Abstract
Cellular FLICE-inhibitory protein (c-FLIPL) is a key regulator of the extrinsic cell death pathway. Although widely regarded as an inhibitor of initiator caspase activation and cell death, c-FLIPL is also capable of enhancing procaspase-8 activation through heterodimerization of their respective protease domains. However, the underlying mechanism of this activation process remains enigmatic. Here, we demonstrate that cleavage of the intersubunit linker of c-FLIPL by procaspase-8 potentiates the activation process by enhancing heterodimerization between the two proteins and vastly improving the proteolytic activity of unprocessed caspase-(C)8. The crystal structures of the protease-like domain of c-FLIPL alone and in complex with zymogen C8 identify the unique determinants that favor heterodimerization over procaspase-8 homodimerization, and induce the latent active site of zymogen C8 into a productive conformation. Together, these findings provide molecular insights into a key aspect of c-FLIPL function that modulates procaspase-8 activation to elicit diverse responses in different cellular contexts.
Keywords: apoptosis, caspase-8, extrinsic pathway, cellular FLICE-inhibitory protein
Apoptosis is a cellular suicide program essential for early development, tissue homeostasis, and immune system maintenance in all metazoans (1, 2). This program is executed by a family of cysteine proteases known as caspases, which are synthesized as latent zymogens, and are activated in a hierarchical manner as initiators and effectors of the cell death process (3). In the extrinsic cell death pathway, ligands such as FasL/CD95L directly engage their cognate death receptors at the cell surface to recruit and activate the initiator caspases (4). This process occurs through homotypic death domain (DD) interactions between the intracellular portion of the Fas receptor and the adaptor protein FADD, and homotypic death effector domain (DED) interactions between FADD and the N terminus of procaspase-8 or procaspase-10. The resulting activation platform constitutes the death-inducing signaling complex (DISC) (5), effectively homo-oligomerizing and activating the protease domains of procaspase-8 and procaspase-10. Once activated and processed to the mature form, these initiator caspases then cleave and activate the effector caspases, which ultimately proteolyze and dismantle the contents of the cell.
During the early stages of death receptor signaling, the key regulatory proteins c-FLIPL, c-FLIPS, and c-FLIPR are also recruited to the DISC, where they modulate activation of procaspase-8 and procaspase-10 (6, 7). The long form of FLIP (c-FLIPL) is highly homologous to procaspase-8 and procaspase-10, possessing the same architecture with tandem DEDs at its N terminus (the prodomain) and a protease-like domain at its C terminus that is catalytically inactive. The short forms (c-FLIPS and c-FLIPR) by contrast primarily contain the N-terminal DEDs. All forms of FLIP are widely accepted as antiapoptotic proteins that compete with procaspase-8 for recruitment into the DISC via their DEDs when expressed at high levels. However, some evidence supports an additional role for c-FLIPL as a procaspase-8 activator that can significantly improve Fas-mediated apoptosis when expressed at much lower and more physiological levels (8). In this proapoptotic context, the protease-like domain of c-FLIPL is thought to heterodimerize with and allosterically activate the protease domains of procaspase-8 and procaspase-10 (8–10). The combined dual regulatory function of c-FLIPL is poorly understood, and it is further confounded by the notion that in some instances c-FLIPL intricately modulates procaspase-8 activation below a certain threshold to elicit nonapoptotic signals that emerge from death receptors (11, 12).
The contrasting cellular fates induced by c-FLIPL suggest a prominent yet complex role for its ability to enhance initiator caspase activation. However, the underlying molecular mechanisms remain poorly defined, especially given the fact that stable association between the isolated protease domains of both proteins has yet to be demonstrated under physiological ionic strength in vitro. Current evidence indicates that prodomain oligomerization at the DISC drives the inherently weak protease domain heterodimerization. Yet, in this context, it is still unclear precisely how the protease domains of procaspase-8 and c-FLIPL are forced together to dramatically enhance initiator caspase activation. Are the interactions regulated in response to procaspase-8 activation, and if so, how? Activated procaspase-8 may cleave itself as well as c-FLIPL. What are the biochemical consequences of these cleavage events? Answers to these important questions remain elusive, and in general, the mechanism of c-FLIPL-mediated procaspase-8 activation beyond the notion of heterodimerization remains to be elucidated. Here, we report a systematic biochemical and structural investigation aimed at uncovering this key aspect of c-FLIPL function.
Results
c-FLIPL Is Preferentially Cleaved by Zymogen and Mature Caspase-(C)8.
