Abstract
The chloride ion, Cl−, is an essential cofactor for oxygen evolution of photosystem II (PSII) and is closely associated with the Mn4Ca cluster. Its detailed location and function have not been identified, however. We substituted Cl− with a bromide ion (Br−) or an iodide ion (I−) in PSII and analyzed the crystal structures of PSII with Br− and I− substitutions. Substitution of Cl− with Br− did not inhibit oxygen evolution, whereas substitution of Cl− with I− completely inhibited oxygen evolution, indicating the efficient replacement of Cl− by I−. PSII with Br− and I− substitutions were crystallized, and their structures were analyzed. The results showed that there are 2 anion-binding sites in each PSII monomer; they are located on 2 sides of the Mn4Ca cluster at equal distances from the metal cluster. Anion-binding site 1 is close to the main chain of D1-Glu-333, and site 2 is close to the main chain of CP43-Glu-354; these 2 residues are coordinated directly with the Mn4Ca cluster. In addition, site 1 is located in the entrance of a proton exit channel. These results indicate that these 2 Cl− anions are required to maintain the coordination structure of the Mn4Ca cluster as well as the proposed proton channel, thereby keeping the oxygen-evolving complex fully active.
Keywords: membrane proteins, oxygen evolution, photosynthesis, manganese enzyme
Photosynthetic water oxidation produces the oxygen required for life on the earth and is catalyzed by Photosystem II (PSII), a multiprotein complex with a total molecular mass of 350 kDa (1). The catalytic center for water splitting, i.e., the oxygen-evolving complex (OEC), is located in the membrane surface of the luminal side of thylakoid membranes and is composed of 4 Mn atoms and 1 Ca atom in the protein matrix of PSII. Oxygen is produced from 2 molecules of water by sequential, light-induced electron-abstracting events at the Mn4Ca cluster that cycle through the Si states with i = 0–4 (2). One or more Cl− ions are required for the water oxidation cycle to proceed, a requirement not often found in other biological systems (3, 4). Depletion of Cl− has been shown to inhibit the S2 → S3 and S3 → S0 transitions (5). The roles of Cl− proposed for OEC include ligation to Mn or Ca atoms (6–8), regulation of the redox potential of the Mn4Ca cluster (9), maintaining a hydrogen bond network (10), and activation of the substrate water (11). In addition, an association of Cl− with amino acid residues in the Mn coordination shell has been proposed (12, 13).
The structure of PSII, including the Mn4Ca cluster, has been analyzed by x-ray crystallography (14–17). These studies, together with polarized extended x-ray absorption fine structure (EXAFS) measurements (18), have provided much information about the structure of the Mn4Ca cluster. However, they did not provide any information about the number and location of Cl− ions within the OEC. The inhibition of particular S-state transitions upon depletion of Cl− has been taken as evidence of the close proximity of Cl− to the Mn4Ca cluster, a proximity that also was suggested by studies of electron spin echo envelope modulation (19, 20) and Fourier transform infrared spectroscopic studies (21). Recent EXAFS studies, however, suggested that Cl− ions do not bind to the Mn4Ca cluster in its S1 state, and that no Cl− is present within a distance of 5 Å from the metal center (13). Thus, the exact location of Cl− ions in PSII remains to be resolved.
The stoichiometry of Cl− associated with PSII is unclear also because of the difficulty of directly detecting Cl−. Lindberg et al. have demonstrated that PSII membranes isolated from spinach grown on a medium containing 36Cl− retained about 1 Cl−/PSII (22). They also proposed that this Cl− site can be either high affinity (Kd = 20 μM) and slowly exchanging (t1/2 = 1 h) or low affinity (Kd = 0.5 mM) and rapidly exchanging (t1/2 < 15 sec) (23). Even in the Cl−-depleted condition, however, the oxygen-evolving activity of PSII membranes reported in these studies remained at 35% of the control level. This result raises the possibility that the Cl− ions associated with PSII were not completely removed from the samples under the conditions used.
The function of Cl− in the OEC can be partially substituted by other anions, with the efficiency being Cl−≈ Br− ≫ NO3−> NO2−> I− (3, 24). This phenomenon can be used to study the Cl− binding in the OEC, and indeed most previous studies have substituted Br− or other anions for Cl− to investigate its binding and function in the OEC. The substitution of Cl− by Br− and I− is particularly useful in studying the coordination structure of Cl− in OEC by x-ray crystallography, because Br− has a K-edge absorption wavelength of 0.92 Å, and I− has significant effects of anomalous dispersion around this wavelength that fall into the x-ray wavelength routinely used for structural studies. Indeed, in a recent study using x-ray crystallography, Murray et al. (25) substituted Br− for Cl− in PSII and identified 2 anion-binding sites in the vicinity of the OEC. In the present study, we substituted Br− or I− for Cl−, using purified PSII dimer from a thermophilic cyanobacterium, Thermosynechococcus vulcanus, and analyzed the crystal structure of the substituted PSII at higher resolutions. Both the difference Fourier maps between native PSII (Cl−-PSII) and PSII crystals with Br− or I− substitutions and the anomalous Fourier maps of PSII with Br− and I− substitutions revealed the presence of 2 anion-binding sites in the vicinity of the Mn4Ca cluster. Because oxygen evolution was inhibited completely by substitution of Cl− with I− and was recovered on re-substitution of I− by either Cl− or Br−, our results indicate that these 2 anion-binding sites represent the Cl−-binding sites in native PSII.
