Abstract
Loss of expression of the TGF-β superfamily coreceptor, the type III TGF-β receptor (TβRIII or betaglycan), occurs in a broad spectrum of human cancers including breast, lung, ovarian, pancreatic, prostate, and renal cell cancer. TβRIII suppresses cancer progression in vivo, at least in part, by reducing cancer cell motility. However, the mechanism by which TβRIII regulates migration is unknown. Here, we demonstrate an unexpected TGF-β signaling independent role for TβRIII in activating Cdc42, altering the actin cytoskeleton and reducing directional persistence to inhibit random migration of both cancer and normal epithelial cells. Functionally, TβRIII through its interaction with the scaffolding protein β-arrestin2, activates Cdc42 and inhibits migration. These studies identify a TGF-β independent homeostatic function for TβRIII in regulating cell migration.
Cell migration is a complex process required for physiological functions including embryonic development and wound healing. Alterations in cell migration are also critical to disease pathogenesis, including inflammatory and vascular diseases, tumor cell invasion, and metastases (1, 2). The ability of cells to migrate is regulated both by the speed and the directionality of migration that can be triggered by external cues (i.e., chemotaxis) or because of the intrinsic property of cells to migrate (i.e., intrinsic persistence) that are in turn regulated by the Rho family of GTPases, integrins, the actin cytoskelton, and microtubules (3, 4). Several migratory processes in development and tissue remodeling occur without any evidence of extrinsic chemotactic signaling, relying instead on intrinsic cell migratory properties (5).
The type III TGF-β receptor (TβRIII/betaglycan), an 849 aa heparan sulfate proteoglycan, is the most abundant and ubiquitously expressed TGF-β superfamily coreceptor. TβRIII has the potential to increase or decrease TGF-β signaling through mechanisms yet to be fully defined (6–8). TβRIII is classically thought to function as a coreceptor, presenting TGF-β superfamily ligands to their respective signaling receptors (8). Recent studies have suggested essential, nonredundant roles for TβRIII in regulating signaling through TβRII and TβRI as well as independently of TβRII and TβRI (9). TβRIII null embryos die on embryonic day 13.5, exhibiting hepatic and cardiovascular defects (10). An essential role for TβRIII has also been demonstrated in mesenchymal transformation during chick embryonic heart development (11, 12) and in mediating TGF-β resistance in intestinal goblet cells (13). We have defined essential roles for the cytoplasmic domain of TβRIII in mediating TGF-β signaling independent of the ligand presentation role (14), along with regulating cell-surface levels of TβRIII and TβRII through interactions with Gα-interacting protein-interacting protein, C terminus (GIPC) (15) and β-arrestin2 (16).
Loss of TβRIII expression has been reported in multiple cancers, with loss of expression correlating with disease progression, advanced stage, or grade and/or a poorer prognosis for patients (17–22). Importantly, increasing or restoring expression of TβRIII in these cancer models decreases cancer cell motility and invasion in vitro (17–21) and angiogenesis, invasion, and metastasis in vivo (17). Thus, the role of TβRIII as a suppressor of cancer cell motility appears to be conserved. Numerous studies have correlated the metastatic potential of tumor cells with their in vitro migratory abilities (22) including TβRIII-mediated inhibition of migration correlating with decreased metastases in vivo (17). Here we investigate the mechanism by which TβRIII inhibits cell migration.
Results
TβRIII Inhibits Directed and Random Cell Migration in Cancer and Epithelial Cells.
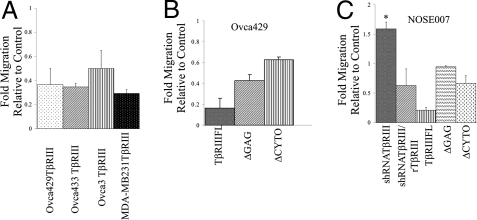
Directional migration can be due to externally directed migration, as during chemotaxis, or due to the intrinsic persistence of cells (5). To determine whether TβRIII-mediated suppression of migration was dependent on an external chemotactic gradient or a defect in the intrinsic ability of the cancer cells to migrate, we examined transwell migration either toward serum (10% FBS, chemotactic gradient) or serum-free media (SFM) (in both chambers, no gradient). In ovarian and breast cancer cell lines, Ovca429 and MDA-MB231, stable expression of TβRIII inhibited migration by 65–70% in the presence of a chemotactic gradient (Fig. 1A and Fig. S1A) and 60–65% in the absence of a chemotactic gradient (Fig. S2A). To rule out that TβRIII-mediated suppression of migration was direct and not acquired during selection, we transiently expressed TβRIII in ovarian cancer cell lines that had either no (Ovca429 or Ovca433) or reduced (Ovca3) TβRIII expression (18), using TβRIII-expressing adenovirus tagged with GFP or GFP-expressing adenovirus alone as a control (Fig. S3 A and B). Expression of TβRIII resulted in a 60–85% reduction in chemotactic migration (Fig. 1A and Fig. S1A) and a 70% reduction in the absence of a chemotactic gradient (Fig. S2A) relative to control cells. These data indicate that TβRIII directly inhibits the intrinsic ability of both breast and ovarian cancer cells to migrate.
Fig. 1.