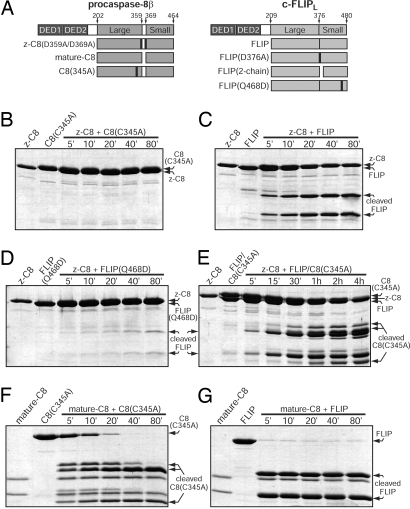
Upon activation of the Fas receptor, c-FLIPL (hereafter referred to as FLIP) is efficiently cleaved between the large and small subunits of its protease-like domain by C8 at the DISC (6, 13). To investigate the functional consequence of FLIP processing in vitro, we reconstituted a C8 activity assay using various enzymes and substrates lacking their prodomains (Fig. 1A). The isolated protease domain of zymogen C8 was rendered uncleavable through mutations of the 2 intersubunit processing sites, D359A and D369A. As anticipated, the single-chain zymogen C8(D359A/D369A) (hereafter referred to as z-C8) exhibited no detectable activity and was unable to cleave the substrate C8(C345A-active site mutant) (Fig. 1B). In sharp contrast, z-C8 acquired robust activity in the presence of FLIP, and readily cleaved FLIP to completion within 40 min (Fig. 1C). These results can be explained by 2 scenarios: preferred substrate specificity for FLIP or FLIP-mediated activation of z-C8 through heterodimerization. To differentiate between these 2 possibilities, we generated FLIP(Q468D), which retains the same cleavage site for C8, but possesses a mutation that is designed to sabotage the putative heterodimerization interface. FLIP(Q468D) was barely proteolyzed by z-C8 (Fig. 1D), confirming the latter explanation. Thus, these results are consistent with published data (8–10), and indicate that FLIP potently activates z-C8 through heterodimerization.
Fig. 1.
Zymogen and mature C8 preferentially cleave FLIP as a substrate over C8(C345A). (A) Schematic representation of purified proteins used throughout this study. (B–E) Time course of C8(C345A) and/or FLIP cleavage by z-C8. (F and G) Time course of C8(C345A) or FLIP cleavage by mature C8.
Using this in vitro system, we asked whether z-C8 activated by FLIP can preferentially cleave FLIP or C8(C345A). When incubated with both substrates simultaneously, z-C8 first cleaved FLIP within 5 min and then visibly processed C8(C345A) after 30 min (Fig. 1E). Similarly, for mature C8, cleavage of FLIP was considerably faster than cleavage of C8(C345A) (Fig. 1 F and G). Thus, our findings identify FLIP as the preferred substrate for both zymogen and mature C8, which correlate well with the observation that FLIP is cleaved before procaspase-8 at activated death receptors in cells (8).
Processed FLIP Significantly Enhances z-C8 Activity.
Next, we asked why FLIP is a preferred substrate for z-C8. One attractive hypothesis is that cleaved FLIP may significantly enhance the proteolytic activity of z-C8, thus providing a positive feedback to procaspase-8 activation. To examine this possibility, we generated 2 variants of the FLIP protease domain, processed (2-chain) and uncleavable (D376A) (Fig. 1A), and assessed their ability to stimulate z-C8 activity by using the fluorogenic substrate Ac-Ile-Glu-Thr-Asp-7-amino-4-trifluoromethylcoumarin (Ac-IETD-AFC). To facilitate heterodimerization between FLIP and z-C8, 10-fold molar excess of FLIP was used in these assays. Although z-C8 alone exhibited an undetectable level of proteolytic activity, it acquired a basal level of activity in the presence of FLIP(D376A) (Fig. 2A). Strikingly, this activity was increased >10-fold in the presence of processed FLIP (Fig. 2A). In a similar experiment, z-C8 was allowed to cleave FLIP into the 2-chain form to various degrees, by incubation at room temperature for 1, 15, and 30 min, and then immediately assayed for proteolytic activity. The result showed that the activity of z-C8 nicely correlates with the degree to which FLIP is processed (Fig. 2B).
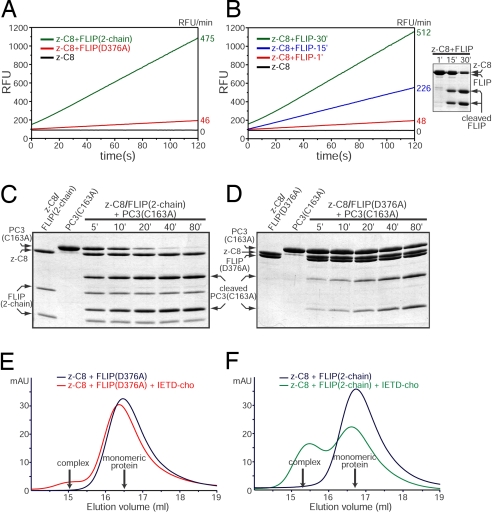
Fig. 2.
Processed FLIP activates z-C8 more efficiently than unprocessed FLIP through enhanced heterodimerization. (A) z-C8 exhibits 10-fold greater activity in the presence of FLIP(2-chain) than with FLIP(D376A). Activity was determined by cleavage of Ac-IETD-AFC. RFU, relative fluorescence units. (B) The degree of FLIP processing directly correlates with its ability to activate z-C8. The SDS/PAGE gel (Right) shows FLIP cleavage achieved at each time point immediately before activity assay. (C and D) Cleavage of catalytically inactive procaspase-3(C163A) by z-C8 in the presence of FLIP(2-chain or D376A). (E and F) FLIP(2-chain) readily forms a tight complex with z-C8 in the presence of inhibitor, whereas FLIP(D376A) does so rather inefficiently, as shown by gel filtration. Note, in the absence of inhibitor (black traces), neither form of FLIP could stably associate with z-C8. AU, absorbance unit.