Results
Substitution of Cl− by Br− and I−.
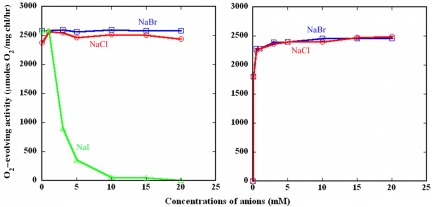
Cl− in the PSII core dimer was replaced by Br− or I− by a washing procedure with solutions containing either Br− or I− instead of Cl− (See Materials and Methods.) The resulting samples were analyzed for their oxygen-evolving activities in the presence of either Br− for the Br−-substituted PSII or I− for the I−-substituted PSII. Fig. 1 shows that, although PSII showed 90% of the activity when measured in a Cl−-free medium because of Cl−contamination of the medium (which was estimated to be ≈30–50 μM), and Br− substitution did not affect the oxygen-evolving activity significantly, I− substitution affected the activity remarkably: the activity was similar to Cl−-PSII at I− concentrations lower than 1 mM but decreased rapidly as the concentration of I− in the measuring medium increased and was inhibited completely in the presence of 15–20 mM I−. This finding is in agreement with previous reports (12) and indicates that the Cl− associated with PSII is replaced completely by I− in the presence of 15–20 mM I−. The activity of PSII with I− substitutions was recovered by washing the I−-substituted PSII with a I−-free medium and supplementing the measuring medium with either Cl− or Br−. A recovery of 70% activity was achieved even in the presence of less than 0.1 mM Cl− or Br− in the measuring buffer, and a full recovery of activity was achieved by 1–3 mM of either of the 2 anions in the concentration. These results indicate that the association of I− with PSII is reversible, and both Cl− and Br− can replace I− to support PSII oxygen evolution with a high affinity. The recovery of activity by simply supplying Cl− or Br− in the measuring medium also indicates that no reduction or release of the Mn atoms occurs upon I− substitution and that I− is inhibited by its reversible binding to the Cl−-binding site. Cl− and Br− were equally effective in recovering the oxygen-evolving activity, in agreement with the previous and present finding that Br− can support oxygen evolution as effectively as Cl−. This finding indicates that Cl−, Br−, and I− bind to the same functional site in PSII.
Fig. 1.
Effects of Br− substitution or I− substitution and Cl− or Br− reconstitution on oxygen evolution of a PSII dimer from T. vulcanus. (Left) Effects of Br− or I− substitution on oxygen evolution measured in the presence of respective anions. O, Cl−-containing PSII; □, PSII with Br− substitution; Δ, PSII with I− substitution. (Right) Recovery of oxygen evolution of PSII with I− substitution by Cl− (O) and Br− (□).
Structural Analysis of PSII with Br− and I− Substitutions.
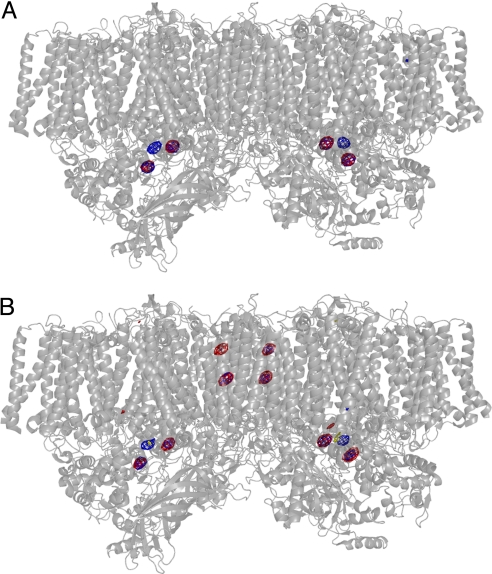
PSII with Br− or I− substitutions was crystallized in the presence of a total concentration of 40 mM of each anion (20 mM NaBr or NaI plus 10 mM CaBr2 or CaI2, see Materials and Methods), ensuring a complete substitution of Cl− by Br− or I−. The crystallization conditions for PSII with Br− or I− substitutions were similar to those for native PSII (Cl−-PSII) (15, 26), and the resulting PSII crystals with Br− and I− substitutions belonged to the same space group and had similar unit cell constants as the native PSII (Cl−-PSII) crystals (Table 1). Diffraction data from PSII with Br− substitutions had a resolution of 3.7 Å, and diffraction data from PSII with I−substitutions had a resolution of 4.0 Å (Table 1). Using the phase information calculated from the PSII structure without Br− or I−, we were able to calculate the difference Fourier maps between Cl−-PSII and PSII with Br− substitution and between Cl−-PSII and PSII with I− substitution. Because the crystallographic data were collected at 0.9 Å for Br−-PSII crystals and at 1.0 Å for I−-PSII crystals, 2 wavelengths with significantly large contributions from the anomalous dispersion effects of Br− and I−, respectively, we also were able to calculate the anomalous Fourier maps of PSII with Br− and I− substitutions. Fig. 2A shows the difference Fourier map (in red) of PSII with Br− substitution minus Cl−-PSII and the anomalous Fourier map (in blue) of PSII with Br− substitution superimposed on the whole structure of PSII dimer. In the difference Fourier map, 2 regions of electron density were observed in the vicinity of the Mn4Ca cluster, suggesting the presence of new electron densities in the Br−-substituted PSII that were not in Cl−-PSII. These 2 regions completely overlapped 2 regions with strong electron density observed in the anomalous Fourier map of Br−, which is calculated from the anomalous contribution of the Br− atoms and thus reflects only the presence of Br−. These results indicate that 2 Br− ions are bound to PSII in the crystals with Br− substitutions. There was an additional region of electron density in the anomalous Fourier map; this density is attributed to the Mn4Ca cluster because of the large anomalous dispersion effect from the 4 Mn atoms in the cluster. On the other hand, no additional signals were observed in the difference Fourier map (Fig. 2A), indicating that no additional Br− was bound to any other sites.
