UBQLN1 | TCTTTTATTA | 27 | — | — | 2 | — | — | — | — | — | — | 10 |
| XIST | GGTGACCACC | 32 | — | — | — | — | — | — | — | — | — | 6 |
| Low in CD34+ cells | ||||||||||||
| APOE | CGACCCCACG | — | 62 | 9 | 7 | 44 | 16 | 4 | — | — | — | — |
| CALM1 | CAGCTTGACG | — | 13 | — | — | 11 | 12 | 8 | 6 | 5 | 4 | 7 |
| CBLB | GTGACCACGG | — | 22 | 18 | 50 | 319 | 51 | 108 | 14 | 17 | 5 | 2,866 |
| CXCR4 | TTAAACTTAA | 3 | — | — | 13 | — | 11 | 35 | 30 | 12 | 89 | 20 |
| HLA-C | GTGCGCTGAG | 11 | 4 | — | 174 | 79 | 173 | 165 | 230 | 226 | 84 | 4 |
| HMOX1 | CGTGGGTGGG | — | 4 | — | 31 | 31 | 17 | — | — | 3 | — | 9 |
| JUNB | ACCCACGTCA | 1 | — | — | 49 | 14 | 24 | 51 | 44 | 10 | 52 | 21 |
| MAP2K2 | CAGGAACGGG | 2 | 11 | 5 | 10 | 8 | 6 | 5 | — | 10 | 11 | — |
| MAP4K3 | CAAATCCAAA | — | 10 | — | 4 | 7 | 213 | 42 | 42 | 31 | 26 | — |
| NFKB2 | GGAAGGGGAG | — | 9 | — | — | 5 | 8 | 13 | 12 | — | 62 | — |
| S100A10 | AGCAGATCAG | 2 | 14 | — | 77 | 63 | 37 | 8 | 18 | 22 | — | 33 |
| TNFRSF1B | ATGGAGCGCA | — | — | — | 38 | 3 | 11 | 4 | — | 13 | — | 26 |
*Please see Table 4 for details.
CD34+ Cell Transcription Map.
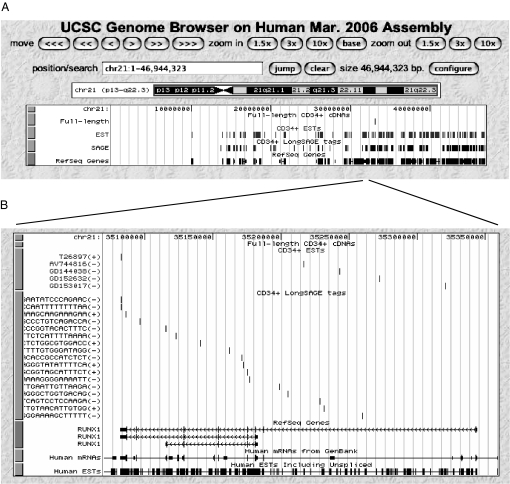
We developed a CD34+ cell transcription map that provides a genomewide view for the transcription activities in CD34+ cells. The map contains detailed mapping information, including the CD34+ transcript-detected genes with their corresponding exon, intron, antisense, promoter, 5′, and 3′ ends, and the CD34+ transcript-mapped intergenic region representing novel transcribed loci (Dataset S10). The map is integrated into the University of California, Santa Cruz (UCSC) human genome browser and can be visualized from the whole-chromosome to the single-base levels (http://projects.bioinformatics.northwestern.edu/wanglab/CD34plus/). By selecting the items listed in the genome browser, the information related to the mapped transcripts can be selected. Comparing the general transcriptional information listed in the browser, the CD34+ cell transcription map shows the commonality and differences between the known transcribed loci and the CD34+-specific transcribed loci. Fig. 1 shows the CD34+ transcription map of chromosome 21.
Fig. 1.
