Abstract
We have asked here how the remarkable variation in maize haplotype structure affects recombination. We compared recombination across a genetic interval of 9S in 2 highly dissimilar heterozygotes that shared 1 parent. The genetic interval in the common haplotype is ≈100 kb long and contains 6 genes interspersed with gene-fragment-bearing Helitrons and retrotransposons that, together, comprise 70% of its length. In one heterozygote, most intergenic insertions are homozygous, although polymorphic, enabling us to determine whether any recombination junctions fall within them. In the other, most intergenic insertions are hemizygous and, thus, incapable of homologous recombination. Our analysis of the frequency and distribution of recombination in the interval revealed that: (i) Most junctions were circumscribed to the gene space, where they showed a highly nonuniform distribution. In both heterozygotes, more than half of the junctions fell in the stc1 gene, making it a clear recombination hotspot in the region. However, the genetic size of stc1 was 2-fold lower when flanked by a hemizygous 25-kb retrotransposon cluster. (ii) No junctions fell in the hypro1 gene in either heterozygote, making it a genic recombination coldspot. (iii) No recombination occurred within the gene fragments borne on Helitrons nor within retrotransposons, so neither insertion class contributes to the interval's genetic length. (iv) Unexpectedly, several junctions fell in an intergenic region not shared by all 3 haplotypes. (v) In general, the ability of a sequence to recombine correlated inversely with its methylation status. Our results show that haplotypic structural variability strongly affects the frequency and distribution of recombination events in maize.
Keywords: Ac, bz locus, genome, hotspots, methylation
There is ample evidence that most recombination in maize takes place in or around genes (1–3) and that little, if any, recombination takes place in the repetitive and methylated retrotransposon DNA that makes up the bulk of the genome (4, 5). The evidence that retrotransposons are largely recombinationally inert is 2-fold: (i) when homozygous, the standard situation in inbreds, they do not contribute significantly to genetic length (4), and (ii) when hemizygous, a common situation in hybrids, no recombination junctions fall in intervals containing them (5).
The unprecedented haplotype diversity recently discovered in maize (6–8) raises several questions regarding the effect of local structural polymorphisms on recombination. In a recent pairwise comparison of 8 bz1 haplotypes, each one consisting of 8 genes spread over a stretch of DNA that averaged ≈90 kb, the percentage of shared sequences ranged from 25% to 84% (9). The lines differed by the existence of many polymorphic insertions in introns and intergenic regions, the main ones being LTR retrotransposons, often arranged in nests (10), miniature inverted repeat transposable elements (MITEs) (11), and Helitron transposons carrying fragments of several genes (12, 13). In addition, high SNP and indel heterozygosity would occur in most intergenic regions of hybrids made from those lines.
This level of structural polymorphisms could affect recombination in multiple ways and, thus, contribute to the 2- to 3-fold variation in estimates of map distances for single genetic intervals that has been reported in different maize mapping populations (14–16). For example, the highly methylated retrotransposon clusters are probably heterochromatic, so retrotransposon hemizygosity could reduce recombination in adjacent genes; the level of small insertion and SNP affects overall sequence homology and, thus, could determine whether recombination occurs in intergenic regions lacking retrotransposons; and Helitrons could lead to the expansion of an interval's genetic length if recombination occurred within the gene fragments they carry.
Cis-effects were demonstrated in a study that examined recombination rates across the a1-sh2 genetic interval in 3 heterozygotes containing the same maize haplotype and different teosinte-derived haplotypes in a common maize background (17). This region had several large insertion/deletion polymorphisms relative to maize, including 2 LTR retrotransposons. The analysis identified up to 3-fold differences in recombination rates and statistical differences in the distribution of recombination junctions across subintervals among haplotypes. Recently, we examined the local effect of intergenic retrotransposons on recombination between 2 genes located at the proximal end of the characterized bz1 haplotypes (18). Recombination in a genetic interval defined by markers in the adjacent genes bz1 and stc1 was compared in heterozygotes between haplotypes that differed by the presence and absence of a large retrotransposon cluster in the bz1-stc1 intergenic region. There was a 2-fold suppressing effect of retrotransposon hemizygosity on recombination across the interval and an even stronger effect in the bz1 and stc1 intervals immediately flanking the cluster.
In the present work, we have analyzed recombination between genes at opposite ends of the characterized bz1 haplotypes (9) in 2 heterozygotes that share 1 common haplotype but differ extensively in the level and type of structural polymorphisms in the intergenic regions. In one heterozygote, most structural polymorphisms are homozygous, although different enough to allow placement of any recombination junctions that might fall within them. In the other one, most structural polymorphisms are hemizygous and, thus, incapable of homologous recombination.
The genetic interval in the 3 haplotypes used in the study ranges in physical length from 62.7 to 111.5 kb. It consists of 5 complete genes plus parts of the 2 genes that mark the opposite ends of the interval and of 6 intergenic regions that contain the 3 main types of intergenic polymorphic insertions discussed above: Helitrons bearing gene fragments, nested and single retrotransposons, and MITEs. The 2 heterozygous configurations studied here have allowed us to: (i) determine whether recombination occurs within gene fragments borne on Helitrons; (ii) examine whether recombination junctions across the ≈100-kb multigenic interval always fall in genes and their immediate vicinity or occasionally fall in intergenic regions apparently devoid of genes; (iii) identify genes that behave as recombination hotspots or coldspots in the interval; (iv) compare recombination frequencies and the distribution of hotspots and coldspots in structurally diverse heterozygotes, and (v) correlate recombination with percentage sequence identity and C-methylation status.