Table 1.
X-ray crystallographic data of PSII crystals from wild-type and Br- and I-PSII
| Data collection | Native PSII-1a | Br-PSII | Native PSII-2a | I-PSII |
|---|---|---|---|---|
| X-ray source | BL41XU | BL41XU | BL41XU | BL41XU |
| Wavelength | 0.9 Å | 0.9 Å | 1.0 Å | 1.0 Å |
| Resolution (Å)b | 50–4.0 | 50–3.7 | 50–4.0 | 50–4.0 |
| Unique reflections | 73143 | 87773 | 68269 | 74602 |
| Rmerge (%)b | 8.2 (71.0) | 7.2 (69.6) | 8.2 (54.2) | 9.1 (71.5) |
| I/σ(I) a | 21.7 (1.8) | 28.8 (2.2) | 22.8 (2.0) | 23.1 (2.2) |
| Completenessb | 94.7 (86.2) | 91.7 (82.6) | 91.7 (77.4) | 99.0 (99.3) |
| Space group | P212121 | P212121 | P212121 | P212121 |
| Unit cell dimensions (Å) | ||||
| a | 130.0 | 129.6 | 128.6 | 128.5 |
| b | 225.6 | 224.6 | 225.7 | 224.7 |
| c | 307.2 | 302.9 | 304.5 | 304.9 |
aNative PSII-1 is the crystal used to calculate the difference Fourier map between Cl− PSII and Br−-substituted PSII. Native PSII-2 is the crystal used to calculate the difference Fourier map between Cl− PSII and I−-substituted PSII.
bNumbers in parenthesis represent the highest-resolution shell.
Fig. 2.
Difference Fourier maps and anomalous Fourier maps of PSII with Br− or I− substitution, superimposed on the structure of PSII dimer. (A) The difference Fourier map of PSII with Br− substitution minus Cl−-PSII and the anomalous Fourier map of PSII with Br− substitution measured at 0.9 Å were superimposed on the PSII dimer structure at 3.0 Å resolution (17) at a view perpendicular to the normal of the membrane plane. Red indicates the difference Fourier map contoured at σ = 6.0; blue: indicates the anomalous Fourier map contoured at σ = 6.0. (B) The difference Fourier map of PSII with I− substitution minus Cl−-PSII and the anomalous Fourier map of PSII with I− substitution measured at 1.0 Å superimposed on the PSII dimer structure at 3.0 Å resolution. Red indicates the difference Fourier map contoured at σ = 6.0; blue indicates the anomalous Fourier map contoured at σ = 5.0.
Fig. 2B shows the difference Fourier map (in red) of PSII with I− substitutions minus Cl−-PSII and the anomalous Fourier map (in blue) of PSII with I− substitutions. There also were 2 regions of electron density in the vicinity of the Mn4Ca cluster in the difference Fourier map that overlapped the 2 regions of electron density observed in the anomalous Fourier map; these 2 regions thus are assigned to the 2 I− ions in the PSII that had I− substitutions (Fig. 2B). The contribution from the Mn4Ca cluster also was observed in the anomalous Fourier map. In addition, there were 3 additional regions of positive signals (1 of which is difficult to see here because of its weak electron density, as discussed later) in both the difference Fourier map and the anomalous Fourier map of PSII with I− substitution (Fig. 2B), indicating the binding of I− to these 3 additional sites, whose detailed positions are described later.
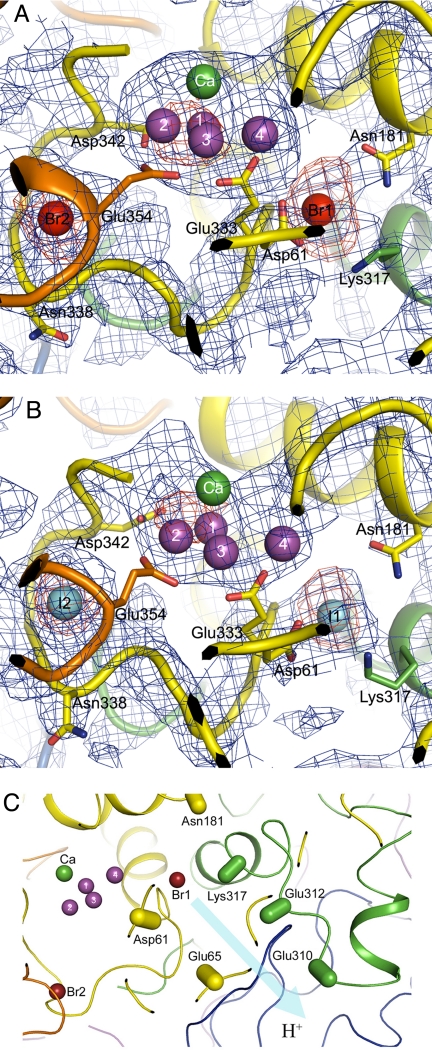
The structures of PSII with Br−and I− substitution were analyzed by the molecular replacement method using the PSII structure (PDB code:2AXT) (17) as the search model and were refined to a Rcryst/Rfree of 0.311/0.363 for PSII with Br− substitutions and a Rcryst/Rfree of 0.289/0.325 for PSII with I− substitutions (Table 2). These R factors can be considered reasonable at the present resolution. Fig. 3 A and B shows enlarged views of the structure around the 2 Br− and I− binding sites, which were superimposed on the B-factor sharpened, composite omit 2Fo-Fc electron density map, together with the anomalous electron density maps attributed to Br− and I−, respectively. The positions of the 2 anion-binding sites were similar for Br− and I−. These 2 halides were separated by ≈14 Å, and both are ≈7 Å from the nearest Mn atom (Table 3). The distance from these 2 sites to the Ca atom is similar also (≈10 Å). Because substitution of Cl− by I− completely inhibits oxygen evolution, and because Br− supports oxygen evolution as effectively as Cl−, we conclude that these 2 sites represent the functional Cl−-binding sites in native PSII.