TβRIII inhibits cell migration in epithelial and cancer cells. (A) Cancer cell lines stably expressing TβRIII with Neo as controls (Ovca429 and MDA-MB231) and transiently expressing TβRIII with GFP as control (Ovca433 and Ovca3) were allowed to migrate for 18–24 h toward 10% FBS (see Materials and Methods). Fold migration relative to controls is presented and data represent the mean ± SE of 3 independent experiments performed in triplicate. The number of cells migrated is presented in Fig. S1A. (B) Ovca429 cells transiently expressing either TβRIII, TβRIIIΔGAG, TβRIIIΔCYTO, or GFP were allowed to migrate for 18–24 h toward 10% FBS. Data represent the mean ± SE of 3 independent experiments performed in triplicate. The number of cells migrated is presented in Fig. S1B. (C) NOSE007 cells in SFM in the top chamber of a transwell were allowed to migrate for 18–24 h toward 10% FBS 72 h after infection with adenovirus-expressing shRNA NTCs, human shRNA-TβRIII alone, human shRNA-TβRIII after infection with rat TβRIII (rTβRIII), or 48 h after infection with TβRIII, TβRIIIΔGAG, TβRIIIΔCYTO and GFP as controls. Fold migration relative to controls (shRNA NTCs for shRNA-TβRIII-infected cells or GFP control) is presented. Data represent the mean ± SE of 3 independent experiments performed in duplicate. *, P = 0.0036, relative to control. The number of cells migrated is presented in Fig. S1 C and D.
Although TβRIII expression is lost in a number of epithelial-derived human cancers, TβRIII is broadly and abundantly expressed in most epithelial cells (23). To determine whether TβRIII might have a homeostatic role in epithelial cells, we investigated the effect of shRNA-mediated silencing of TβRIII expression in normal ovarian surface epithelial cells (NOSE007), with a non-targeting control (NTC) shRNA as control (Fig. S3 D and E). Reducing TβRIII expression in NOSE007 cells increased cell migration both in the presence (Fig. 1C) and absence of a 10% FBS gradient (Fig. S2B), whereas rescuing loss of TβRIII by expression of rat TβRIII in shRNA-TβRIII-infected cells (Fig. S3F) decreased NOSE007 cell migration (Fig. 1C). In a reciprocal manner, increasing TβRIII expression in NOSE007 cells (Fig. S3C) resulted in an 80% suppression in chemotactic migration (Fig. 1C and Fig. S1D), similar to the effect of restoration of TβRIII expression in cancer cells. These data indicate that TβRIII inhibits the intrinsic ability of epithelial cells to migrate. Because reduced TβRIII expression in NOSE007 cells increased cell migration, and TβRIII has previously defined roles in regulating epithelial to mesenchymal transition (EMT) during development (11, 12) and in cancer models (21), we investigated whether reduced TβRIII expression induced EMT in NOSE007 cells. Consistent with a role in inducing EMT, shRNA-mediated silencing of TβRIII expression in NOSE007 cells resulted in an increase in expression of mesenchymal markers, including N-cadherin and Vimentin (Fig. S4A), an increase in expression of the EMT mediator, the transcription factor Slug (Fig. S4A), and an increase in NOSE007 cell invasiveness (Fig. S4B). Other markers of EMT, including reduced E-cadherin expression and induction of Snail expression could not be assessed because these markers could not be detected in NOSE007 cells.
To determine the structural requirements for TβRIII-mediated inhibition of migration, we transiently expressed TβRIII, TβRIIIΔGAG (lacking the glucosaminoglycan chains), TβRIIIΔCYTO (lacking the cytoplasmic domain), or GFP alone as control in either Ovca429 or NOSE007 cells (Fig. S3 B and C) and assessed their ability to migrate. Although full-length TβRIII suppressed migration by 84% (Fig. 1B and Fig. S1B), TβRIIIΔGAG and TβRIIIΔCYTO suppressed migration by 60% and 40% in Ovca429 cells, respectively (Fig. 1B and Fig. S1B) and by 6% and 34% in NOSE007 cells, respectively (Fig. 1C and Fig. S1D). These data indicate that both the cytoplasmic domain and the glucosaminoglycan (GAG) modifications in the extracellular domain are important for TβRIII-mediated inhibition of migration and suggest that the mechanism of suppression might be similar between epithelial cells and epithelial-derived cancer cells.
TβRIII Regulates the Actin Cytoskeleton of Cancer and Epithelial Cells.
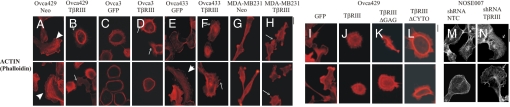
The migration of cells is intricately linked to the dynamic organization and reorganization of the actin cytoskeleton. To examine if TβRIII-mediated inhibition of migration was associated with changes in the underlying actin cytoskeleton, we examined the actin organization in either stable cell lines (Ovca429Neo, Ovca429TβRIII, MDA-MB231Neo, and MDA-MB231TβRIII) or in cancer cell lines transiently expressing TβRIII (Ovca3 and Ovca433). Subconfluent Ovca429Neo cells stained with rhodamine-conjugated phalloidin exhibited an elongated morphology with actin-rich filaments extending into the lamellipodia, and extensive membrane ruffling in some cells (Fig. 2, arrowheads). Ovca429TβRIII cells were less elongated and more rounded with the actin concentrated in a ring-like structure and exhibited limited lamellipodial structures (Fig. 2B). Seventy percent of Ovca429TβRIII cells contained numerous filopodia/microspike-like projections extending across the entire cell surface (Fig. 2B, open arrows and Fig. S5A), whereas 6% of Ovca429Neo cells exhibited this phenotype (Fig. 2A and Fig. S5A). Although there were some inherent differences in the actin cytoskeleton between cell lines, with Ovca3 control cells exhibiting a highly cuboidal morphology with smooth edges (Fig. 2C) and MDA-MB231Neo cells exhibiting a spindle-like morphology (Fig. 2G), in all cases restoration of TβRIII expression resulted in an increase in microspike/filopodia-like structures across the entire cell surface (Fig. 2 and Fig. S5A). To determine whether TβRIII had the same structural requirements for regulating the actin cytoskeleton for inhibiting migration, we examined actin organization in Ovca429 cells transiently expressing TβRIII, TβRIIIΔGAG, TβRIIIΔCYTO, or GFP alone. Because all constructs also expressed GFP, only cells expressing GFP were analyzed. Similar to the phenotype observed in Ovca429 cells with stable TβRIII expression, transient TβRIII expression also altered the actin cytoskeleton with increased cell-rounding and filopodia/microspike-like structures across the entire cell surface in 90% of single cells (Fig. 2 I and J and Fig. S5B). In comparison, cells expressing either TβRIIIΔGAG or TβRIIIΔCYTO recapitulated the TβRIII-induced actin cytoskeletal phenotype in only 42% and 22% of the cells, respectively (Fig. 2 K and L and Fig. S5B). The remainder of the cells expressing TβRIIIΔGAG or TβRIIIΔCYTO exhibited fewer filopodia/microspike-like structures and exhibited observable lamellipodial structures (Fig. 2 K and L).