Results
Derivation and Analysis of the bz1-m2(D1) Ac7077 Chromosome.
To analyze the effect of Helitrons and retrotransposon insertions on the number and distribution of exchanges across a genetic interval containing a mix of genes and polymorphic insertions, we followed a similar strategy to that described in ref. 18. Basically, we took advantage of a Ds mutable allele reporter whose expression depends on the presence of an Ac element at a second locus that is separated from the reporter by the length of the genetic interval under study. Crossing over between the transposable elements results in derivatives with a stable phenotype and a recombinant arrangement of flanking markers (Fig. 1). In this instance, the Ds mutable allele is bz1-m2(D1), and the locus where Ac resides is tac7077. In W22 (18), tac7077 is separated from bz1 by ≈100 kb of DNA that contain 5 genes and a mixture of large and small insertions in the intergenic regions. This locus was defined initially by the insertion of an Ac transposon (Ac7077) in its 5′ UTR and, subsequently, by the isolation of a full-length cDNA clone by using the Ac-adjacent sequence as probe (19).
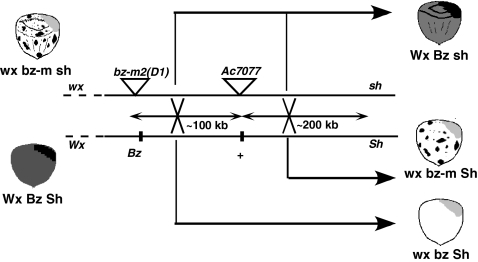
Fig. 1.
Recovery of recombination products in the bz1-Ac7077 and Ac7077-sh1 intervals. The cartoon depicts the parental (Left) and recombinant (Right) phenotypes. The contrasting kernel phenotypes are: Bz, purple; bz-m, spotted; and bz, bronze; Wx, nonwaxy and wx, waxy, which stain dark and light, respectively, with an I-KI solution, as illustrated at the top right of each kernel (although no staining is necessary when scoring bz Sh seed); Sh, plump; and sh, shrunken. Recombination in the bz-m2(D1)-Ac7077 interval gives rise to wx bz Sh and Wx Bz sh reciprocal recombinants. Recombination in the Ac7077-sh1 interval gives rise to wx bz-m Sh and Wx Bz sh reciprocal recombinants. The physical distance between Ac7077 and bz in B73 is from ref. 4; that between sh and Ac7077 is from the maize genome sequencing project (www.maizesequence.org). These will vary from inbred to inbred and are shown for relative purposes only. Table 1 shows the seed populations screened for the Ac7077/W22 and Ac7077/B73 heterozygotes and the number of recombinants isolated in each class.
Our 2 main objectives at the outset of this study were to analyze the recombinational properties of the gene fragments borne on Helitrons and to determine the extent to which insertion polymorphisms affected recombination across the length of a relatively gene-rich ≈100-kb segment of maize genomic DNA. Thus, we anticipated that a comparison of recombination in heterozygotes between McC and the contrasting haplotypes W22, which it resembles (18), and B73, from which it differs greatly (6), should generate this information (Fig. 2). A chromosome carrying bz1-m2(D1) linked in-cis to the distal Ac7077 element was derived for this study, and the composition of its bz1-tac7077 region was determined by cloning it into a BAC vector (20) and sequencing it. In bz1-m2(D1) Ac7077, referred to hereafter as the Ac7077 haplotype, Ds and Ac are separated by 98.9 kb (Fig. 2). The proximal (left) 81 kb are identical in sequence to McC and comprise the genes bz1, stc1, rpl35A, tac6058, and hypro1, their respective intergenic regions, and a 66.7-kb segment between hypro1 and znf that includes the 5.9-kb HelA and 2.6-kb HelB Helitrons (12) plus a 53-kb retrotransposon cluster consisting of Opie2, Huck1, and Ji-6 nested sequences (10). However, close to the transcription start of znf, the sequences diverge, and the distal 17.9 kb of the Ac7077 haplotype closely resemble B73, rather than McC, in their MITE and retrotransposon make-up. The znf-tac7077 intergenic region in Ac7077 and B73 contains a 12.4-kb Grande1 retrotransposon and 3 previously unannotated small insertions, including a 286-bp MITE into which Grande1 has inserted and a 360-bp mPIF element (21). Thus, as expected from its origin as a Ds derivative of the bz1-m2(Ac) allele (18, 22), the bz1-m2(D1) allele is borne on a predominantly McC bz1 haplotype, but the recombination event that coupled it with Ac7077 led to a reshuffling of the McC haplotype's distal end.
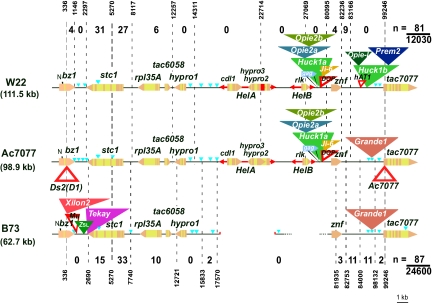
Fig. 2.