Table 2.
Refinement statistics of Br−-substituted and I−-substituted PSIIA
| Refinement | Br−-PSII | I−-PSII |
|---|---|---|
| Resolution (Å) | 34.7–3.7 | 20–4.0 |
| Rcryst /Rfree a | 0.311/ 0.363 | 0.289/ 0.325 |
| No. of chainsb | 40 | 40 |
| No. of residuesb | 5,404 | 5,404 |
| No. of prosthetic moleculesb | 142 | 142 |
| Average B-factor (Å2) | ||
| Overall structure | 171.3 | 168.5 |
| Br1/Br2 or I1/I2 | 152.8/182.3 | 158.8/156.9 |
| Rms deviations | ||
| Bond angles (°) | 3.6 | 3.7 |
| Bond lengths (Å) | 0.033 | 0.033 |
aRcryst=Σ||Fobs|-|Fcalc||/Σ|Fobs|. Rfree is the cross-validation of the R-factor for the test-set reflections omitted in refinements.
bTotal molecule counts for 2 PSII complexes in an asymmetric unit.
Fig. 3.
Location of the 2 anion-binding sites in PSII. (A) Composite omit Fo-Fc map (blue) and anomalous map (red) of PSII with Br− substitution contoured at σ = 1.0 and 4.0, respectively, superimposed on the structure of the Mn4Ca cluster and its surrounding regions. The structure of PSII with Br−substitution was shown by the molecular replacement method with the structure of the PDB code 2AXT as the search model. Color codes for the residues are as follows: yellow, D1; green, D2; orange, CP43; purple, Mn atoms. (B) Composite omit Fo-Fc map (blue) and anomalous map (red) of I−-substituted PSII contoured at σ = 1.0 and 4.0, respectively, superimposed on the structure of the Mn4Ca cluster and its surrounding regions. The structure of PSII with I− substitution was shown by the molecular replacement method. The color codes for the residues are the same as in A. (C) Location of Br1 relative to the proton exit channel proposed in (16, 27–29). The residues of PsbO are drawn in blue, and the residues of other subunits are in the same colors as in A. The directions of atoms from Cα to Cβ in the residues are indicated by capsule-shaped objects.
Table 3.
Distances of the 2 anion-binding sites from the Mn4Ca-cluster. The number of Mn atoms corresponds to those labeled in Fig. 3
| Distances (Å) | |
|---|---|
| Mn4-Br1 | 6.4 |
| Mn4-I1 | 7.4 |
| Mn2-Br2 | 7.3 |
| Mn2-I2 | 6.5 |
| Br1-Br2 | 14.6 |
| I1-I2 | 13.9 |
| Ca-Br1 | 9.4 |
| Ca-I1 | 10.1 |
| Ca-Br2 | 11.1 |
| Ca-I2 | 9.6 |
Anion-binding site 1 (Br1) is surrounded by D2-Lys-317, D1-Asn-181, and D1-Glu-333, with distances of ≈3 Å to the side chain nitrogen of D2-Lys-317, ≈4.2 Å to the side chain nitrogen of D1-Asn-181, and 3.5 Å to the main chain nitrogen of D1-Glu-333. Thus, Br1 (and I1) may be coordinated by these residues. Among these residues, the side chain of D1-Glu-333 is known to coordinate a Mn atom in the Mn4Ca cluster. D1-Asp-61 also seems to be located near Br1, but its side chain oxygen is 5.8 Å from Br1, suggesting that it may not provide a ligand directly to the anion-binding site but may provide a second coordination shell to this site. The Br2 binding site is surrounded by CP43-Glu-354 and D1-Asn-338, with distances of 3.6 Å to the main chain nitrogen of CP43-Glu-354 and 3.2 Å to the main chain nitrogen of D1-Asn-338, suggesting that the Br2 (and I2) may be coordinated by these 2 residues. The side chain of CP43-Glu-354 is known to coordinate the Mn4Ca cluster. The main chain nitrogen of D1-Asp-342 is 6.7 Å apart from Br2 (I2), suggesting that it may not be involved directly in the coordination of Br2 (I2) but may constitute the second coordination shell. The 2 Cl atoms are relatively far from to their first coordination shell, suggesting that the 2 anions may be coordinated by the surrounding amino acid residues through aqua-ion or hydrogen bonding.