Fig. 2.
TβRIII alters the actin cytoskeletal organization of epithelial and cancer cells. (A–L) We plated 5–8 × 104 of the indicated cell lines expressing the indicated proteins on coverslips and stained for actin by using rhodamine phalloidin. For transiently expressing TβRIII-IRES-GFP and control-GFP, cells were fixed and stained 48 h after infection and GFP-expressing cells were examined. Closed arrows indicate distinct lamellipodia in control cells and open arrows indicate rounded cells with multiple microspikes in TβRIII-expressing cells. Scale bar = 8.6 μm. (M and N) Subconfluent NOSE007 cells were infected with adenovirus expressing control shRNA NTCs (M) or shRNA-TβRIII (N). Cells were fixed and stained with actin-phalloidin-conjugated Alexa-488 72 h after infection and dsRed-expressing cells were examined. Open arrows indicate lamellipodia formation in shRNA-TβRIII cells. Scale bar = 21.6 μm.
To determine whether shRNA-mediated knockdown of endogenous TβRIII in NOSE007 resulted in changes in the actin cytoskeleton, we examined the actin organization in NOSE007 by using either shRNA-TβRIII or NTC shRNA as a control. shRNA-TβRIII and the NTC shRNA were both coexpressed with dsRed, allowing for specific inspection of cells expressing the appropriate shRNA. Because NOSE007 cells are epithelial cells lines (24), the majority of single NTC shRNA-infected cells were cuboidal with peripheral actin bundling, reminiscent of an epithelial-like morphology, and lacked significant stress fibers or lamellipodia formation (Fig. 2M). In contrast, the majority of shRNA-TβRIII-infected cells exhibited a robust increase in both stress fibers and lamellipodia formation (Fig. 2N, open arrows). Quantification of stress-fiber density by incorporating a line profile across the cytoplasm to identify stress fibers by their increased fluorescence relative to areas devoid of stress fibers revealed a 2- to 3-fold increase in stress fibers in shRNA-TβRIII cells compared with levels in NTC shRNA-infected cells (Fig. S5C). The shRNA-TβRIII-mediated increase in stress fibers and lamellipodia is consistent with the increased migration observed in transwell assays (Fig. 1C).
Components of the cytoskeleton including actin, microtubules, and nonmuscle myosin II are required for sustained directional migration and polarity in a broad range of cell types (25–28). We assessed the relative contribution of these components on TβRIII-mediated inhibition of migration by treating Ovca429Neo and Ovca429TβRIII cells with either cytochalasin D, an inhibitor of actin polymerization; blebbistatin, an inhibitor of nonmuscle myosin II; or nocodazole to disrupt microtubules. Cytochalasin D potently decreased transwell migration in both Ovca429Neo and Ovca429TβRIII cells, whereas nocodazole had a more modest effect in both Ovca429Neo and Ovca429TβRIII cells, and blebbistatin had little to no effect on migration (Fig. S5D). In addition, no significant difference was observed in the microtubule organization between Ovca429Neo and Ovca429TβRIII cells. These data support a role for TβRIII in inhibiting cellular migration largely through its effects on the actin cytoskeleton in both cancer and epithelial cells.
TβRIII Suppresses Migration and Regulates the Cytoskeleton Independent of TβRII, ALK5, and Smad2.
Our previous work suggested that TβRIII's tumor-suppressor effects could be partially mediated by the soluble form of TβRIII (sTβRIII), possibly by sequestering TGF-β (17, 20, 21). Here we demonstrate that TβRIII inhibits the migration of cancer cell lines that migrate either in a TGF-β-responsive (i.e., MDA-MB231 cells) (29) or in a TGF-β-nonresponsive manner (i.e., Ovca429 cells) (24) (Fig. 1), suggesting that the effects of TβRIII on cell migration might be independent of its effects on TGF-β signaling. TβRIII expression reduced TGF-β-stimulated Smad2 phosphorylation in Ovca429 cells (Fig. S6A), similar to previous observations in breast cancer cells (17). To assess whether decreasing or increasing TGF-β signaling altered TβRIII-mediated inhibition of migration, we used the TβRI/ALK5 inhibitor, SB431542, to inhibit TβRI/ALK5-mediated Smad2 phosphorylation and dominant-negative TβRII (TβRIIΔCyto) to reduce TβRII/TβRI signaling (Fig. S6 B and C). Neither SB431542 or TβRIIΔCyto altered the ability of TβRIII to suppress migration in either Ovca429 or MDA-MB231 cells (Fig. S6D), nor did they alter TβRIII-dependent changes in the actin cytoskeleton. In addition, expression of constitutively active ALK5, which bypasses the ligand requirement, did not attenuate the ability of TβRIII to suppress migration (Fig. S6E). We also found that neither sTβRIII or a pan-TGF-β (TGFb1,2,3) neutralizing antibody had a significant effect on the migration of Ovca429Neo or Ovca429TβRIII cells, respectively (Fig. S6 F and G). Finally, to determine the contribution of Smad2 on TβRIII-mediated inhibition of migration, we introduced siRNA to Smad2 in Ovca429Neo and Ovca429TβRIII cells and examined migration (Fig. S6H). We found that siRNA-mediated silencing of Smad2 expression did not alter TβRIII-mediated inhibition of migration (Fig. S6H). Taken together, these data strongly support a role for TβRIII in inhibiting cell migration, independent of sTβRIII and the TGF-β/TβRII/TβRI/Smad signaling pathway.