Distribution of recombination junctions in the Ds2(D1)-Ac7077 genetic interval among Sh bz recombinants from bz-m2(D1) Ac7077/Bz-W22 + (Upper) and bz-m2(D1) Ac7077/Bz-B73 + (Lower) heterozygotes. Each haplotype is identified by the name of the genetic line, followed by the physical size of the interval under study, in parentheses. The 9S centromere (proximal side) is to the left; the 9S telomere (distal side), to the right. Genes are shown as pentagons pointing in the direction of transcription; exons are in bronze, and introns in yellow. There are 7 genes in the genetic interval: bz1, stc1, rpl35A, tac6058, hypro1, znf, and tac7077. The same symbolism is used for gene fragments carried by Helitrons (Hels), which are represented as bidirectional arrows below the line for each haplotype. The vacant sites for HelA and HelB in B73 are provided as reference points and marked with short vertical strokes. Dashed lines represent deletions. Retrotransposons are indicated by solid triangles of different colors. DNA transposons, including Ds2(D1) and Ac7077, are indicated by open triangles of red color. Small insertions are indicated in light blue color. Only the genes have been drawn to scale. For analysis, the interval was subdivided into a series of roughly corresponding segments defined by the same polymorphic sites, where possible, or by polymorphisms located close to each other in each heterozygote. The locations of the different polymorphisms are numbered according to the bz-m2(D1) Ac7077 sequence, starting with the NotI site in the first exon of bz1 as position 1. The nature of the polymorphisms at each location (SNPs or indels) is identified in Tables S1 and S2. The number of recombination junctions in each subinterval is shown above (W22) and below (B73) the unique haplotype in each heterozygote. See Results for additional details.
The sequences of the genes in the bz1-m2(D1) Ac7077 interval of McC, W22, and B73 show the typical percentage of divergence of maize alleles and no particular haplotypic pattern. Thus, allelic comparisons show bz1 and stc1 to be more polymorphic between McC and W22 than between McC and B73 (1.5% vs. 1.2% and 1.7% vs. 1.5% divergence, respectively), rpl35A and tac6058 to be less so (0.0% vs. 0.8% and 0.0% vs. 1.1% divergence, respectively), and hypro1 to be equally polymorphic (1.3% divergence). The znf allele found in the Ac7077 haplotype is closer to B73 than to W22 (0.8% vs. 1.3% divergence).
Recombination Between bz1-m2(D1) and Ac7077.
All 3 haplotypes used in the recombination experiment were first introduced into a W22 inbred background (18). Heterozygotes between haplotypes sharply different in overall structure were synthesized by crossing bz1-m2(D1) Ac7077 with either Bz1-W22 or Bz1-B73 (Fig. 2). bz1-m2(D1) Ac7077 and Bz1-W22 lack a retrotransposon cluster in the bz1-stc1 intergenic region, share the same 5.9-kb and 2.6-kb Helitrons and the 53-kb retrotransposon cluster in the hypro1-znf intergenic region, the latter accounting for half of the interval's physical length, and differ mainly in the makeup of the retrotransposons in the znf-tac7077 intergenic region. In contrast, bz1-m2(D1) Ac7077 and Bz1-B73 differ by the presence in one, but not the other, of the 26-kb retrotransposon cluster in the bz1-stc1 intergenic region, the 2 Helitrons, and the 53-kb retrotransposon cluster in the hypro1-znf intergenic region and resemble each other only in the znf-tac7077 intergenic region, where they share the Grande1 retrotransposon inserted in a MITE and 2 other small insertions.
The experiment set up to identify recombinants in the bz1-m2(D1)-Ac7077 interval is diagrammed in Fig. 1. The bz1-m2(D1) allele produces a spotted seed phenotype in the presence of Ac7077 and a bronze phenotype in its absence. The contrasting W22 and B73 Bz1 haplotypes produce a purple phenotype. The interval is flanked by the outside markers wx1 and sh1, so individuals in which the bz1-m2(D1) reporter has been separated from Ac7077 by crossing over can be easily identified as stable bronze derivatives with recombined flanking markers. Heterozygotes between wx1 bz1-m2(D1) Ac7077 sh1 and either Wx1 Bz1-B73 + Sh1 or Wx1 Bz1-W22 + Sh1 were hand-pollinated with a wx1 bz1-sh1-X2 tester. The tester stock carries a deletion of the entire bz1-sh1 region (23) and is useful for recovering selections in a hemizygous condition that facilitates their molecular analysis. Recombination between bz1-m2(D1) and Ac7077 will produce wx bz Sh and Wx Bz sh seed as reciprocal products, and that between Ac7077 and sh1 will produce wx bz-m Sh and Wx Bz sh seed as reciprocal products. Hence, the sum of bz Sh and bz-m Sh seed should approximately equal the number of Bz sh seed, and, as shown in Table 1, it does in both heterozygotes. However, the size of the bz1-sh1 interval, estimated from the sum of the last 2 columns in the table, is significantly larger in the Ac7077/W22 heterozygote than in the Ac7077/B73 heterozygote (2.75 vs. 2.08 cM), most of the difference being attributable to the bz Sh class. This class provides a rough estimate of recombination in the bz1-Ac7077 interval, which was shown to be structurally more similar in the Ac7077 and W22 haplotypes (Fig. 2). Most bz Sh selections were wx, as expected from the high chiasma interference in the wx1-bz1-sh1 region (24). Subsequent molecular analysis of the wx bz Sh selections confirmed that most arose by recombination between bz1-m2(D1) and Ac7077, although ≈10% of successfully tested selections represented coincidental Ds or Ac excision and recombination in the bz1-sh1 interval (8/89 in Ac7077/W22 and 14/101 in Ac7077/B73).