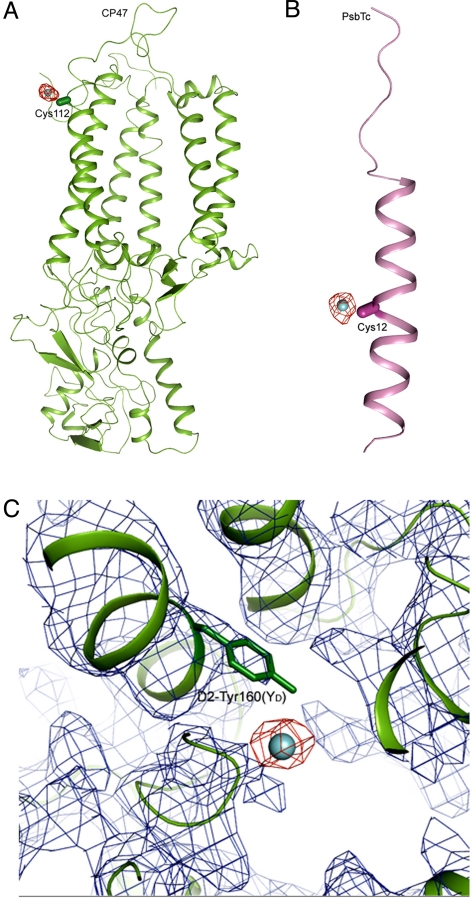
Additional sites found to bind I− in I−-substituted PSII were identified as CP47-Cys-112, PsbTc-Cys-12, and D2-Tyr-160 (YD) (Fig. 2B and Fig. 4). The electron density of I− associated with D2-Tyr-160 was weaker than at the other 2 sites. The distances between I− and CP47-Cys-112 and between I− and PsbTc-Cys-12 are rather short, suggesting a direct binding of I− to the side chain of these 2 Cys residues. These sites may not be involved in oxygen evolution and thus may not be relevant to the functional Cl−-binding sites (see Discussion).
Fig. 4.
Additional binding sites of I− in I−-substituted PSII. (A) Fo-Fc omit map of PSII with I− substitution (red) contoured at σ = 3.0, superimposed on the structure of CP47 (green) obtained by the molecular replacement method, showing the association of I− with CP47-Cys-112. (B) Fo-Fc omit map of PSII with I− substitution (red) contoured at σ = 3.0, superimposed on the structure of PsbTc (pink), showing the association of I− with PsbTc-Cys-12. The capsule-shaped objects show the direction of atoms from Cα to Cβ in the 2 Cys residues. (C) 2Fo-Fc map (blue, σ = 1.0) and omit Fo-Fc map (red, σ = 2.0) of PSII with I− substitution around the region of D2-Tyr-160(YD), together with the structure of D2 (green) in this region.
Discussion
We demonstrated here that there are 2 anion-binding sites on either side of the Mn4Ca cluster at a similar distance from the nearest Mn atom and from the Ca atom. We conclude that these 2 sites bind the functional Cl− ions required for oxygen evolution because substitution by I− completely abolished oxygen evolution and the inhibited activity could be recovered simply by replacing I− with Cl− or Br−. These results indicate that in our PSII functional Cl− was replaced effectively by Br− and I−. The 2 sites revealed here are consistent with a very recent report showing 2 Br-binding sites around the Mn4Ca cluster (25). Our structural analysis was performed at a higher resolution (i.e., 3.7 Å−4.0 Å vs. 4.21−4.45 Å). We also used I− as a substitute for Cl−, and this substitution inhibited oxygen evolution completely. This finding allowed us to correlate these binding sites directly to oxygen evolution.
Because of possible radiation damage caused by the X-rays used, we cannot completely exclude the direct binding of Cl− to the Mn4Ca cluster. However, the 2 Cl−-binding sites identified here have significant functional consequences. The similar locations of these 2 Cl−-binding sites and their closeness to the Mn4Ca cluster suggest that they both may be important for the normal function of the OEC. This importance is supported by the fact that the 2 anion-binding sites are surrounded by residues that either are coordinated directly with the Mn4Ca cluster or are in close proximity to the cluster. Because 1 of the residues coordinated with Cl2 is the main chain of CP43-Glu-354, and its side chain is known to coordinate the Mn4Ca cluster, Cl− may be required to maintain the structure needed for the correct coordination of this residue with the Mn4Ca cluster. The removal of Cl− from this site or the replacement of Cl− by I− may affect the effective coordination of this residue to Mn. A similar situation is found in the Cl1-binding site, because 1 of the residues coordinated with this site is D1-Glu-333, whose side chain is coordinated with the Mn4Ca cluster. Furthermore, recent studies have identified a possible proton exit channel from the Mn4Ca cluster that involves D2-Lys-317, D1-Asp-61 as well as D1-Asn-181 (16, 27–29). Cl1 is located in the entrance of the proton exit channel from the metal cluster (Fig. 3C). Cl or Br may be required to maintain the structure of this channel, and the binding of I− instead of Cl− or Br− at this site may block the proton exit pathway either because of a larger ion radius (2.20 Å for I− vs. 1.81 Å for Cl− or 1.96 Å for Br−) or because of a protein conformational change induced by the binding of the heavier I− anion. Alternatively, because I− is heavier than Cl−, binding of I− at this site might lower the flexibility of the protein matrix. That flexibility might be required for conformational changes that are necessary for the functional S-state transitions to occur.