TβRIII Inhibits Random Migration by Reducing Directional Persistence.
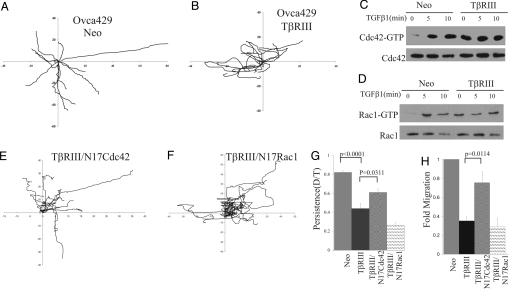
To explore the mechanism by which TβRIII functions to inhibit migration, we tracked the movement of individual Ovca429TβRIII or control Ovca429Neo cells by using live cell imaging. Ovca429Neo cells exhibited a polarized morphology (Fig. 3 and Movie S1) with less protrusions, a distinct leading, and a trailing edge in the vast majority of the single cells imaged. In contrast, Ovca429TβRIII cells were not polarized and had multiple small protrusions and microspike-like structures consistent with the actin cytoskeletal phenotype (Fig. 3 and Movie S2). Analysis of cell tracks of Ovca429Neo and Ovca429TβRIII cells over a 2 h period (Fig. 3 A and B) revealed that, although the overall velocity of migration of Ovca429Neo (0.89 ± 0.06 μm/min) and Ovca429TβRIII (0.95 ± 0.11 μm/min) was not significantly different, Ovca429TβRIII cells (Fig. 3B) consistently made more directional changes compared with Ovca429Neo cells (Fig. 3A), which tended to persist in one direction. Indeed, the directionality ratio [the linear distance from the starting point to the end point (D)/the total distance traversed by the cell (T)] (5) was reduced 2-fold in TβRIII cells (0.82 ± 0.019 in Ovca429Neo cells vs. 0.43 ± 0.04 in Ovca429TβRIII, P < 0.0001; Fig. 3G). These data indicate that TβRIII functions to inhibit cellular migration not by decreasing the velocity of migration but by decreasing directional persistence.
Fig. 3.
TβRIII inhibits directional persistence, motility, and mediates alterations in the actin cytoskeleton through constitutive activation of Cdc42. (A and B) Migratory behavior of Ovca429Neo (A) (Movie S1) and Ovca429TβRIII (B) (Movie S2) cells tracked over a 120-min period. The origins of migration were superimposed at 0, 0. The tracks of representative individual cells during the 120-min period are presented. (C and D) Ovca429Neo and Ovca429TβRIII cell lines were serum-starved for 16 h followed by stimulation with TGF-β1 (200 pM) for the times indicated, and pull-down assays for active Cdc42 (C) and Rac1 (D) were performed. Five percent of lysate was assessed for effects on total Cdc42 and Rac1 levels. A representative experiment of 4 replicates is presented. Quantification of active Cdc42 or Rac1 is presented in Fig. S7 A and B. (E and F) The tracks of representative individual Ovca429TβRIII cells 24 h after infection with adenovirus-expressing GFP-tagged N17Cdc42 (E; see Movie S3) or GFP-tagged N17Rac1 (F) are presented. (G) Directional persistence was calculated under the indicated conditions from data acquired during live cell imaging. Data represent the mean ± SE for 3 independent experiments, n = 20. (H) Transwell migration toward a 10% FBS gradient was assessed 48 h after infecting Ovca429TβRIII cells with adenovirus-expressing GFP-tagged N17Cdc42 or GFP-tagged N17Rac1. Fold migration relative to Ovca429Neo control is presented. Data represent the mean ± SE for 5 independent experiments.
TβRIII Inhibits Directed Migration and Regulates the Actin Cytoskeleton by Means of Constitutive Activation of Cdc42.
Actin cytoskeletal organization and directional migration are both regulated at multiple levels, including Rho, GTPases, Cdc42, and Rac1 (3, 5, 30). In addition, TGF-β1 has been previously demonstrated to activate Cdc42 and Rac1 in a Smad2-independent fashion (31). To determine whether TβRIII inhibits directional migration through altering the basal or TGF-β1-stimulated levels of activated Rho, Rac1, or Cdc42 in Ovca429 cells, levels of GTP-bound (i.e., activated) Rho, Rac1, and Cdc42 were assessed in basal unstimulated and TGF-β1-stimulated Ovca429Neo and Ovca429TβRIII cells. Ovca429Neo cells had little to no activated Cdc42 or Rac1 in the basal unstimulated state (Fig. 3 C and D), whereas TGF-β1-stimulated activation of both Cdc42 and Rac1 (Fig. 3 C and D). In contrast, in Ovca429TβRIII cells, both Cdc42 and Rac1 (Fig. 3 C and D) were constitutively activated under basal conditions (4-fold for Cdc42-GTP and 11-fold for Rac1-GTP; Fig. S7 A and B). In addition, exogenous TGF-β1 did not significantly increase the level of activation of Cdc42 and Rac1, suggesting that they were already maximally activated (Fig. 3 C and D and Fig. S7 A and B). Similar effects of TβRIII on constitutive activation of Cdc42 and Rac1 were also observed in the MDA-MB231 breast cancer cells (Fig. S8). These effects were specific because no significant differences between Ovca429Neo and Ovca429TβRIII in basal or TGF-β1-stimulated RhoA activation levels were observed. Taken together, these data indicate that TβRIII-expressing cells have constitutively high levels of activated Rac1 and Cdc42.