Table 1.
Seed selections from heterozygotes bz1-m2(D1) Ac7077 and either W22 or B73 Bz1+
| Heterozygous haplotype | Seed population | Recombinant seed class phenotypes |
|||
|---|---|---|---|---|---|
| bz Sh | bz-m Sh | (bz+b-zm) Sh | Bz sh | ||
| Ac7077/W22 | 38,200 | 307 | 229 | 536 | 515 |
| Percentage | 0.80 | 0.60 | 1.40 | 1.35 | |
| 95% C.L. | 0.74–0.86 | 0.55–0.65 | 1.32–1.48 | 1.27–1.43 | |
| Ac7077/B73 | 30,090 | 126 | 177 | 303 | 323 |
| Percentage | 0.42 | 0.59 | 1.01 | 1.07 | |
| 95% C.L. | 0.37–0.47 | 0.53–0.65 | 0.93–1.09 | 0.99–1.16 | |
C.L., confidence limits.
Distribution of Recombination Junctions.
The recombination junctions in an approximately equal number of wx bz Sh selections from each heterozygote were placed within the Ds2(D1)-Ac7077 interval by PCR-genotyping a series of indel polymorphisms across the interval and sequencing the PCR products when they contained multiple polymorphisms. Supporting information (SI) Tables S1 and S2 identify the position and nature of the polymorphisms and summarize the marker data for each recombinant in the Ac7077/W22 and Ac7077/B73 series, respectively. The results from placing 168 junctions are shown graphically in Fig. 2. The Ds2(D1)-Ac7077 interval has been subdivided into a series of approximately corresponding intervals in the 2 heterozygotes. They are: the bz1 gene, the bz1-stc1 intergenic region, and the stc1 gene—3 segments compared in detail previously (18); the rpl35A-tac6058 segment; the hypro1 gene; the 2 Helitrons and the 53-kb retrotransposon cluster in the hypro1-znf intergenic region of the Ac7077/W22 heterozygote; the znf gene, and the znf-tac7077 intergenic region. It is important to note that the 81 recombinants analyzed in Ac7077/W22 come from a population approximately half the size of the one that generated the 87 recombinants analyzed in Ac7077/B73.
The data summarized in Fig. 2 allow us to refine the estimate of the Ds2(D1)-Ac7077 genetic distance to 1.35 and 0.70 cM, respectively, in Ac7077/W22 and Ac7077/B73. The stc1 gene, including ≈1 kb upstream, is a clear recombination hotspot (25) in both heterozygotes. It accounts for 70% (58 of 81) and 55% (48 of 87) of recombinants, respectively, in the Ac7077/W22 and Ac7077/B73 heterozygotes and measures close to 1 cM, ≈1/3 of the bz1-sh1 distance, in the former. However, as reported previously (18), the distribution of recombination junctions within stc1 differs in the 2 heterozygotes, being relatively much lower (15 of 48 vs. 31/58) in the segment of the stc1 gene that borders the hemizygous 25-kb retrotransposon cluster in Ac7077/B73. In fact, in this heterozygote, no recombination junctions at all were recovered in the 29 kb that lie between Ds2(D1) and the 3′ end of the stc1 gene at the left (proximal) end of the Ds2(D1)-Ac7077 interval. A number of recombinants proportionate to the 2 gamete populations screened occurred in the rpl35A-tac6058 segment of each heterozygote, but none occurred in hypro1 in either heterozygote, making the latter a genic recombination coldspot in the interval (26).
No recombination junctions fell within either Helitron, although their sequences are almost 100% identical in Ac7077 and W22. Only a 680-bp deletion in the hypro2 gene fragment of W22's HelA and a single SNP in the rlk gene fragment of HelB distinguish both haplotypes. Therefore, the gene fragments carried by Helitrons do not contribute to genetic length. Not surprisingly, no recombination junctions fell within the 53-kb retrotransposon cluster found in the hypro1-znf intergenic region of Ac7077 and W22, despite a >99% sequence identity. Therefore, no recombination occurs within this large retrotransposon block that makes up approximately half of the physical size of the bz1-tac7077 genetic interval in Ac7077 and W22. In the Ac7077/W22 heterozygote, 4 junctions fell within znf and 9 fell in a 0.9-kb segment immediately downstream, which contains 280 bp of the znf 3′ UTR and is 98.2% identical between the 2 haplotypes. However, no junctions were observed in the rest of the znf-tac7077 intergenic region, which differs in size (16 kb in Ac7077 vs. 29.7 kb in W22) and retrotransposon cluster content and shares only short stretches of interrupted homology in these 2 haplotypes.
Unexpectedly, a large number of junctions fell in the znf-tac7077 intergenic region of Ac7077/B73 heterozygotes: 24 of 87. This region is 99.5% identical in the 2 haplotypes, so very few polymorphisms are available to map the 24 junctions. However, 2 SNPs, 1 indel, and the Ac7077 insertion enabled us to divide the intergenic region into 3 segments. Of the 24 junctions, 11 fell in a 1.2-kb interval right after znf, 11 fell in a 14-kb interval that contains Grande1 and 2 small insertions, and 2 fell in a 1.1-kb interval just upstream of Ac7077, which contains an mPIF MITE. Other than parts of the znf 3′ UTR and tac7077 5′ UTR, no genes are known to be present in these segments, so some recombination may occur in maize intergenic regions. Supporting this observation, 2 junctions were also found in a 1.7-kb segment of the hypro1-znf intergenic region of Ac7077/B73 heterozygotes that contains a Tourist Zm-3 MITE in both haplotypes.