CD34+ cell transcription map in chromosome 21. (A) The positions of full-length cDNA, EST, and longSAGE tags in chromosome 21 of hg18. The RefSeq in the bottom line represents the known human genes. The map is integrated in the UCSC human genome browser. (B) A zoom-in view of the mapped transcripts at a locus containing AML1/RUNX1, the gene important for early hematopoiesis. Of the 5 ESTs mapped to this gene, 1 maps to the 3′ end but in an antisense orientation, and 4 map to the first intron; of the 17 mapped longSAGE tags, 2 map to the 3′ end, and 15 map to different introns of which 5 are in antisense orientation.
Discussion
Our study used normal CD34+ transcript sequences of full-length cDNA, EST, and SAGE tags collected by the Sanger sequencing system (30). By analyzing the integrated CD34+ transcript sequences with the current genome information, our study provides a comprehensive view on the CD34+ transcriptome. Data from the study show that early hematopoiesis is an orchestrated process involving over half of the human genes and indicate that systems approaches will be required to fully reveal the genetic base of hematopoiesis.
Two issues need to be considered for this study. One is the nature of CD34+ cells, and the other is the comprehensiveness of the data. CD34+ cells are not a homogeneous but a heterogeneous population covering stem cells, earlier multipotent progenitors, and later lineage-restricted progenitors. Using more specific markers, CD34+ cells can be classified into narrower differentiation stages. Although it would be ideal to analyze the cells at more specific stages, the increased rarity of such cells restricts their practical usage for large-scale transcriptome studies. Developing new approaches, such as single cell-based assays for transcript isolation, and collecting sequences by using next-generation DNA sequencers that demand less input material than the Sanger sequencer could improve the situation. Like transcriptome studies in most human cell types, our current study does not cover the entire CD34+ transcriptome. This is illustrated by the absence of certain genes known to play roles in early hematopoiesis. For example, multiple microRNAs are involved in regulating hematopoiesis, but many are not included in the microRNA dataset from the current study (23). Our current study targeted only the poly(A)+ mRNAs. Increasing evidence shows that different types of transcripts exist, such as the regulatory small RNAs (31). Those transcripts do not contain poly(A) tails [poly(A)−] (32) and cannot be detected by the poly(A)+-based approach. In addition, at a given sequencing scale, certain functionally important genes expressed at lower abundance will be under the threshold of detection. The next-generation sequencers provide much higher throughput capacity. Their application should increase transcriptome coverage.
Methods
De Novo Collection of CD34+ 3′ ESTs.
Bone marrow CD34+ cells of 3 healthy donors were purchased from AllCells. 3′ ESTs were collected by using the GLGI method (18, 19). A total of 10,000 CD34+ SAGE tags identified in a previous study (12) were selected as the sense primers for the GLGI reactions on the basis of the following conditions: i. Each tag should map to the human genome sequences. This will provide a minimal guarantee that the SAGE tag is likely to be from transcripts expressed from the human genome. ii. There should be no poly(A) track (>7 consecutive A's) 200 bp downstream of the tag-mapped location (33). This restriction will help to exclude the SAGE tags from the cDNA generated by internal oligo(dT) priming. iii. It should not map to known human mRNA sequences. This will increase the chance of identifying novel transcripts.
Sources of Existing Transcript Sequences from Human CD34+ and Other Cell Types.
Full-length cDNA, ESTs, and SAGE tags from normal human CD34+ cells were downloaded from NCBI Entrez (http://www.ncbi.nim.nih.gov/Entrez). SAGE data from other cell types were downloaded from NCBI GEO (http://www.ncbi.nim.nih.gov/geo). Statistical SAGE data comparisons were performed using the IDEG6 program (http://telethon.bio.unipd.it/bioinfo/IDEG6/) under the cutoff of P < 0.05 and fold change ≥3 between datasets.
Reference Databases Used for the Analyses.