In parallel to the fluorogenic assay, we also examined the ability of z-C8 to cleave a natural substrate, full-length procaspase-3(C163A), in the presence of processed or uncleavable FLIP. Similar to results shown in Fig. 2A, z-C8 in the presence of processed FLIP, but not FLIP(D376A), promoted rapid and substantial processing of procaspase-3(C163A) (Fig. 2 C and D). Together, these data unambiguously demonstrate that unprocessed FLIP is capable of activating z-C8 to a basal level, but after being processed, FLIP can dramatically elevate the proteolytic activity of z-C8.
Processed FLIP Exhibits Enhanced Heterodimerization with z-C8.
The ability of processed FLIP to markedly enhance z-C8 activity suggests that it might heterodimerize with z-C8 more efficiently than uncleavable FLIP. To examine this possibility, we used gel filtration to detect the interaction between these proteins. At physiological ionic strength, no stable interaction was detected between z-C8 and either version of FLIP. Previous studies showed that weak association between these protease domains can be strengthened by addition of the C8 inhibitor Ac-IETD aldehyde (Ac-IETD-cho) (9, 10). When this strategy was used, a small fraction of total protein was found to form a stable heterodimer between z-C8 and FLIP(D376A) (Fig. 2E). In sharp contrast, processed FLIP exhibited a markedly enhanced ability to stably associate with z-C8 (Fig. 2F). These findings indicate that cleavage of FLIP significantly improves its ability to form a heterodimer with z-C8. These data (along with data from Fig. 2 A–D) further indicate that improved heterodimerization between processed FLIP and z-C8 in the absence of inhibitor is inherently weak, but efficiently transforms the latent active site of z-C8 into a productive conformation to promote high catalytic activity; stabilization of the competent active site through the tightly bound inhibitor Ac-IETD-cho in turn locks heterodimeric interactions between both proteins to form an artificially stable complex in solution (Fig. 2F).
Structure of the Protease-Like Domain of FLIP.
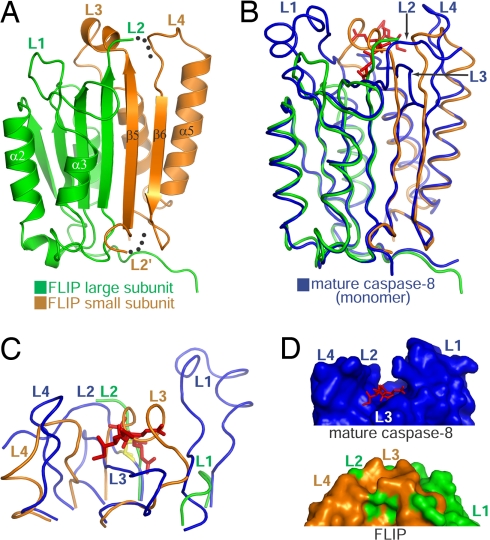
To understand the structural basis of FLIP function, we crystallized unprocessed FLIP (residues 209–480) and determined its structure at 2.2-Å resolution (Fig. 3; Table S1). The protease-like domain of FLIP exhibits 28% sequence identity and 44% sequence similarity with the corresponding domain of C8. As anticipated, this region of FLIP adopts a well-defined caspase fold, consisting of a large subunit and a small subunit linked together by a flexible intersubunit sequence that is largely disordered in the structure (Fig. 3A). The core structural elements of FLIP comprise a central, 6-stranded β-sheet, with 2 α-helices residing on one side and 3 α-helices on the other (Fig. 3A). Compared with the structure of mature C8 (14, 15), the FLIP monomer can be superimposed over a single protomer of the C8 dimer with a rmsd of 1.8 Å over 175 Cα atoms (Fig. 3B). Despite the resemblance of the overall fold, important structural differences are evident in the 4 loops (L1–L4) that comprise the active site pocket of caspases (Fig. 3C). Specifically, L3, a critical loop that forms the base of the active site groove and takes part in numerous interactions with substrate, is replaced by an α-helix that protrudes into and occupies the catalytic groove. Also, L1 is considerably shortened in FLIP. The combined effect of these structural changes in FLIP results in the absence of an active site groove observed in the mature form of C8 (Fig. 3D). These structural features are fully consistent with the fact that FLIP itself is not a protease, but rather an important regulator of proteases.
Fig. 3.
Structure of unprocessed FLIP. (A) Ribbon diagram of FLIP. The loops shaping the active site are labeled L1–L4. The L2 and L2′ portions leading to the disordered and uncleaved intersubunit linker are identified by dotted lines. (B) Structural overlay of FLIP and a single protomer of the mature C8 dimer bound to inhibitor (red stick model). (C) Overlay of the active site of mature C8 with the corresponding region in FLIP. For clarity, this view is oriented differently than in A and B (≈180° y-axis rotation). The active site cysteine in L2 of C8 is highlighted in yellow and the bound inhibitor shown in red. (D) Surface representation of the active site of mature C8 and the corresponding region in FLIP (same orientation as C).