To determine whether the constitutive activation of Cdc42 and/or Rac1 contributed to the reduction in directional persistence in TβRIII cells, we examined the effect of blocking Cdc42 or Rac1 activation with either dominant-negative Cdc42 or Rac1 (N17Cdc42 and N17Rac1) on directional persistence. N17Cdc42 and N17Rac1 were both coexpressed with GFP (32), allowing for imaging of only infected cells. Live cell analysis of cell tracks from Ovca429Neo and Ovca429TβRIII cells revealed that N17Cdc42 increased the directional persistence of Ovca429TβRIII cells (Fig. 3 E–G and Movie S3). In contrast, N17Rac1 further reduced the directional persistence (Fig. 3 E–G) and the velocity of migration of TβRIII cells. The specific effect of dominant-negative Cdc42 on TβRIII-mediated inhibition of directional persistence (Fig. 3 E and G), and the similar actin cytoskeletal phenotype obtained with expression of either TβRIII or constitutively active Cdc42 (33), both suggested that TβRIII may be altering the actin cytoskeleton and inhibiting migration through constitutive activation of Cdc42. Consistent with this hypothesis, we find that expression of N17Cdc42 significantly attenuated TβRIII-mediated inhibition of migration (Fig. 3H) and suppressed TβRIII's effects on the actin cytoskeleton, with a majority of cells exhibiting a more polarized morphology (Fig. S7 F–I; 39% of TβRIII/N17Cdc42 exhibited multiple filopodia/microspike-like structures vs. 90% cells expressing TβRIII alone). In contrast, although N17Rac1 effectively blocked Rac1 activation (Fig. S7E), N17Rac1 had little to no effect on migration (Fig. 3H and Fig. S7E); and despite exhibiting a variety of qualitative changes in the actin cytoskeleton it continued to exhibit cells with filopodia/microspike-like structures across the entire cell surface, similar to TβRIII-expressing cells (Fig. S7 F–I). The specific ability of dominant-negative Cdc42 to reverse the effects of TβRIII on migration, directional persistence, and the actin cytoskeleton strongly support activation of Cdc42 as the mechanism for TβRIII-mediated suppression of migration.
TβRIII's Interaction with the Scaffolding Protein β-arrestin2 Regulates Migration, the Actin Cytoskeleton, and Activation of Cdc42.
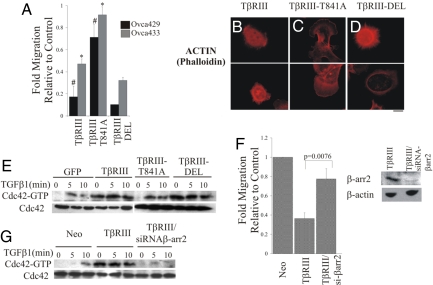
TβRIII interacts with the PDZ domain-containing protein, GIPC (15), and the scaffolding protein, β-arrestin2 (16), through discrete motifs in the cytoplasmic domain of TβRIII. Because loss of TβRIII's short cytoplasmic domain impairs the ability of TβRIII to suppress migration (Fig. 1 B and C) we investigated whether TβRIII mediates its effects on migration by means of interaction with these scaffolding proteins. Ovarian cancer cell lines Ovca429 and Ovca433 were transiently infected with either GFP control, TβRIII- or TβRIII-T841A-expressing adenovirus (unable to bind β-arrestin2), (16) or TβRIII-DEL-expressing adenovirus (unable to bind GIPC) (15) (Fig. S3 A and B), and effects on migration were assessed. TβRIII and TβRIII-DEL potently inhibited migration in both cell lines (Fig. 4A). In contrast, TβRIII-T841A was largely unable to mediate this TβRIII function in either ovarian cancer cell line (Fig. 4A). In addition, although transient expression of TβRIII or TβRIII-DEL resulted in cell-rounding and filopodia/microspike-like structures across the entire cell surface (Fig. 4 B–D), TβRIII-T841A was unable to induce this phenotype (Fig. 4 B–D) with cells maintaining their membrane ruffling and lamellipodial structures observed in control cells in the absence of TβRIII expression. We next determined the ability of TβRIII-T841A to regulate levels of active GTP-bound Cdc42 relative to TβRIII and TβRIII-DEL. TβRIII and TβRIII-DEL both constitutively and maximally activated Cdc42, 3.6-fold and 4-fold, respectively (Fig. 4E and Fig. S7C), relative to unstimulated Ovca429 controls in the absence of TGF-β (Fig. 4E). In contrast, TβRIII-T841A failed to constitutively activate Cdc42 (Fig. 4E and Fig. S7C), whereas TGF-β was able to stimulate Cdc42 activation in the presence of TβRIII-T841A (Fig. 4E and Fig. S7C), suggesting a role for the β-arrestin2 interaction motif mediated in these TβRIII functions. To further investigate a role for the TβRIII/β-arrestin2 interaction we determined the effect of siRNA-mediated β-arrestin2 silencing on the ability of TβRIII to suppress migration and Cdc42 activation. siRNA-mediated silencing of β-arrestin2 expression (Fig. 4F Inset) in Ovca429TβRIII cells significantly attenuated TβRIII-mediated inhibition of migration (Fig. 4F) and suppressed TβRIII-mediated Cdc42 activation (Fig. 4G and Fig. S7D). These data demonstrate that the TβRIII/β-arrestin2 interaction is required for TβRIII-mediated activation of Cdc42, alterations in the actin cytoskeleton, and suppression of migration.
Fig. 4.