Methylation Analysis.
In general, recombinogenic maize genes and single-copy sequences are not methylated at CG or CNG residues (4, 27, 28), whereas the recombinationally inert repetitive retrotransposons are (4, 29–31). To examine whether the ability of a sequence in the Ds2(D1)-Ac7077 interval to recombine was correlated with its methylation status, we carried out an analysis of C-methylation using methylation-sensitive restriction endonucleases, which detect C-methylation in either a CG or CNG sequence context, and bisulfite sequencing, which detects any methylated C. We focused on: stc1 and hypro1, a genic recombination hotspot and coldspot, respectively; the nonrecombining gene-fragment-carrying Helitrons; the bz1-stc1 intergenic region, which shows no recombination in either heterozygote, and the znf-tac7077 intergenic region, which showed a surprising amount of recombination in the Ac7077/B73 heterozygote.
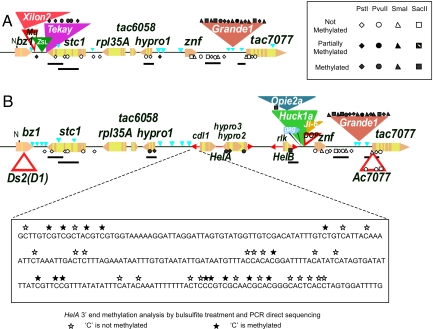
Methylation across the interval was assayed by restricting genomic and BAC DNAs with 4 C-methylation-sensitive restriction enzymes, PstI, PvuII, SmaI, and SacII, hybridizing Southern blots with individual probes from stc1, hypro1, the rlk gene fragment of HelB, single-copy sequences just downstream of znf and upstream of tac7077 in the znf-tac7077 intergenic region, and analyzing the observed band patterns against that expected from the DNA sequence of the respective BAC clones (Figs. S1–S3). The methylation status of the 3′ (left) end of HelA was assayed by bisulfite sequencing (32). The results of the B73 and Ac7077 analysis, summarized in Fig. 3, reveal a strong correlation between the methylation status of a sequence and its ability to recombine. Thus, none of the assayed sites in the recombination hotspot stc1 gene or its upstream region were methylated. All CG and CNG sites assayed in the nonrecombining HelA and HelB Helitrons and Tekay retrotransposon were methylated. Sites in the recombination coldspot hypro1 were methylated in Ac7077, the common haplotype in the 2 heterozygotes, but not in B73. Last, sites in the recombination-proficient znf-tac7077 intergenic region of Ac7077/B73 heterozygotes showed 2 distinct methylation patterns in both haplotypes: Most of those immediately adjacent to znf and tac7077 were not methylated, whereas all of those within the Grande1 retrotransposon were.
Fig. 3.
Summary of C-methylation analysis in B73 (A) and Ac7077 (B) haplotypes. The results from methylation-sensitive restriction endonucleases are represented with a series of geometric symbols, whose meaning is described in the box at the top right of the figure. The probes used to hybridize the blots are shown as bars beneath each haplotype. The C-methylation status at the 3′ end of HelA in Ac7077 was determined by bisulfite sequencing. Unmethylated sites are represented as open stars and methylated sites, as filled stars. The C-methylation status of W22 sites between Ds2(D1) and HelB was essentially identical to Ac7077.
Discussion
Rates of recombination for specific chromosome segments are known to vary greatly in maize (14–16). In this and a recent study (18), we have addressed the issue of how the remarkably variable genome structure of maize affects recombination. We previously demonstrated that the presence of a large intergenic retrotransposon block in hemizygous condition reduced recombination in the adjacent genes 2- to 4-fold (18). Here, we have examined the distribution of recombination junctions in an ≈100-kb genetic interval that contains several genes mixed with intergenic Helitron and retrotransposon insertions, either in homozygous or hemizygous condition, and have compared recombination across the interval in 2 highly dissimilar heterozygotes.
We synthesized 2 heterozygotes that had 1 haplotype in common, but differed in the other. The common haplotype was Ac7077, and the unique one was either W22 or B73 (Fig. 2). In terms of the large insertion polymorphisms in the intergenic regions, the interval of Ac7077 can be divided in 2: the proximal 80 kb, which includes 2 gene-fragment-bearing Helitrons and a 3-level nested retrotransposon cluster, are shared with W22 but not with B73; conversely, the distal 20 kb, which includes a single retrotransposon, are shared with B73 but not with W22. This arrangement allows us to determine in what homozygous genic or intergenic regions recombination occurs and to examine the overall effect of insertion hemizygosity on recombination across the interval.
Given that recombination in maize takes place mostly in genes, we were particularly interested to test whether Helitrons contributed to genetic length. Helitrons are recently discovered transposons that presumably move by a rolling-circle mode of transposition (33). In maize, many Helitrons have trapped host gene fragments of variable length and mobilized them around the genome (34, 35). Like retrotransposons, Helitrons are highly polymorphic and contribute to the remarkable haplotype variability of modern maize (9, 12, 13, 36). In terms of abundance, they are estimated to comprise ≈1.4% of the sequenced B73 genome (C. Du, personal communication). To determine whether the gene fragments in Helitrons recombined, we paired Ac7077 and W22, haplotypes that share the same 2 Helitrons. Together, these Helitrons make up ≈10 kb of the hypro1-znf intergenic region and contain fragments from 4 genes (12). The Helitrons in the 2 haplotypes are structurally homozygous, although polymorphic, enabling us to determine whether any recombination junctions fall within them, and, as seen in Fig. 2, none did. Methylation analysis revealed that all CG and CNG sites assayed in the nonrecombining Helitrons were methylated (Fig. 3), in agreement with what has been observed with retrotransposons (4, 29–31). We conclude, then, that recombination within Helitrons is not likely to contribute to the variability in recombination rates that has been observed in maize. Because HelA and HelB are found in the same intergenic region as a 53-kb retrotransposon cluster, our experimental setup does not allow us to determine whether Helitron hemizygosity in Ac7077/B73 heterozygotes affects recombination in nearby genes.