Structure of z-C8 Bound to Processed FLIP.
To decipher the molecular basis of procaspase-8 activation by FLIP, we sought to determine the structure of z-C8 bound to the protease-like domain of FLIP. A prerequisite for crystallization is formation of a stable heterodimer between z-C8 and FLIP. Using high protein concentrations and prolonged preincubation in the presence of Ac-IETD-cho, we were able to convert the vast majority of z-C8 into a stable heterodimeric complex with processed FLIP at physiological pH and ionic strength (Fig. S1). The complex was subsequently crystallized, and the structure was determined at 1.9-Å resolution (Fig. 4; Table S1).
Fig. 4.
Structure of processed FLIP in complex with z-C8. (A) Ribbon diagram of the heterodimeric complex. Here, and in the following figures, the active site cysteine in L2 of z-C8 is highlighted in yellow and the bound inhibitor is shown as a red stick model. (B) Structural overlay of isolated unprocessed FLIP with processed FLIP bound to z-C8. For clarity, this view is oriented differently than in A and C (≈180° z-axis rotation). (C) Structural overlay of the heterodimeric complex with the mature C8 dimer (same orientation as A). Three significant regions of contact between FLIP and z-C8 are indicated: the β6 interface (Fig. 5), the closed L2′ loop of z-C8 (Fig. 6), and the loop–bundle region (L2′ of one monomer engages L2 and L4 of the adjacent monomer; Fig. 7). Another region of contact, not visible here, is shown in Fig. S3.
In the heterodimeric complex, z-C8 and processed FLIP associate with each other primarily through their respective β6 strands in an antiparallel orientation (Fig. 4A). The structure of FLIP in this complex is nearly identical to that of the isolated, unprocessed FLIP, with an rmsd of 0.55 Å over 198 Cα atoms (Fig. 4B). The structural similarity covers the entire molecule, including the L3 loop that protrudes into the putative active site groove. This analysis indicates that FLIP does not undergo significant conformational changes on heterodimerization with z-C8 or after proteolytic processing. z-C8 in this complex is bound to the inhibitor Ac-IETD-cho and trapped in an active conformation that is virtually identical to that observed in the mature C8 structure (14, 15), with an rmsd of 0.46 Å over 222 Cα atoms (Fig. 4C). Heterodimerization results in the burial of ≈3,440 Å2 of exposed surface area.
The β6-Strand Dimerization Interface.
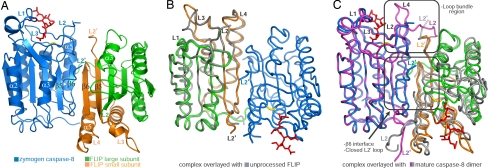
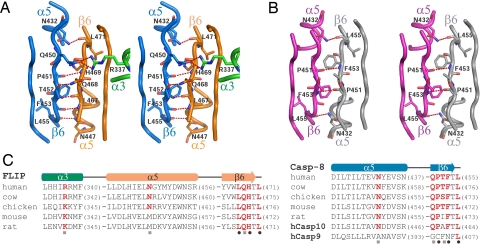
The primary interface between z-C8 and FLIP occurs through specific and antiparallel association of their β6-strands (Fig. 4A). At the center of this interface are 8 intermolecular H bonds, including 2 main-chain interactions between carbonyl oxygen atoms of Pro-451 and Phe-453 in z-C8 and amide nitrogen atoms of His-469 and Leu-467 in FLIP, respectively (Fig. 5A). FLIP also uses the side chains of Arg-337 and Gln-468 to engage Gln-450 and Thr-452 in z-C8 (Fig. 5A).
Fig. 5.
The β6-strand dimerization interface. (A) Stereoview of the β6 interface between z-C8 (blue) and FLIP (green and orange). (B) Stereoview of the β6 interface in the mature C8 dimer, which is predicted to reflect the zymogen procaspase-8 homodimer. (C) Sequence alignment of regions from FLIP and C8 involved in heterodimerization at the β6 interface. Conserved residues that form H bonds between the two proteins are colored red. Residues involved in main-chain (black circles) and side-chain (gray squares) interactions are noted.
Although zymogen procaspase-8 homodimerization is energetically unfavorable and currently undefined, the β6 interface of this activation intermediate is likely to resemble the mature C8 dimer (Fig. 4C) (14, 15). This interface involves 4 H bonds, including a pair of main-chain interactions within β6-strands, between carbonyl oxygen of Pro-451 and amide nitrogen of Phe-453, and 2 additional interactions immediately adjacent to the β6-strands, between carbonyl oxygen of Leu-455 and the side chain of Asn-432 (Fig. 5B). Compared with the β6-strand interface of the z-C8/FLIP heterodimer, the putative zymogen C8 homodimer contains only half as many H bonds in this region. Thus, the increased H-bond capacity endowed by FLIP at this interface likely represents an important factor that favors heterodimerization over zymogen procaspase-8 homodimerization to promote enhanced caspase activation.