TβRIII-mediated Cdc42 activation, inhibition of migration, and alterations in the actin cytoskeleton are mediated through the interaction of TβRIII with β-arrestin2. (A) Transwell migration toward a 10% FBS gradient was assessed 48 h after infecting Ovca429 or Ovca433 cells with adenovirus-expressing GFP, full-length TβRIII (TβRIII), TβRIII unable to bind β-arrestin2 (TβRIII-T841A), or TβRIII unable to bind GIPC (TβRIII-DEL). Fold migration relative to GFP control is presented. Data represent the mean ± SE for 4 independent experiments for Ovca429 and 2 independent experiments for Ovca433, each done in triplicate. (#, P = 0.0189 and *, P = 0.0440). (B–D) The indicated cells were fixed and stained for actin using rhodamine phalloidin. GFP-expressing cells were examined and representative examples are presented. Scale bar = 6.6 μm. (E and G) Cells were serum starved and stimulated with TGF-β1 (200 pM) for the indicated times and pull-down assays for active Cdc42 conducted. Quantification is presented in Fig. S7C andFig. S7D, respectively. (F) Ovca429TβRIII cells were subjected to siRNA-mediated silencing of β-arrestin2 (Inset) and transwell migration performed toward 10% FBS. Fold migration relative to Neo controls are presented. Data represent the mean ± SE of 2 independent experiments.
Discussion
TβRIII has a well-established role as a TGF-β superfamily coreceptor (6–8, 12, 34). We and others have determined that TβRIII expression is lost or reduced in a broad spectrum of epithelial-derived human cancers (17–21, 35). Here, we uncover a biological function for TβRIII in regulating cell migration by specifically suppressing directional persistence. TβRIII mediates these effects through β-arrestin2-mediated Cdc42 activation, which alters actin cytoskeleton organization. Surprisingly, these effects appear to be independent of TβRIII's TGF-β coreceptor function as TβRIII inhibits the migration of cells that are resistant to TGF-β's effects on migration, TβRIII inhibits both random and chemotactic migration, and TβRIII-mediated inhibition of migration is not altered by either activators or inhibitors of TGF-β signaling.
While loss of TβRIII during cancer progression did not significantly affect cell survival and proliferation, TβRIII expression potently inhibited cancer cell migration and invasion in vitro and in vivo (17–21). Hence, TβRIII may suppress tumor progression, at least in part, through its effects on cell migration and invasion. In addition, the effects of TβRIII on epithelial cell motility suggest a homeostatic role for TβRIII in limiting epithelial cell motility. During cancer progression, loss of TβRIII expression may relieve this inhibition, resulting in increased motility and facilitating cancer cell invasion and metastases. Indeed, we have demonstrated that restoration of TβRIII expression specifically suppresses invasion and metastasis in a murine model of breast cancer (17).
Although TβRIII-mediated inhibition of migration is likely one mechanism by which TβRIII functions as a putative tumor suppressor, TβRIII could also function to suppress cancer progression through its role as a TGF-β coreceptor, enhancing TGF-β ligand function, and enhancing their tumor-suppressor functions. Thus, loss of TβRIII expression would not only abrogate TβRIII-mediated inhibition of migration but would also facilitate resistance to the tumor-suppressor effects of TGF-β. This potential dual mechanism reinforces the role of TβRIII as a suppressor of cancer progression and as a putative tumor suppressor.
Cdc42 has been shown to both suppress and promote cell migration. However, most of these studies relied on globally blocking or activating Cdc42, although the regulated spatial and temporal localization of Cdc42 activity are likely to have a significant role. Although physiological activation of Cdc42 usually results in increased migration, here we demonstrate that constitutive Cdc42 activation by TβRIII inhibits migration, and blocking this constitutive Cdc42 activation increases migration. To reconcile this discrepancy, we hypothesize that in normal epithelial cells, TβRIII globally activates Cdc42, altering the spatial and temporal localization of Cdc42 activity in response to migratory stimuli. Indeed, this may be an important mechanism by which TβRIII serves to protect the epithelial phenotype and suppress cancer progression. Consistent with this hypothesis, EMT- (21) or transformation- (17–21) mediated-increases in migration are associated with loss of TβRIII expression. Whereas blocking TβRIII-mediated constitutive Cdc42 activation increases migration, constitutive activation of Cdc42 in the absence of TβRIII has no effect on migration. TβRIII also mediated increases in Rac1 activation, however, blocking this constitutive Rac1 activation with dominant-negative Rac1 had no effect on TβRIII-mediated effects on the actin cytoskeleton or migration, further supporting a specific role for Cdc42. Because the presented studies have focused on assessing alterations in global levels of active Cdc42 or Rac1, ongoing studies are investigating the effects of TβRIII on the localization of Cdc42 and Rac1 activity.
The results obtained with the TβRIII mutant unable to bind β-arrestin2 and siRNA-mediated β-arrestin2 silencing demonstrate that TβRIII's effects on Cdc42 are mediated through β-arrestin2. β-arrestin2 has been demonstrated to have a trafficking and desensitization role for TβRIII/betaglycan (16) and has recently emerged as a regulator of directional migration (36). In terms of how β-arrestin2 might be mediating TβRIII-stimulated Cdc42 activation to regulate the actin cytoskeleton and migration, β-arrestin2 might be directly scaffolding TβRIII to Cdc42 to regulate its interaction with guanine nucleotide exchange factors and/or GTPase activating proteins. In addition, this interaction could be altering TβRIII signaling at specific locations within the cell (i.e., the trailing edge) or scaffolding TβRIII with other signaling molecules involved in cytoskeletal reorganization to promote actin reorganization (36). Our results with another TβRIII mutant, TβRIIIΔGAG, suggest a significant contribution from the GAG modifications in the extracellular domain as well. We expect this to be by means of mechanisms distinct from Cdc42 activation. The GAG modification of TβRIII could function to regulate migration while altering the adhesive properties of cells to the extracellular matrix, similar to the function of other proteoglycans and coreceptors (37, 38). Alternatively, the GAG modifications could serve to bind and sequester ligands regulating migration. The mechanism by which the GAG modifications and the extracellular domain of TβRIII regulate TβRIII-mediated inhibition of migration are currently under investigation.