The RefSeq mRNA sequences of “REVIEWED” and “VALIDATED” were downloaded from http://www.genome.ucsc.edu. The “SAGEmap reliable” database was downloaded from http://www.ncbi.nlm.nih.gov/projects/SAGE/. The Gene Ontology database was downloaded from http://www.geneontology.org/. The transcription factor genes were downloaded from http://dbd.mrc-lmb.cam.ac.uk/DBD/. The genes in signal transduction pathways were downloaded from http://www.genome.jp/kegg/pathway.html. Kinase genes were downloaded from http://kinase.com/human/kinome/. microRNA precursor sequences were downloaded from miRbase under “hairpin sequences” (http://microrna.sanger.ac.uk/sequences/). To identify SAGE tag-detected microRNAs, the hairpin sequences were extended with 30-bp genomic sequences at both 5′ and 3′ ends to increase the chance of finding the CATG site (27). Reference SAGE tags were then extracted next to the identified CATG sites. The CAGE database was downloaded from http://gerg01.gsc.riken.jp/cage/hg17prmtr/. The database of transcription start sites was downloaded from http://dbtss.hgc.jp/. Antisenses were downloaded from http://natsdb.cbi.pku.edu.cn. Noncoding transcripts were downloaded from http://research.imb.uq.edu.au/RNAdb.
Determining Alternative Splicing and Adenylation.
Each 3′ EST was examined for the poly(A) signal 10–30 bases upstream from the 3′ end in the order of AATAAA, ATTAAA, TATAAA, AGTAAA, AAGAAA, AATATA, AATACA, CATAAA, GATAAA, AATGAA, TTTAAA, ACTAAA, AATAGA (26). The 3′ ESTs were mapped directly to the RefSeq mRNA sequences. The 3′ ESTs that ended within ±10 bp of the mapped RefSeq sequences were classified to represent the 3′ ends, and those mapped farther upstream were classified to represent alternative spliced sequences. To identify SAGE tag-detected alternatively spliced transcripts, 14- or 21-bp reference SAGE tags were extracted after all CATG sites in the RefSeq sequences.
Genome Mapping.
Full-length cDNA sequences and ESTs were mapped directly to hg18, and longSAGE tags were mapped to the reference longSAGE tags extracted from all CATG sites in hg18. The mapping is chromosome based and integrated into the UCSC genome browser with its all selectable features.
Supplementary Material
Acknowledgments.
We thank Connie J. Eaves (BC Cancer Agency, Vancouver, BC) for providing CD34+ LongSAGE tag data. We appreciate the thoughtful comments and criticisms of Kenneth Boheler and Macelo Bento Soares. This research was supported by National Institutes of Health grant R01HG002600 (to S.M.W.), by the Daniel F. and Ada L. Rice Foundation (S.M.W.), by a Career Development Award from Evanston Northwestern Healthcare Research Institute (to S.M.W.), by National Institutes of Health Grants R01 HG001696 (to M.Q.Z.) and CA84405 (to J.D.R.), and by the University of Chicago (J.D.R.).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Data deposition: The 3′ EST data generated from the study were deposited in NCBI dbEST with accession number GD135551-161348. The genome mapping information is listed at http://projects.bioinformatics.northwestern.edu/wanglab/CD34plus.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903390106/DCSupplemental.
References
- 1.Orkin SH. In: Stem Cell Biology. Marshak DR, Gardner R, Gottlieb D, editors. Plainview, NY: Cold Spring Harbor Lab Press; 2001. pp. 289–301. [Google Scholar]
- 2.Simmons DL, Satterthwaite AB, Tenen DG, Seed B. Molecular cloning of a cDNA encoding CD34+, a sialomucin of human hematopoietic stem cells. J Immunol. 1992;148:267–271. [PubMed] [Google Scholar]
- 3.Satterthwaite AB, Burn TC, Le Beau MM, Tenen DG. Structure of the gene encoding CD34+, a human hematopoietic stem cell antigen. Genomics. 1992;12:788–794. doi: 10.1016/0888-7543(92)90310-o. [DOI] [PubMed] [Google Scholar]
- 4.Burt RK, et al. Clinical applications of blood-derived marrow-derived stem cells for nonmalignant diseases. JAMA. 2008;299:925–936. doi: 10.1001/jama.299.8.925. [DOI] [PubMed] [Google Scholar]
- 5.Zon L. Intrinsic extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453:307–313. doi: 10.1038/nature07038. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y, Peterson KR, Stamatoyannopoulos G, Papayannopoulou T. Human CD34+ cell EST database: single-pass sequencing of 402 clones from a directional cDNA library. Exp Hematol. 1996;24:605–612. [PubMed] [Google Scholar]