Sequence analysis reveals that the residues involved in heterodimerization at the β6-strand interface are highly conserved (Fig. 5C). All residues involved in main-chain interactions between FLIP and z-C8 are invariant, and most residues involved in side-chain interactions are conserved. The conservation of amino acids within and surrounding the β6 interface indicates that the specificity of interactions between procaspase-8 and FLIP is likely maintained across different species. This observation also provides a plausible explanation as to why FLIP heterodimerizes with and activates procaspase-10, which possesses the conserved elements required for heterodimerization, but is unable to do so with procaspase-9, which is largely divergent in the same regions (Fig. 5C).
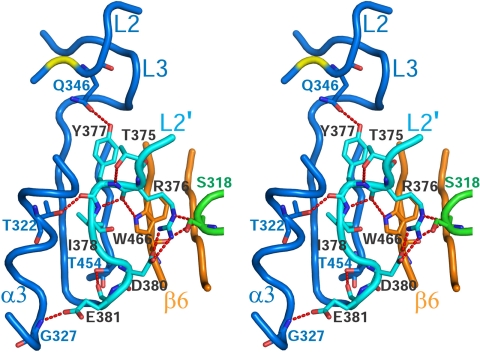
The L2′ Loop of z-C8 Exists in a Closed Conformation.
Maturation cleavage in the intersubunit sequence of each procaspase-8 zymogen allows translocation of their L2 and L2′ loops from the center of the dimer to the periphery, where they stabilize the active site conformation and solidify mature C8 dimerization through a pair of trans L2′-L2 or loop–bundle interactions (Fig. 4C; see below) (3, 14, 15). In the heterodimer between z-C8 and processed FLIP, the intersubunit sequence of z-C8 is uncleaved; consequently the L2′ portion of this sequence is oriented in the opposite direction and tethered in the center of the complex, resulting in a closed conformation (Fig. 4C and Fig. 6). This conformation is similar to the closed configuration previously observed in the structure of the latent procaspase-7 zymogen (16, 17). However, unlike the predominantly van der Waals contacts observed in procaspase-7 (16, 17), the interactions that secure the closed L2′ loop in this complex mainly involve a network of 11 intra- and intermolecular H bonds (Fig. 6). Arg-376 of z-C8 appears to play a central role, mediating 3 H bonds to Ser-318 and Trp-466 of FLIP. Within this network, z-C8 also forms an intramolecular H bond between Tyr-377 of L2′ and Gln-346 of L2, which may contribute to the stability of the active site by properly positioning the catalytic residue Cys-345 for substrate catalysis.
Fig. 6.
Shown in stereo, the L2′ of z-C8 (cyan) resides in a closed conformation and directly interacts with FLIP (green and orange).
To determine whether the closed L2′ configuration can be accommodated during zymogen procaspase-8 homodimerization, we generated a dimeric model of our z-C8 structure using the mature C8 structure as a template (14, 15). This model reveals significant multiple steric clashes at the homodimeric interface incurred by the closed L2′ loop (Fig. S2). In particular, Asp-380 within L2′ of one monomer occupies the same space as Val-391 of the adjacent monomer; this clash likely impedes proper arrangement of the L3 loop and consequently disrupts formation of the active site. Also, the close proximity of Arg-376 within L2′ to Tyr-377 from the opposing L2′ would negate the stabilizing role of the L2 loop mediated by Tyr-377. Thus, the closed L2′ configuration may be a unique feature in the FLIP/z-C8 complex, contributing additional binding energy between both proteins and possibly facilitating productive arrangement of active site loops.
Additional Interfaces Between FLIP and z-C8.
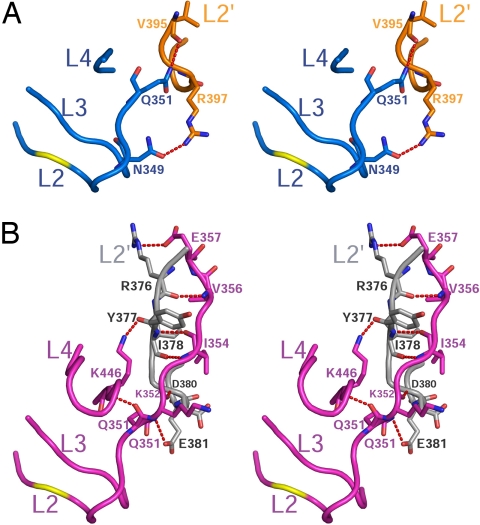
As described above, intersubunit cleavage of each unprocessed C8 allows the released L2′ of one monomer to engage L2 and L4 of the adjacent monomer to form 2 sets of loop–bundle interactions (Fig. 4C). In this manner, the L2′ loop functions as a critical structural element to facilitate formation of functional active sites (16–18). In the structure of z-C8 bound to processed FLIP, the L2′ loop of FLIP is mostly disordered (Fig. 4). Accordingly, Arg-397 and Val-395 within the remaining portion of L2′ of FLIP form only 2 H bonds with Asn-349 and Gln-351 in the L2 loop of z-C8 (Fig. 7A). Consequently, L4 within z-C8 is not stabilized and is disordered in the structure. By comparison, the fully processed mature C8 dimer forms an array of H bonds within the loop–bundle, which stabilizes homodimerization, as well as a productive conformation at each active site (Fig. 7B). The absence of these extensive interactions in the FLIP/z-C8 complex explains why protease domain heterodimerization is significantly weaker than mature C8 homodimerization (Fig. 2F, in the absence of inhibitor), and it lends insight into why the heterodimer activity is not as robust as the mature C8 homodimer (Fig. 1 E–G).