The frequency of recombination across the entire bz1-Ac7077 interval was significantly lower in the Ac7077/B73 heterozygote than in the Ac7077/W22 heterozygote (0.70 vs. 1.35 cM). The main reason for the difference is that recombination within the stc1 gene, a clear recombination hotspot in the interval, is much lower in the former. The stc1 gene and its upstream region measure 0.96 cM in the Ac7077/W22 heterozygote, but only 0.39 cM in the Ac7077/B73 heterozygote, most likely because hemizygosity for the large retrotransposon cluster adjacent to stc1 in the B73 haplotype reduces recombination in the adjacent intervals (18). The most distal retrotransposon in the cluster, Tekay, is actually inserted within the 3′ UTR of the stc1-B73 allele, leading to transcription termination within the 5′ LTR (C. Lin, personal communication). We have shown here that this retrotransposon is heavily methylated, but the stc1 sequences next to it are not. The whole 25-kb cluster is probably heterochromatic and may interfere with proper homologous alignment of the adjacent stc1 and bz1 regions (18).
Some level of recombination was detected in all of the genes in the interval, except for hypro1, which behaves as a genic recombination coldspot in both heterozygotes. This observation supports the conclusion of Yao et al. (5) from their analysis of recombination in the a1-sh2 region that not all maize genes are recombination hotspots. Peculiarly, the hypro1 gene was found to be methylated in Ac7077 and W22 but not B73. A possible reason for this difference is that in the first 2, the gene lies adjacent to a large intergenic region that contains methylated Helitrons and a 53-kb retrotransposon cluster, but in B73, it lies adjacent to a short intergenic region that contains no large insertions. Possibly, recombination across the stc1-znf interval would be higher in a heterozygote between B73 and a structurally closer haplotype, such as Mo17 or A188 (9).
The initial conclusion that little, if any, recombination takes place within homozygous retrotransposons was based on the observation that the ratio of genetic to physical distance between 2 markers derived from the same haplotype and separated by a 94-kb retrotransposon nest was close to the genome's average (4). Here, we have examined recombination between markers in different haplotypes that share a 53-kb retrotransposon cluster of >99% sequence identity. The use of SNP and indel polymorphisms on either side of the cluster enabled us to determine that no recombination junctions fell within it, confirming that most retrotransposons are probably recombinationally inert. They are heavily methylated and probably organized into a condensed chromatin (37) that would not be readily accessible to the cell's recombination machinery.
Perhaps the most unexpected outcome of this study was the relatively large number of junctions that fell in the znf-tac7077 intergenic region of Ac7077/B73 heterozygotes (Fig. 2). Although this region is 99.5% identical in the 2 haplotypes, we were able to divide it into 3 subintervals through the use of polymorphic markers. Of the 24 junctions, 11 fell in a 1.2-kb interval right after znf, which contains the long znf 3′ UTR, and 11 others fell in a 14-kb interval that contains Grande1 and 2 small insertions but no known genes. There are no polymorphisms in the interval to formally rule out recombination within the homozygous 12.4-kb Grande1 retrotransposon, but its heavily methylated status relative to the rest of the znf-tac7077 intergenic region (Fig. 3) would suggest that, like other retrotransposons, Grande1 does not recombine. From the discussion above and the observation that the sequence of the znf-tac7077 intergenic region in W22 is very different, it appears that some recombination in maize can occur in chromosomal segments apparently lacking genes, an observation that supports earlier conclusions based on more limited sequence data (5, 28). Not surprisingly, no recombination occurred in the highly dissimilar corresponding segment in the znf-tac7077 intergenic region of Ac7077/W22 heterozygotes, so differential recombination in intergenic regions also contributes to the overall variation in recombination rates observed in maize. Coupled with the differences observed in the bz1-stc1 segment, this comparison highlights the fact that haplotypic structural variability will strongly affect the frequency and distribution of recombination events in maize.
Materials and Methods
Genetic Lines.
All of the stocks used in this study carry bz1 haplotypes introgressed into the common genetic background of the inbred W22 and, except for the bz1-m2(D1) Ac7077 stock, have been described previously (18). The bronze1 alleles and the aleurone phenotypes of the various stocks are as follows. bz1-m2(D1) (bronze in the absence of Ac; spotted, in its presence) harbors a 3.3-kb Dissociation (Ds) element at position 755–762 in the second exon of the Bz1-McC allele (22, 38). bz1-m2(D1) Ac7077 (spotted): a version of bz1-m2(D1) carrying a transposed Ac element in the 5′ UTR of the distal tac7077 gene (19), at position 216489–216496 of GenBank accession no. AF391808. This stock was derived by recombining the bz1-m2(D1) allele, an McC haplotype derivative, with the nearby Ac7077 insertion present in a Bz1 haplotype of unknown origin. A 145-kb NotI BAC clone containing the entire bz1-m2(D1) Ac7077 region was isolated and sequenced as described below. bz1-sh1-X2 (bronze, shrunken): an X-ray-induced deletion of a large chromosomal fragment that includes the bz1 and sh1 loci (39) and, therefore, the entire bz1-tac7077 region under study. Bz1-W22 (purple): the normal allele of the color-converted version of the inbred W22 (40). Bz1-B73 (purple): the normal allele of the inbred B73 introgressed into W22 (18).