The distance of the 2 anion-binding sites from the Mn4Ca cluster are consistent with a recent EXAFS study that found that Cl− is not present within 5 Å of the Mn4Ca cluster (13), suggesting that a direct interaction of the anion with the metal cluster is rather unlikely. Also, because these anions are rather far from the nearby residues, no covalent bond is likely to exist for their coordination. This evidence suggests that Cl− is bound rather weakly to the PSII, which explains the easy release of Cl− from binding sites in the PSII and the easy substitution of Cl− by Br− or I−, and vice versa. This weak bonding may represent a normal feature of halide binding in proteins.
The 3 additional I−-binding sites found here can be excluded for binding of functional Cl− for several reasons. First, Br− did not bind to these sites in Br−-substituted PSII. Because Br− can fully support oxygen evolution, this lack of binding indicates that functional Cl− cannot be related with these residues. Second, all 3 sites are distant from the Mn4Ca cluster, making a direct interaction of these residues with the OEC unlikely. Finally, oxygen evolution is still active even in the deletion mutant of cyanobacteria lacking the PsbTc subunit (30) and in site-directed mutants of YD in which binding of Cl− would become impossible if these residues do coordinate Cl− in native PSII. In fact, the association of I− with YD is consistent with previous results that YD is iodinated in the dark (31). The weak electron density of I− associated with YD may reflect the fact that the crystals were grown in the dark for several days, during which time part of YD+ would have decayed to YD, resulting in a partial iodination of this residue. Iodination of another tyrosine residue, YZ, under illumination also was reported previously (31, 32). However, because the crystals were grown in the complete darkness, we did not observe electron density attributable to I− at the YZ site.
The observation of the 3 additional I−-binding sites not relevant to oxygen evolution suggests that the substitution of Cl− by I− in our experimental conditions was rather complete and reinforces our conclusion that the 2 anion-binding sites surrounding the Mn4Ca cluster identified here represent the true Cl−-binding sites in native PSII.
Materials and Methods
Purification and Crystallization of PSII.
PSII dimer was purified from the thermophilic cyanobacterium Thermosynechococcus vulcanus according to methods described by Shen et al. (26, 33) and was suspended in a medium containing 20 mM MES (pH 6.0), 20 mM NaCl, and 3 mM CaCl2 (medium A). For substitution of Cl− by Br− or I−, the PSII samples were diluted to a concentration of 1 mg chlorophyll (chl)/mL with a medium containing 20 mM MES (pH 6.0), 20 mM NaBr, and 10 mM CaBr2 (medium B, for Br substitution) or 20 mM MES (pH 6.0), 20 mM NaI, and 10 mM CaI2 (medium C, for I substitution), mixed with the same volume of precipitation buffer (containing 26% PEG 1450 in medium B or C), and incubated for 30 min on ice in the dark. After incubation, PSII was precipitated by centrifugation at 15,000 × g for 15 min, washed again with medium B or C, and finally suspended in either medium B or C at a concentration of 4.0 mg chl/mL.
Crystallization of PSII dimer was performed using the hanging drop vapor diffusion method. PSII samples were mixed with the same volume of crystallization buffer containing 20 mM MES (pH 6.0), 26% glycerol, 7% to 10% PEG 1450, 80 mM MgSO4, 0.02% n-dodecyl-β-D-maltoside, 20 mM NaCl, and 10 mM CaCl2 for native PSII (Cl-PSII) (26). For PSII with Br or I substitution, 20 mM NaCl and 10 mM CaCl2 in the crystallization buffer were replaced by 20 mM NaBr and 10 mM CaBr2 or by 20 mM NaI and 10 mM CaI2, respectively. Crystallization was carried out in complete darkness at 20 °C. Crystals grew to a full size of 1.0 × 0.5 × 0.1 mm in 3 to 5 days, and the crystals were harvested and transferred to a cryoprotectant solution as described previously (15, 26), with NaBr and CaBr2 or with NaI and CaI2, replacing NaCl and CaCl2, for Br-PSII and I-PSII, respectively. The crystals were flash-cooled in a nitrogen gas stream at 100 K.
X-Ray Diffraction Measurements and Structure Analysis.
X-ray diffraction experiments were performed at BL41XU of SPring-8 (34) at 100K. The wavelengths used were 0.9 Å for Cl-PSII and Br-PSII and 1.0 Å for I-PSII. Diffraction patterns were recorded with a CCD detector Quantum 315 (ADSC) every 0.6° with an exposure time of 4 sec and an x-ray flux of 1.0–1.8 × 1011 photons/sec. The diffraction data were processed with HKL2000 (35).
The structures of PSII with Br− substitution and I− substitution were solved by the molecular replacement method using as a search model a PSII monomer that was generated from the 3.0-Å structure (PDB code: 2AXT) (17), with the program AMORE from CCP4 suite (36). The refinement processes were carried out using Program CNS version 1.2 (37). Rigid-body refinement was performed first to optimize the position of the 2 PSII monomers in the asymmetric unit; based on that refinement, the non-crystallographic symmetry (NCS) matrices were defined and used in the following refinement process. The initial model was improved using simulated annealing refinement to reduce the search-model bias. The structural models were modified according to the electron density map using the graphic program Coot (38). The locations of iodine and bromine atoms were defined using their anomalous Fourier maps. The Fo-Fc omit maps calculated from the refined PSII structures without Br or I atoms were generated with the program CNS version 1.2. The model geometry and quality statistics were calculated with the PROCHECK program (39).