In summary, the current findings define a role for TβRIII in activating members of the Rho GTPase family by means of its interaction with β-arrestin2 to inhibit directional migration independent of its function as a TGF-β coreceptor, while defining a potential mechanism for the putative tumor-suppressor function of TβRIII.
Materials and Methods
Cell Culture, Transfection, Adenoviral Infection, and Reagents.
Human ovarian cancer cell lines Ovca3, Ovca429, Ovca433, and normal spontaneously immortalized ovarian epithelial cell line NOSE007 were cultured in RPMI medium 1680 (Life Technologies, Invitrogen), with 10% FBS at 37 °C in a humidified incubator. Ovarian cancer stable cells lines, Ovca429Neo, Ovca429TβRIII, and breast cancer stable cell lines MDA-MB231TβRIII and MDA-MB231Neo were derived as characterized previously (17, 18). Transient transfections and adenoviral infections were performed as previously described (21). Additional antibodies and adenoviral constructs including siRNA β-arrestin2 and siRNA Smad2 are described in SI Methods.
TGF-β Binding and Cross-Linking.
TGF-β binding and cross-linking studies were performed as previously described (17, 21).
Transwell Migration and Invasion Assays.
Cells (25,000–50,000) were seeded in the upper chamber of a transwell filter, coated both at the top and bottom with 30 μg/mL fibronectin or a 24-well Matrigel-coated transwell (BD Biosciences), to assess cell migration and invasion, respectively. Cells were allowed to migrate for 18 to 24 h at 37 °C through the fibronectin toward the lower chamber, either containing media plus 10% FBS or SFM as appropriate. Cells on the upper surface of the filter were removed and the cells that migrated to the underside of the filter were fixed and stained using the 3 Step Stain Set (Richard-Allan Scientific). Use of pharmacological reagents is described in SI Methods. Each assay was set up in duplicate, and each experiment was conducted at least 3 times with 4 random fields from a 10× magnification analyzed for each membrane.
Live Cell Imaging and Immunoflourescence.
For live cell random migration analysis, cells were plated on 35-mm glass bottom dishes (MatTek Corporation) at a density of 104 cells per well in regular culture medium and placed in a temperature- and CO2-controlled chamber. Time-lapse recording started either 24 h or 48 h (for infected cells) after plating. Images were collected at 1-min intervals over 120 min. Microscope description, calculation of motility parameters, including migration path, distance, rate, and directional persistence were obtained from time-lapse movies and are described in detail in SI Methods. Immunofluorescence for actin staining was performed as previously described (21) and in SI Methods.
Cdc42 and Rac1 pull-down assays were conducted as described (39) and in SI Methods.
Statistical tests were conducted for the datasets described in figures and figure legends. All data were analyzed using 2-tailed, unpaired student's t test, and with a minimum of a 95% confidence interval.
Supplementary Material
Acknowledgments.
We thank Tam How for purification of the adenovirus used in these studies. We also thank Dr. Sam Johnson and the Duke University Microscopy Core Facility for their expert assistance with the microscopy aspects of these studies. The adenovius constructs, N17Cdc42 and N17Rac1, were generous gifts from Dr. George E. Davis (University of Missouri-Columbia, Columbia, MO). This work was funded by National Institutes of Health/National Cancer Institute Grant R01-CA106307 (to G.C.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812879106/DCSupplemental.
References
- 1.Lauffenburger DA, Horwitz AF. Cell migration: A physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 2.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Nobes CD, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiner OD. Regulation of cell polarity during eukaryotic chemotaxis: The chemotactic compass. Curr Opin Cell Biol. 2002;14:196–202. doi: 10.1016/s0955-0674(02)00310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pankov R, et al. A Rac switch regulates random versus directionally persistent cell migration. J Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang XF, et al. Expression cloning and characterization of the TGF-beta type III receptor. Cell. 1991;67:797–805. doi: 10.1016/0092-8674(91)90074-9. [DOI] [PubMed] [Google Scholar]
- 7.Lopez-Casillas F, et al. Structure and expression of the membrane proteoglycan betaglycan, a component of the TGF-beta receptor system. Cell. 1991;67:785–795. doi: 10.1016/0092-8674(91)90073-8. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Casillas F, Wrana JL, Massague J. Betaglycan presents ligand to the TGF beta signaling receptor. Cell. 1993;73:1435–1444. doi: 10.1016/0092-8674(93)90368-z. [DOI] [PubMed] [Google Scholar]
- 9.Lewis KA, et al. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature. 2000;404:411–414. doi: 10.1038/35006129. [DOI] [PubMed] [Google Scholar]
- 10.Stenvers KL, et al. Heart and liver defects and reduced transforming growth factor beta2 sensitivity in transforming growth factor beta type III receptor-deficient embryos. Mol Cell Biol. 2003;23:4371–4385. doi: 10.1128/MCB.23.12.4371-4385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown CB, Boyer AS, Runyan RB, Barnett JV. Requirement of type III TGF-beta receptor for endocardial cell transformation in the heart. Science. 1999;283:2080–2082. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- 12.Kirkbride KC, Townsend TA, Bruinsma MW, Barnett JV, Blobe GC. Bone morphogenetic proteins signal through the transforming growth factor-beta type III receptor. J Biol Chem. 2008;283:7628–7637. doi: 10.1074/jbc.M704883200. [DOI] [PubMed] [Google Scholar]