Selection and Characterization of bz1-Ac7077 Recombinants.
The mutations wx1 (waxy endosperm) and sh1 (shrunken endosperm), located ≈25 cM proximal and 2 cM distal to bz1, respectively, were used as flanking markers. The wx1-sh1 region exhibits high chiasma interference (24), so double cross-overs in the region are rare. The wx1 bz1-m2(D1) Ac7077 sh1/Wx1 Bz1-B73 + Sh1 and wx1 bz1-m2(D1) Ac7077 sh1/Wx1 Bz1-W22 + Sh1 heterozygotes were hand-pollinated with a wx1 bz1-sh1-X2 stock. Sh bz recombinants were selected as single seed with a plump, solid bronze phenotype in ears segregating spotted and purple seed. The selections were grown in the greenhouse, back-crossed to wx1 bz1-sh1-X2 to confirm heritability, and characterized molecularly as described below.
DNA Extraction, PCR, and Sequencing.
Leaf DNA from all selections was made by a CTAB extraction procedure and used for subsequent PCR amplification. PCR was performed by using Qiagen Taq polymerase and the primers listed in Table S3. The PCR products were run on either 1% agarose gels or 8% polyacrylamide gel based on their size and the polymorphisms to be discriminated. For sequencing, PCR products were purified by isopropanol precipitation and 70% ethanol washing. The PCR amplification primers were also used to directly sequence purified PCR products by using ABI BigDye Terminator V3.1 reagent (Applied Biosystems). DNA sequencing was carried out in an ABI 3730xl DNA analyzer.
BAC Isolation and Sequencing.
NotI BAC clones of the bz genomic region of the bz1-m2(D1) Ac7077 stock were isolated as previously described (20). The BAC clones were sequenced by the shotgun sequencing strategy, assembled, analyzed, and annotated as described (9). The GenBank accession no. for the Ac7077 haplotype BAC sequence is FJ624873.
Southern Blot Analysis.
Restriction-digested genomic DNA (10 μg) was resolved in 0.8% agarose gels and transferred to Hybond XL nylon membranes (GE Healthcare). The membranes were hybridized with random-primer-labeled 32P probes from various genes. Conditions for hybridization, high stringency washing, and exposure to X-ray film were standard.
Methylation Analysis by Bisulfite Sequencing.
Bisulfite treatment of DNA was performed with the EZ DNA Methylation kit (Zymo Research). Purified DNA was used for PCR amplification by primers designed through the MethPrimer program (University of California, San Francisco). The PCR product was then cleaned and directly sequenced with the same primer in an ABI 3730xl DNA analyzer.
Supplementary Material
Acknowledgments.
We thank Chunguang Du and Changfa Lin for sharing unpublished data; Remy Bruggman for masking repetitive sequences that aided in the identification of new MITEs; Krystyna Dooner for assistance in the genetic experiments; Marc Probasco for greenhouse plant care; Jo Messing for access to the ABI-3730xl DNA analyzer; and Jun Huang, Yubin Li, and Qinghua Wang for valuable comments on the manuscript. This work was supported by National Science Foundation Grant MCB 05-23103 (to H.K.D.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The Ac7077 haplotype sequence reported in this paper has been deposited in the GenBank database (accession no. FJ624873).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902972106/DCSupplemental.