Measurement of Oxygen-Evolving Activities.
Oxygen evolution was measured with a Clark-type oxygen electrode at 30 °C under continuous, saturating light with 0.5 mM phenyl-p-benzoquinone and 0.5 mM potassium ferricyanide as electron acceptors. To remove Cl− from PSII samples, the samples were diluted with a Cl−-free medium containing 20 mM MES (pH 6.0) and 5 mM CaSO4 (for supplying Ca2+ to PSII) at a concentration of 1 mg chl/mL, mixed with the same volume of precipitation buffer containing 20 mM MES (pH 6.0), 26% PEG 1450, and 5 mM CaSO4. After incubation for 30 min, the PSII samples were precipitated by centrifugation at 15,000 × g for 15 min and were re-suspended in the Cl−-free medium. This washing step was repeated, and the final PSII samples were suspended in 20 mM MES (pH 6.0) at 5 mM CaSO4 at a concentration of 0.5 mg chl/mL. For measuring the dependency of oxygen evolution on concentrations of Cl−, Br−, or I−, samples were incubated with NaCl, NaBr, or NaI at indicated concentrations for 30 min in the dark on ice, and the activity was measured in the presence of the same concentrations of NaCl, NaBr, or NaI. For measuring the recovery of oxygen evolution of PSII when I− was replaced by Cl− or Br−, the samples with I− substitution were washed twice with 20 mM MES (pH 6.0) and 5 mM CaSO4 and were suspended in the same medium at a concentration of 0.5 mg chl/mL. The samples were supplemented with NaCl or NaBr at indicated concentrations and were incubated in the dark for 30 min. Then oxygen evolution was measured in the presence of the same concentrations of NaCl or NaBr.
Acknowledgments.
We thank N. Shimizu and the staff at SPring-8 (BL41XU, BL44XU) for their assistance in using the beamlines. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (Structures of Biological Macromolecular Assemblies), a Grant-in-Aid for Creative Scientific Research, a GCOE program on Pico-biology, and a Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structures of Br-substituted and I-substituted PSII have been deposited with the PDB under accession numbers of 3A0B and 3A0H, respectively.
References
- 1.Shen JR, Henmi T, Kamiya N. In: Photosynthetic Protein Complexes, a Structural Approach. Fromme F, editor. Weinheim: Wiley-VCH; 2008. pp. 83–106. [Google Scholar]
- 2.Kok B, Forbush B, McGloin M. Cooperation of charges in photosynthetic oxygen evolution I. A linear four step mechanism. Photochem Photobiol. 1970;11:457–475. doi: 10.1111/j.1751-1097.1970.tb06017.x. [DOI] [PubMed] [Google Scholar]
- 3.van Gorkom HJ, Yocum CF. In: Photosystem II: The Light-Driven Water: Plastoquinone Oxidoreductase. Wydrzynski TJ, Satoh K, editors. Dordrecht: Springer; 2005. pp. 307–328. [Google Scholar]
- 4.Popelková H, Yocum CF. Current status of the role of Cl− ion in the oxygen-evolving complex. Photosynth Res. 2007;93:111–121. doi: 10.1007/s11120-006-9121-5. [DOI] [PubMed] [Google Scholar]
- 5.Wincencjusz H, van Gorkom HJ, Yocum CF. The photosynthetic oxygen evolving complex requires chloride for its redox state S2→S3 and S3→S0 transitions but not for S0→S1 or S1→S2 transitions. Biochemistry. 1997;36:3663–3670. doi: 10.1021/bi9626719. [DOI] [PubMed] [Google Scholar]
- 6.Yachandra VK, Sauer K, Klein MP. Manganese cluster in photosynthesis: Where plants oxidize water to dioxygen. Chem Rev (Washington, DC) 1996;96:2927–2950. doi: 10.1021/cr950052k. [DOI] [PubMed] [Google Scholar]
- 7.Vrettos JS, Limburg J, Brudvig GW. Mechanism of photosynthetic water oxidation: Combining biophysical studies of photosystem II with inorganic model chemistry. Biochim Biophys Acta. 2001;1503:229–245. doi: 10.1016/s0005-2728(00)00214-0. [DOI] [PubMed] [Google Scholar]
- 8.Britt RD, et al. Recent pulsed EPR studies of the Photosystem II oxygen-evolving complex: Implications as to water oxidation mechanisms. Biochim Biophys Acta. 2004;1655:158–171. doi: 10.1016/j.bbabio.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Boussac A, Rutherford AW. Electron transfer events in chloride-depleted photosystem II. J Biol Chem. 1994;269:12462–12467. [PubMed] [Google Scholar]
- 10.Olesen K, Anderasson LE. The function of the chloride ion in photosynthetic oxygen evolution. Biochemistry. 2003;42:2025–2035. doi: 10.1021/bi026175y. [DOI] [PubMed] [Google Scholar]
- 11.McEvoy JP, Brudvig GW. Structure-based mechanism of photosynthetic water oxidation. Physical Chemistry Chemical Physics. 2004;6:4754–4763. [Google Scholar]
- 12.Hasegawa K, Kimura Y, Ono TA. Chloride cofactor in the photosynthetic oxygen-evolving complex studied by Fourier transform infrared spectroscopy. Biochemistry. 2002;41:13839–13850. doi: 10.1021/bi026595n. [DOI] [PubMed] [Google Scholar]