- 13.Deng X, Bellis S, Yan Z, Friedman E. Differential responsiveness to autocrine and exogenous transforming growth factor (TGF) beta1 in cells with nonfunctional TGF-beta receptor type III. Cell Growth Differ. 1999;10:11–18. [PubMed] [Google Scholar]
- 14.Blobe GC, et al. Functional roles for the cytoplasmic domain of the type III transforming growth factor beta receptor in regulating transforming growth factor beta signaling. J Biol Chem. 2001;276:24627–24637. doi: 10.1074/jbc.M100188200. [DOI] [PubMed] [Google Scholar]
- 15.Blobe GC, Liu X, Fang SJ, How T, Lodish HF. A novel mechanism for regulating transforming growth factor beta (TGF-beta) signaling. Functional modulation of type III TGF-beta receptor expression through interaction with the PDZ domain protein, GIPC. J Biol Chem. 2001;276:39608–39617. doi: 10.1074/jbc.M106831200. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, et al. Beta-arrestin 2 mediates endocytosis of type III TGF-beta receptor and down-regulation of its signaling. Science. 2003;301:1394–1397. doi: 10.1126/science.1083195. [DOI] [PubMed] [Google Scholar]
- 17.Dong M, et al. The type III TGF-beta receptor suppresses breast cancer progression. J Clin Invest. 2007;117:206–217. doi: 10.1172/JCI29293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hempel N, et al. Loss of betaglycan expression in ovarian cancer: Role in motility and invasion. Cancer Res. 2007;67:5231–5238. doi: 10.1158/0008-5472.CAN-07-0035. [DOI] [PubMed] [Google Scholar]
- 19.Turley RS, et al. The type III transforming growth factor-beta receptor as a novel tumor suppressor gene in prostate cancer. Cancer Res. 2007;67:1090–1098. doi: 10.1158/0008-5472.CAN-06-3117. [DOI] [PubMed] [Google Scholar]
- 20.Finger EC, et al. TbetaRIII suppresses non-small cell lung cancer invasiveness and tumorigenicity. Carcinogenesis. 2008;29:528–535. doi: 10.1093/carcin/bgm289. [DOI] [PubMed] [Google Scholar]
- 21.Gordon KJ, Dong M, Chislock EM, Fields TA, Blobe GC. Loss of type III transforming growth factor beta receptor expression increases motility and invasiveness associated with epithelial to mesenchymal transition during pancreatic cancer progression. Carcinogenesis. 2008;29:252–262. doi: 10.1093/carcin/bgm249. [DOI] [PubMed] [Google Scholar]
- 22.Partin AW, Schoeniger JS, Mohler JL, Coffey DS. Fourier analysis of cell motility: Correlation of motility with metastatic potential. Proc Natl Acad Sci USA. 1989;86:1254–1258. doi: 10.1073/pnas.86.4.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. J Clin Oncol. 2005;23:2078–2093. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez GC, et al. Regulation of invasion of epithelial ovarian cancer by transforming growth factor-beta. Gynecol Oncol. 2001;80:245–253. doi: 10.1006/gyno.2000.6042. [DOI] [PubMed] [Google Scholar]
- 25.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 26.Wu X, Kodama A, Fuchs E. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell. 2008;135:137–148. doi: 10.1016/j.cell.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glasgow JE, Daniele RP. Role of microtubules in random cell migration: Stabilization of cell polarity. Cell Motil Cytoskeleton. 1994;27:88–96. doi: 10.1002/cm.970270110. [DOI] [PubMed] [Google Scholar]
- 28.Lo CM, et al. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol Biol Cell. 2004;15:982–989. doi: 10.1091/mbc.E03-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL. p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J Cell Sci. 2002;115:3193–3206. doi: 10.1242/jcs.115.15.3193. [DOI] [PubMed] [Google Scholar]
- 30.Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421:753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- 31.Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol Biol Cell. 2002;13:902–914. doi: 10.1091/mbc.01-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayless KJ, Davis GE. The Cdc42 and Rac1 GTPases are required for capillary lumen formation in three-dimensional extracellular matrices. J Cell Sci. 2002;115:1123–1136. doi: 10.1242/jcs.115.6.1123. [DOI] [PubMed] [Google Scholar]
- 33.Hall A. RhoGTPases and the actin cytoskeleton. Science. 1998;23:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 34.Waite KA, Eng C. From developmental disorder to heritable cancer: It's all in the BMP/TGF-beta family. Nat Rev Genet. 2003;4:763–773. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]
- 35.Copland JA, et al. Genomic profiling identifies alterations in TGFbeta signaling through loss of TGFbeta receptor expression in human renal cell carcinogenesis and progression. Oncogene. 2003;22:8053–8062. doi: 10.1038/sj.onc.1206835. [DOI] [PubMed] [Google Scholar]
- 36.DeFea KA. Stop that cell! Beta-arrestin-dependent chemotaxis: A tale of localized actin assembly and receptor desensitization. Annu Rev Physiol. 2007;69:535–560. doi: 10.1146/annurev.physiol.69.022405.154804. [DOI] [PubMed] [Google Scholar]
- 37.Kirkbride KC, Ray BN, Blobe GC. Cell-surface co-receptors: Emerging roles in signaling and human disease. Trends Biochem Sci. 2005;30:611–621. doi: 10.1016/j.tibs.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Ruoslahti E, Yamaguchi Y. Proteoglycans as modulators of growth factor activities. Cell. 1991;64:867–869. doi: 10.1016/0092-8674(91)90308-l. [DOI] [PubMed] [Google Scholar]
- 39.Sahai E, Olson MF, Marshall CJ. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 2001;20:755–766. doi: 10.1093/emboj/20.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.