Fig. 7.
C8 loop–bundle interactions. (A) Stereoview of the L2′ of FLIP (orange) forming partial loop–bundle interactions with z-C8 (blue). (B) Stereoview of loop–bundle interactions in the fully processed mature C8 dimer. The L2′ of 1 monomer (gray) engages the active site of the adjacent monomer (magenta).
In addition to the interfaces described above, the N terminus of the protease-like domain of FLIP interacts with α4/α5 of z-C8. The main-chain amide of Glu-241 and the side-chain carboxylate of Glu-242 in FLIP H-bond with the side chains of Glu-430 and Tyr-433, respectively (Fig. S3). The main-chain carbonyl oxygen of Gln-237 in FLIP also accepts a pair of charge-stabilized H bonds from Arg-417 in z-C8. These interactions appear to be unique to the heterodimeric complex and are absent from the mature C8 homodimer.
Discussion
How does c-FLIPL facilitate activation of procaspase-8? The current view is that prodomain interactions of the two proteins at the DISC force together the weakly associated protease domains to mediate enhanced initiator caspase activation. However, beyond this notion, the mechanistic basis of this critical process is poorly understood. In this study, we report several closely linked findings. First, we demonstrate that z-C8 preferentially cleaves FLIP over itself (Fig. 1). The accelerated cleavage of FLIP by z-C8 was mainly due to FLIP-mediated activation of z-C8 through heterodimerization (Fig. 1D). Second, we show that, compared with unprocessed FLIP, processed FLIP markedly elevated the proteolytic activity of z-C8 (Fig. 2 A–D). The underlying explanation for this observation was found to be the enhanced ability of processed FLIP to form a heterodimeric complex with z-C8 (Fig. 2 E and F). Each property (preferred heterodimerization over z-C8 homodimerization, preferred cleavage of FLIP by z-C8 over itself, and enhancement of z-C8 activity by processed FLIP) directly engenders the subsequent property. The net consequence of these preferences is marked acceleration of procaspase-8 activation to the mature and fully processed form under conditions where FLIP is not effectively sequestered by other signaling proteins (see below and Fig. S4). Thus, based on our findings, proteolytic cleavage of FLIP contributes an additional layer of positive regulation to procaspase-8 activation by further lowering the energetic barrier to heterodimerization. In contrast to our work, a previous study has suggested that FLIP processing is not required for procaspase-8 activation; however, those experiments were performed in nonphysiological conditions that did not directly compare the effects of processed versus unprocessed FLIP (10), as demonstrated in our report. Indeed, our overall conclusion is supported by the observation that the uncleavable form of FLIP compared with the normal form exhibits a weaker ability to facilitate C8 autoprocessing in vitro and promote apoptosis in cell culture (8).
It is important to note that our in vitro studies were performed with recombinant forms of FLIP and C8 lacking their DEDs. In this context, the interaction between the isolated protease domains of FLIP(2-chain and D376A) and z-C8 is such low affinity that heterodimerization cannot be detected at all by gel filtration. Rather, stable association between both proteins is induced artificially only by addition of the active site inhibitor Ac-IETD-cho (Fig. 2F). This observation likely reflects the enhanced ability of FLIP to transiently convert the latent procaspase-8 into an active conformation when both proteins are anchored to the DISC and clustered together by their prodomains. During apoptosis, we predict the relatively weak protease domain heterodimerization that facilitates transient activation (and subsequent cleavage of FLIP and procaspase-8; Fig. 1E) also ensures rapid dissociation of FLIP protease domain to allow stable homodimerization of fully processed mature C8 and its release from the DISC to occur. Rapid dissociation would also allow processed FLIP protease domain to transiently but potently activate all neighboring molecules of procaspase-8 it encounters at the DISC to accelerate C8 autoproteolysis. Thus, although full-length FLIP and procaspase-8 do stably associate at the DISC through their prodomains during death receptor signaling, their protease domains do not necessarily form a stable heterodimeric enzyme with unique properties.
In light of inherently weak protease domain interactions described, how does FLIP specifically recognize zymogen C8 to produce an active conformation? Our structural data provide detailed molecular insights. In one region, the unprocessed intersubunit linker (closed L2′) of z-C8 is unexpectedly fixed in the central groove between the two proteins (Fig. 4C and Fig. 6). This loop is uniquely held in place through a network of H bonds that also contact FLIP and stabilize the active site of z-C8. In another region, the N terminus of the protease domain of FLIP forms additional H bonds with α4/α5 of z-C8 (Fig. S3). These interactions likely contribute additional binding energy that favors association with FLIP. Aside from these contacts, the most critical and highly conserved determinant of heterodimerization resides at the β6-strand interface (Fig. 5 A and C). Importantly, FLIP has an improved capacity to H-bond with z-C8 within this region as compared with the putative zymogen C8 homodimer. Overall, these structural features may greatly facilitate heterodimerization with FLIP over zymogen procaspase-8 homodimerization to promote enhanced initiator caspase activation.