- 7.Mao M, et al. Identification of genes expressed in human CD34+(+) hematopoietic stem/progenitor cells by expressed sequence tags efficient full-length cDNA cloning. Proc Natl Acad Sci USA. 1998;95:8175–8180. doi: 10.1073/pnas.95.14.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang QH, et al. Cloning functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 2000;10:1546–1560. doi: 10.1101/gr.140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips RL, et al. The genetic program of hematopoietic stem cells. Science. 2000;288:1635–1640. doi: 10.1126/science.288.5471.1635. [DOI] [PubMed] [Google Scholar]
- 10.Zhou G, et al. The pattern of gene expression in human CD34+(+) stem/progenitor cells. Proc Natl Acad Sci USA. 2001;98:13966–13971. doi: 10.1073/pnas.241526198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomes I, et al. Novel transcription factors in human CD34+ antigen-positive hematopoietic cells. Blood. 2002;100:107–119. doi: 10.1182/blood.v100.1.107. [DOI] [PubMed] [Google Scholar]
- 12.Venezia TA, et al. Molecular signatures of proliferation quiescence in hematopoietic stem cells. PLoS Biol. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georgantas RW, et al. Microarray serial analysis of gene expression analyses identify known novel transcripts overexpressed in hematopoietic stem cells. Cancer Res. 2004;64:4434–4441. doi: 10.1158/0008-5472.CAN-03-3247. [DOI] [PubMed] [Google Scholar]
- 14.Kimura K, et al. Diversification of transcriptional modulation: large-scale identification characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, et al. A modified polymerase chain reaction-long serial analysis of gene expression protocol identifies novel transcripts in human CD34+ bone marrow cells. Stem Cells. 2007;25:1681–1689. doi: 10.1634/stemcells.2006-0794. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Lee S, Zhou G, Wang SM. High-throughput GLGI procedure for converting a large number of serial analysis of gene expression tag sequences into 3′ complementary DNAs. Genes Chromosomes Cancer. 2002;33:252–261. doi: 10.1002/gcc.10017. [DOI] [PubMed] [Google Scholar]
- 17.Kim YC, Jung YC, Xuan Z, Zhang MQ, Wang SM. Pan-genome isolation of low abundant transcripts through SAGE tag mis-priming. FEBS Lett. 2006;580:6721–6729. doi: 10.1016/j.febslet.2006.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Gene Ontology Consortium. Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akala O, Clarke MF. Hematopoietic stem cell self-renewal. Curr Opin Genet Dev. 2006;16:496–501. doi: 10.1016/j.gde.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 21.Kikushige Y, et al. Human Flt3 is expressed at the hematopoietic stem cell the granulocyte/macrophage progenitor stages to maintain cell survival. J Immunol. 2008;180:7358–7367. doi: 10.4049/jimmunol.180.11.7358. [DOI] [PubMed] [Google Scholar]
- 22.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 23.Georgantas RW, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression function: A circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai X, Hagedorn CH, Cullen R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J, He F, Hu S, Yu J. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge X, Wu Q, Jung YC, Chen J, Wang SM. A large quantity of novel human antisense transcripts detected by LongSAGE. Bioinformatics. 2006;22:2475–2479. doi: 10.1093/bioinformatics/btl429. [DOI] [PubMed] [Google Scholar]
- 28.Mattick JS, Majunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–29. doi: 10.1093/hmg/ddl046. (Spec No 1) [DOI] [PubMed] [Google Scholar]
- 29.Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol Cell Biol. 2006;26:7167–7177. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang SM. Long-short-long games in transcript identification: The length matters. Curr Pharm Biotechnol. 2008;9:362–367. doi: 10.2174/138920108785915166. [DOI] [PubMed] [Google Scholar]
- 31.Affymetrix ENCODE Transcriptome Project; Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Q, et al. Poly A− transcripts expressed in HeLa cells. PLoS ONE. 2008;3:e2803. doi: 10.1371/journal.pone.0002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nam DK, et al. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci USA. 2002;99:6152–6156. doi: 10.1073/pnas.092140899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.