- 13.Haumann M, et al. Bromide does not bind to the Mn4Ca complex in its S1 state in Cl−-depleted and Br−-reconstituted oxygen-evolving photosystem II: Evidence from X-ray absorption spectroscopy at the Br K-edge. Biochemistry. 2006;45:13101–13107. doi: 10.1021/bi061308r. [DOI] [PubMed] [Google Scholar]
- 14.Zouni A, et al. Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Angstrom resolution. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]
- 15.Kamiya N, Shen JR. Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7 Å resolution. Proc Natl Acad Sci USA. 2003;100:98–103. doi: 10.1073/pnas.0135651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 17.Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- 18.Yano J, et al. Where water is oxidized to dioxygen: Structure of the photosynthetic Mn4Ca cluster. Science. 2006;314:821–825. doi: 10.1126/science.1128186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clemens KL, Force DA, Britt RD. Acetate binding at the photosystem II oxygen evolving complex: An S(2)-state multiline signal ESEEM study. J Am Chem Soc. 2002;124:10921–10933. doi: 10.1021/ja012036c. [DOI] [PubMed] [Google Scholar]
- 20.Yu H, Aznar CP, Xu XZ, Britt RD. Evidence that azide occupies the chloride binding site near the manganese cluster in photosystem II. Biochemistry. 2005;44:12022–12029. doi: 10.1021/bi0505767. [DOI] [PubMed] [Google Scholar]
- 21.Cooper IB, Barry BA. Perturbations at the chloride site during the photosynthetic oxygen-evolving cycle. Photosynth Res. 2007;92:345–356. doi: 10.1007/s11120-007-9147-3. [DOI] [PubMed] [Google Scholar]
- 22.Lindberg K, Wydrzynski T, Vänngård T, Andréasson LE. Slow release of chloride from 36Cl-labeled photosystem II membranes. FEBS Lett. 1990;264:153–155. [Google Scholar]
- 23.Lindberg K, Andréasson LE. A one-site, two-state model for the binding of anions in photosystem II. Biochemistry. 1996;35:14259–14267. doi: 10.1021/bi961244s. [DOI] [PubMed] [Google Scholar]
- 24.Kelly PM, Izawa S. The role of chloride ion in Photosystem II I. Effects of chloride ion on Photosystem II electron transport and on hydroxylamine inhibition. Biochim Biophys Acta. 1978;502:198–210. doi: 10.1016/0005-2728(78)90042-7. [DOI] [PubMed] [Google Scholar]
- 25.Murray JW, et al. X-ray crystallography identifies two chloride binding sites in the oxygen evolving centre of Photosystem II. Energy and Environmental Science. 2008;1:161–166. [Google Scholar]
- 26.Shen JR, Kamiya N. Crystallization and the crystal properties of the oxygen-evolving photosystem II from Synechococcus vulcanus. Biochemistry. 2000;39:14739–14744. doi: 10.1021/bi001402m. [DOI] [PubMed] [Google Scholar]
- 27.Ishikita H, Saenger W, Loll B, Biesiadka J, Knapp EW. Energetics of a possible proton exit pathway for water oxidation in photosystem II. Biochemistry. 2006;45:2063–2071. doi: 10.1021/bi051615h. [DOI] [PubMed] [Google Scholar]
- 28.Murray JW, Barber J. Identification of a calcium-binding site in the PsbO protein of photosystem II. Biochemistry. 2006;45:4128–4130. doi: 10.1021/bi052503t. [DOI] [PubMed] [Google Scholar]
- 29.Murray JW, Barber J. Structural characteristics of channels and pathways in photosystem II including the identification of an oxygen channel. J Struct Biol. 2007;159:228–237. doi: 10.1016/j.jsb.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 30.Iwai M, Katoh H, Katayama M, Ikeuchi M. PSII-Tc protein plays an important role in dimerization of photosystem II. Plant Cell Physiol. 2004;45:1809–1816. doi: 10.1093/pcp/pch207. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi Y, Takahashi MA, Satoh K. Identification of the site of iodide photooxidation in the photosystem II reaction center complex. FEBS Lett. 1986;208:347–351. [Google Scholar]
- 32.Ikeuchi M, Koike H, Inoue Y. Iodination of D1 (herbicide-binding protein) is coupled with photooxidation of 125I− associated with Cl−-binding site in Photosystem-II water-oxidation system. Biochim Biophys Acta. 1988;932:160–169. [Google Scholar]
- 33.Shen JR, Inoue Y. Binding and functional properties of two new extrinsic components, cytochrome c-550 and a 12 kDa protein, in cyanobacterial photosystem II. Biochemistry. 1993;32:1825–1832. doi: 10.1021/bi00058a017. [DOI] [PubMed] [Google Scholar]
- 34.Kawamoto M, Kawano Y, Kamiya N. The bio-crystallography beamline (BL41XU) at SPring-8. Nuclear Instruments & Methods in Physics Research, SectionA. 2001;467–468:1375–1379. [Google Scholar]
- 35.Otwinowski Z, Minor M. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276(part A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Navaza J. AMoRe: An automated package for molecular replacement. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 37.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]