One structural element that may contribute to procaspase-8 activation is the L2′ loop of FLIP, which is largely disordered, but still forms 2 H bonds with the active site of z-C8 (Fig. 7A). These contacts, in conjunction with an H bond directed to the active site via the closed L2′ loop of z-C8, may play a decisive role (Fig. 6). Allosteric regulation emanating from the β6 interface may also enable proper ordering of the active site to achieve catalytic competency (Fig. 5A). Recent studies of caspase-1 and caspase-7 revealed that compounds targeting the dimerization interface, ≈15 Å away from the active site, can propagate structural changes that ultimately deform the active sites (19, 20). Perhaps the most convincing observation from our work in support of this conjecture emerges from the fact that the β6 interface is the most conserved element among all of the regions of contact between FLIP and z-C8 (Fig. 5C). This region forms the majority of heterodimerization contacts, possibly stabilizing the β6-strand and neighboring elements in a productive configuration. Together, these structural insights along with our biochemical analysis give rise to a coherent molecular explanation of FLIP-mediated activation of procaspase-8 (Fig. S4).
In the context of nonapoptotic signaling, the transient nature of FLIP and z-C8 protease domain heterodimerization may contribute to the multifaceted function of FLIP as well (Fig. S4). Current studies indicate that cleavage of FLIP by procaspase-8 also promotes recruitment of TRAF2, RIP1, and the regulatory subunit of the IKK complex to FLIP to activate NF-κB (21–23). Under nonapoptotic conditions, unprocessed FLIP may activate procaspase-8 to a moderate level such that only FLIP itself and possibly other substrates located nearby are cleaved. Because of weak protease domain heterodimerization, processed FLIP may then interact with TRAF2 and RIP1 in a manner that occludes its ability to potentiate procaspase-8 activation. Our observation that FLIP is preferentially cleaved by procaspase-8 suggests that this early event may function as a critical switch point to ensure activation of other signaling pathways without eliciting autoproteolysis of initiator caspases, which would likely lead to cell death. This model, although speculative, suggests that FLIP-mediated activation of procaspase-8 can be modulated in a graded and controlled manner required to induce NF-κB and other nonapoptotic signaling pathways. Future studies should be directed at investigating how other factors are integrated with FLIP to modulate different signaling pathways emerging from a protease normally associated with cell death.
Materials and Methods
Protein Preparation.
Variants of C8 (202–464) and FLIP (209–480) were cloned into pET21b and pET15b (Novagen), respectively. All were expressed in Escherichia coli BL21(DE3) and purified by Ni-NTA (Qiagen). The N-His tag of all FLIP variants was removed by thrombin treatment. All were further fractionated by using Source-15Q and Superdex-200 (GE Healthcare). FLIP was further treated with trypsin to generate FLIP(2-chain). The FLIP(2-chain)/z-C8 complex was assembled by incubating in an equimolar ratio for 1 h at 25 °C in the presence of 10 mM DTT followed by treatment with 2:1 molar ratio of Ac-IETD-cho (BD Biosciences). Preparation of procaspase-3(C163A) has been previously described (18).
C8 Activity Assays and Gel Filtration Analysis.
All assays were performed in 20 mM Hepes (pH 7.5), 150 mM NaCl, and 10 mM DTT at 25 °C. For Fig. 1, substrate concentrations were fixed at 12.8 μM, while enzyme concentrations of z-C8 and mature C8 were fixed at 6.4 and 3.2 μM, respectively. For Fig. 2 C and D, z-C8 and FLIP(2-chain or D376A) were fixed at 6.4 μM, whereas PC3(C163A) was fixed at 12.8 μM. Reactions were stopped by boiling in loading buffer at noted time points and products were visualized by SDS/PAGE. For Fig. 2A, z-C8 was mixed with FLIP(2-chain or D376A) to a final concentration of 0.2 and 2 μM, respectively, and incubated for 5 min before addition of Ac-IETD-AFC (final concentration of 200 μM; BD Biosciences). Ac-IETD-AFC hydrolysis was assessed by using a Hitachi F2500 fluorescence spectrophotometer (excitation 400 nm; emission 505 nm). For Fig. 2B, z-C8 and regular FLIP were preincubated at 2.2 and 22 μM, respectively, at 25 °C for 1 min, 15 min, and 30 min. These samples were then immediately diluted ≈11-fold and assayed for activity using Ac-IETD-AFC. A portion of the preincubated material was boiled in loading buffer at noted time points and visualized by SDS/PAGE. For Fig. 2 E and F, z-C8 and FLIP(2-chain or D376A) were mixed to a final concentration of 4 μM for each protein in the presence or absence of 16.6 μM Ac-IETD-cho at 4 °C; samples were then immediately applied to Superdex-200 and fractionated at 4 °C.
Crystallization and Structure Determination.