References
- 1.Dooner HK, et al. A molecular genetic analysis of insertion mutations in the bronze locus in maize. Mol Gen Genet. 1985;200:240–246. [Google Scholar]
- 2.Wessler S, Varagona R. Molecular basis of mutations at the waxy locus of maize: Correlation with the fine structure genetic map. Proc Natl Acad Sci USA. 1985;82:4177–4181. doi: 10.1073/pnas.82.12.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dooner HK, Hsia A-P, Schnable PS. In: Handbook of Maize: Domestication, Genetics, and Genome. Bennetzen JL, Hake SC, editors. New York: Springer Science; 2009. pp. 377–403. [Google Scholar]
- 4.Fu H, Zheng Z, Dooner HK. Recombination rates between adjacent genic and retrotransposon regions differ by two orders of magnitude. Proc Natl Acad Sci USA. 2002;99:1082–1087. doi: 10.1073/pnas.022635499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao H, et al. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc Natl Acad Sci USA. 2002;99:6157–6162. doi: 10.1073/pnas.082562199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu H, Dooner HK. Intraspecific violation of genetic colinearity and its implications in maize. Proc Natl Acad Sci USA. 2002;99:9573–9578. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song R, Messing J. Gene expression of a gene family in maize based on noncollinear haplotypes. Proc Natl Acad Sci USA. 2003;100:9055–9060. doi: 10.1073/pnas.1032999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell. 2005;17:343–360. doi: 10.1105/tpc.104.025627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Dooner HK. Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc Natl Acad Sci USA. 2006;103:17644–17649. doi: 10.1073/pnas.0603080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.SanMiguel P, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 11.Feschotte C, Zhang Y, Wessler SR. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Am Soc Microbiol Press: Washington, DC; 2002. pp. 1147–1158. [Google Scholar]
- 12.Lai J, Li Y, Messing J, Dooner HK. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA. 2005;102:9068–9073. doi: 10.1073/pnas.0502923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgante M, et al. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 14.Beavis WD, Grant D. A linkage map based on information from four F2 populations of maize. (Zea mays L) Theor Appl Genet. 1991;82:636–644. doi: 10.1007/BF00226803. [DOI] [PubMed] [Google Scholar]
- 15.Williams CG, Goodman MM, Stuber CW. Comparative recombination distances among Zea mays L. inbreds, wide crosses and interspecific hybrids. Genetics. 1995;141:1573–1581. doi: 10.1093/genetics/141.4.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fatmi A, Poneleit CG, Pfeiffer TW. Variability of recombination frequencies in the Iowa Stiff Stalk Synthetic (Zea mays L. ) Theor Appl Genet. 1993;86:859–866. doi: 10.1007/BF00212613. [DOI] [PubMed] [Google Scholar]
- 17.Yao H, Schnable PS. Cis-effects on meiotic recombination across distinct a1-sh2 intervals in a common Zea genetic background. Genetics. 2005;170:1929–1944. doi: 10.1534/genetics.104.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dooner HK, He L. Maize genome structure variation: Interplay between retrotransposon polymorphisms and genic recombination. Plant Cell. 2008;20:249–258. doi: 10.1105/tpc.107.057596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowperthwaite M, et al. Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell. 2002;14:713–726. doi: 10.1105/tpc.010468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu H, Dooner HK. A gene enriched BAC library for cloning large allele-specific fragments from maize: Isolation of a 240-kb contig of the bronze region. Genome Res. 2000;10:866–873. doi: 10.1101/gr.10.6.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, et al. P instability factor: An active maize transposon system associated with the amplification of Tourist-like MITEs and a new superfamily of transposases. Proc Natl Acad Sci USA. 2001;98:12572–12577. doi: 10.1073/pnas.211442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClintock B. Topographical relations between elements of control systems in maize. Carnegie Inst Wash Yrbk. 1962;61:448–461. [Google Scholar]
- 23.Mottinger JP. The effects of X rays on the bronze and shrunken loci in maize. Genetics. 1970;64:259–271. doi: 10.1093/genetics/64.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dooner HK. Genetic fine structure of the bronze locus in maize. Genetics. 1986;113:1021–1036. doi: 10.1093/genetics/113.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lichten M, Goldman AS. Meiotic recombination hotspots. Annu Rev Genet. 1995;29:445–476. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 26.Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 27.Eggleston WB, Alleman M, Kermicle JL. Molecular organization and germinal instability of R-stippled maize. Genetics. 1995;141:347–360. doi: 10.1093/genetics/141.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timmermans MC, Das OP, Messing J. Characterization of a meiotic crossover in maize identified by a restriction fragment length polymorphism-based method. Genetics. 1996;143:1771–1783. doi: 10.1093/genetics/143.4.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennetzen JL, Schrick K, Springer PS, Brown WE, SanMiguel P. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- 30.Rabinowicz PD, et al. Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat Genet. 1999;23:305–308. doi: 10.1038/15479. [DOI] [PubMed] [Google Scholar]
- 31.Stam M, et al. The regulatory regions required for B′ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics. 2002;162:917–930. doi: 10.1093/genetics/162.2.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobsen SE, Sakai H, Finnegan EJ, Cao X, Meyerowitz EM. Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr Biol. 2000;10:179–186. doi: 10.1016/s0960-9822(00)00324-9. [DOI] [PubMed] [Google Scholar]
- 33.Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci USA. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lal SK, Giroux MJ, Brendel V, Vallejos CE, Hannah LC. The maize genome contains a Helitron insertion. Plant Cell. 2003;15:381–391. doi: 10.1105/tpc.008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta S, Gallavotti A, Stryker GA, Schmidt RJ, Lal SK. A novel class of Helitron-related transposable elements in maize contain portions of multiple pseudogenes. Plant Mol Biol. 2005;57:115–127. doi: 10.1007/s11103-004-6636-z. [DOI] [PubMed] [Google Scholar]
- 36.Du C, Caronna J, He L, Dooner HK. Computational prediction and molecular confirmation of Helitron transposons in the maize genome. BMC Genomics. 2008;9:51. doi: 10.1186/1471-2164-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ananiev EV, Phillips RL, Rines HW. Complex structure of knob DNA on maize chromosome 9. Retrotransposon invasion into heterochromatin. Genetics. 1998;149:2025–2037. doi: 10.1093/genetics/149.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dooner HK, English J, Ralston E, Weck E. A single genetic unit specifies two transposition functions in the maize element Activator. Science. 1986;234:210–211. doi: 10.1126/science.234.4773.210. [DOI] [PubMed] [Google Scholar]
- 39.Mottinger J. Unstable mutants of bronze induced by premeiotic X-ray treatment in maize. Theor Appl Genet. 1973;43:190–195. doi: 10.1007/BF00306570. [DOI] [PubMed] [Google Scholar]
- 40.Ralston EJ, English J, Dooner HK. Sequence of three bronze alleles of maize and correlation with the genetic fine structure. Genetics. 1988;119:185–197. doi: 10.1093/genetics/119.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.