Crystals were grown in a hanging drop by mixing ≈10 mg/mL protein with an equal volume of well buffer. FLIP alone crystallized in 0.1 M Mes (pH 6.5), 18% PEG, 5000 monomethyl ether, and 0.1 M ammonium sulfate. The processed FLIP/z-C8 complex crystallized in 0.9 M sodium dihydrogen phosphate, 0.8 M dipotassium hydrogen phosphate, 0.1 M N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) (pH 10.5), and 0.2 M lithium sulfate. Crystals were equilibrated in well buffer containing 20% glycerol (vol/vol) and flash frozen in liquid nitrogen. Native datasets were collected at National Synchrotron Light Source Beamline X25 and processed with Denzo and Scalepack (24). Both structures were solved by molecular replacement using PHASER (25). Models used were C8 from Protein Data Bank entry 1I4E (26) and the refined structure of the FLIP monomer. The FLIP and FLIP/z-C8 structures were refined against 2.2- and 1.9-Å native data, respectively, using O (27) and COOT (28).
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grant 2 R01 CA90269 (to Y.S.). J.W.Y. is a Damon Runyon Fellow supported by Damon Runyon Cancer Research Foundation Grant DRG-1905-06.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3H11 and 3H13).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812453106/DCSupplemental.
References
- 1.Horvitz HR. Nobel lecture. Worms, life and death. Biosci Rep. 2003;23:239–303. doi: 10.1023/b:bire.0000019187.19019.e6. [DOI] [PubMed] [Google Scholar]
- 2.Danial NN, Korsmeyer SJ. Cell death: Critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 3.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 4.Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 5.Kischkel FC, et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irmler M, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 7.Golks A, Brenner D, Fritsch C, Krammer PH, Lavrik IN. c-FLIPR, a new regulator of death receptor-induced apoptosis. J Biol Chem. 2005;280:14507–14513. doi: 10.1074/jbc.M414425200. [DOI] [PubMed] [Google Scholar]
- 8.Chang DW, et al. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 2002;21:3704–3714. doi: 10.1093/emboj/cdf356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Micheau O, et al. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J Biol Chem. 2002;277:45162–45171. doi: 10.1074/jbc.M206882200. [DOI] [PubMed] [Google Scholar]
- 10.Boatright KM, Deis C, Denault JB, Sutherlin DP, Salvesen GS. Activation of caspases-8 and -10 by FLIP(L) Biochem J. 2004;382:651–657. doi: 10.1042/BJ20040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu JW, Shi Y. FLIP and the death effector domain family. Oncogene. 2008;27:6216–6227. doi: 10.1038/onc.2008.299. [DOI] [PubMed] [Google Scholar]
- 12.Budd RC, Yeh WC, Tschopp J. cFLIP regulation of lymphocyte activation and development. Nat Rev Immunol. 2006;6:196–204. doi: 10.1038/nri1787. [DOI] [PubMed] [Google Scholar]
- 13.Scaffidi C, Schmitz I, Krammer PH, Peter ME. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem. 1999;274:1541–1548. doi: 10.1074/jbc.274.3.1541. [DOI] [PubMed] [Google Scholar]
- 14.Blanchard H, et al. The three-dimensional structure of caspase-8: An initiator enzyme in apoptosis. Structure. 1999;7:1125–1133. doi: 10.1016/s0969-2126(99)80179-8. [DOI] [PubMed] [Google Scholar]
- 15.Watt W, et al. The atomic-resolution structure of human caspase-8, a key activator of apoptosis. Structure. 1999;7:1135–1143. doi: 10.1016/s0969-2126(99)80180-4. [DOI] [PubMed] [Google Scholar]
- 16.Chai J, et al. Crystal structure of a procaspase-7 zymogen: Mechanisms of activation and substrate binding. Cell. 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 17.Riedl SJ, et al. Structural basis for the activation of human procaspase-7. Proc Natl Acad Sci USA. 2001;98:14790–14795. doi: 10.1073/pnas.221580098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiozaki EN, et al. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 19.Hardy JA, Lam J, Nguyen JT, O'Brien T, Wells JA. Discovery of an allosteric site in the caspases. Proc Natl Acad Sci USA. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheer JM, Romanowski MJ, Wells JA. A common allosteric site and mechanism in caspases. Proc Natl Acad Sci USA. 2006;103:7595–7600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kataoka T, Tschopp J. N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol Cell Biol. 2004;24:2627–2636. doi: 10.1128/MCB.24.7.2627-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dohrman A, et al. Cellular FLIP (long form) regulates CD8+ T cell activation through caspase-8-dependent NF-kappa B activation. J Immunol. 2005;174:5270–5278. doi: 10.4049/jimmunol.174.9.5270. [DOI] [PubMed] [Google Scholar]
- 23.Golks A, Brenner D, Krammer PH, Lavrik IN. The c-FLIP-NH2 terminus (p22-FLIP) induces NF-κB activation. J Exp Med. 2006;203:1295–1305. doi: 10.1084/jem.20051556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 26.Xu G, et al. Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex. Nature. 2001;410:494–497. doi: 10.1038/35068604. [DOI] [PubMed] [Google Scholar]
- 27.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.