Abstract
Purple bacteria have thus far been considered to operate light-driven cyclic electron transfer chains containing ubiquinone (UQ) as liposoluble electron and proton carrier. We show that in the purple γ-proteobacterium Halorhodospira halophila, menaquinone-8 (MK-8) is the dominant quinone component and that it operates in the QB-site of the photosynthetic reaction center (RC). The redox potentials of the photooxidized pigment in the RC and of the Rieske center of the bc1 complex are significantly lower (Em = +270 mV and +110 mV, respectively) than those determined in other purple bacteria but resemble those determined for species containing MK as pool quinone. These results demonstrate that the photosynthetic cycle in H. halophila is based on MK and not on UQ. This finding together with the unusual organization of genes coding for the bc1 complex in H. halophila suggests a specific scenario for the evolutionary transition of bioenergetic chains from the low-potential menaquinones to higher-potential UQ in the proteobacterial phylum, most probably induced by rising levels of dioxygen 2.5 billion years ago. This transition appears to necessarily proceed through bioenergetic ambivalence of the respective organisms, that is, to work both on MK- and on UQ-pools. The establishment of the corresponding low- and high-potential chains was accompanied by duplication and redox optimization of the bc1 complex or at least of its crucial subunit oxidizing quinols from the pool, the Rieske protein. Evolutionary driving forces rationalizing the empirically observed redox tuning of the chain to the quinone pool are discussed.
Keywords: electron transport, evolution, photosynthesis
Chemiosmotic energy-converting mechanisms in all life on this planet are variations on a strikingly conserved theme. Electrons derived from reduced substrates enter a chain of membrane-integral and/or associated enzymes and are channeled on toward terminal electron-accepting substrates. Some of the free energy available during individual electron-transfer steps is stored in a transmembrane proton motive gradient. Mitchell (1) proposed a general mechanism for coupling electron transfer to proton translocation, which accounts for the majority of these systems. In this mechanism, electrons travel through 2 types of carrier “arms” back and forth across the energetic membrane. In an “electrogenic arm,” electrons move alone across the membrane from the positively to the negatively charged side, building up an electric field. Via a “neutral arm,” electrons travel in the opposite direction, along with protons. Placing electrogenic and neutral arms in series leads to net proton translocation across the membrane.
Without exception, the chemiosmotic neutral arms involve diffusion of small, lipophilic molecules across the energetic membrane. In all bioenergetic systems except methanogenesis, these molecules are quinones that diffuse in the lipid bilayer. The bulk of these diffusing quinones electrochemically connecting redox enzymes is called the quinone pool, to distinguish them from quinones present as fixed enzyme cofactors or having completely different metabolic functions. A plethora of chemical types of pool-quinones, such as ubi-, plasto-, mena-, rhodo-, caldariella- or sulfolobus-quinones (to cite only the best-studied cases) have been identified so far individually in different species or coexisting in single organisms (2–4).
Menaquinone (MK) is the most widely distributed quinone on the phylogenetic tree of species and, in particular, is the only quinone in early branching archaeal and bacterial phyla (5, 6). MK thus appears to represent the ancestral type of quinone in bioenergetic systems. Within this bulk of MK-species, isolated islands are distinguishable on the phylogenetic tree formed by species where MK has been complemented or substituted by other types of quinones such as plastoquinone (PQ) in the cyanobacterial/plastidic lineage, ubiquinone (UQ) in α/β/γ- proteobacteria and consequently in mitochondria, or caldariellaquinone (CQ) in the sulfolobales. These islands contain predominantly facultative or obligate aerobic organisms. Despite their strongly diverging chemical structures, all these “more recent” quinones feature identical redox midpoint potentials that are significantly more positive (by 150 mV) than that of MK. The fact that high-potential quinones (HPQ) have only appeared in species permanently or periodically exposed to high concentrations of oxygen suggests that high O2 tensions and/or the concomitant increase in ambient redox potential are evolutionarily unfavorable for organisms using MK-based energy-converting chains. Reduced MK indeed becomes rapidly and noncatalytically oxidized in the presence of O2, whereas the HPQH2 are relatively insensitive toward oxygen and thereby minimize the bioenergetically unproductive leakage of reducing equivalents to O2. These considerations notwithstanding, not all phyla containing aerobic species have abandoned MK as pool-quinones (7) such as, for example, Gram-positive bacteria or chloroflexaceae. Because of their lower reducing power, the HPQ would be poor electron donors to downstream enzymes of a MK-based bioenergetic chain. Compensating this loss in driving force, all redox cofactors in bioenergetic chains containing HPQ as pool-quinones, are found to have (again by ≈150 mV) more positive redox potentials than in their MK-based analogues (5, 7, 8). Bioenergetic systems in prokaryotes therefore either contain a MK-based “low-potential (LP) chain” or use UQ, PQ, or CQ within a “high-potential (HP) chain” in which all redox potentials are upshifted on average by 150 mV. Correspondingly, the Em-spans for individual electron-transfer steps are conserved between LP and HP chains.
The most plausible rationalization for the transition from LP to HP bioenergetic chains thus consists in a functional adaptation to the increasing oxidation state of the environment (5, 6) brought about by the photosynthetic injection of O2 into the biosphere ≈2.5 billion years ago. The substantial chemical differences between UQ, PQ, or CQ indicate that the respective transitions occurred independently in different parts of the phylogenetic tree. For the specific case of the MK/UQ transition in the proteobacterial lineage, the change in operating redox potential of bioenergetic chains must have happened after the divergence of the α/β/γ- from the earlier-branching δ- and ε-subgroups because the δ- and ε-subgroups both exclusively contain MK whereas α-, β- and γ-proteobacteria possess either UQ and MK or UQ only (2, 9, 10).
In line with the fact that UQ is the dominant quinone species in α/β/γ-proteobacteria, all electron transport chains characterized so far in representatives from these subgroups have been found to be of the HP type (8, 11). The only exception to this rule are the γ-proteobacterial enterobacteria such as Escherichia coli and closely related species that apparently are able to switch between UQ- and MK-based bioenergetic chains in response to varying growth conditions (12).
The overall shift in potentials of whole bioenergetic chains necessarily involves numerous independent mutational events and therefore represents a complex evolutionary transition, the proceedings of which are presently unknown. In this work, we describe the identification of a LP photosynthetic electron transfer chain based on a MK-pool in the purple γ-proteobacterium Halorhodospira halophila. The genomic organization, in this species, of the genes coding for the subunits of the cytochrome (cyt) bc1 complex, a key energy-coupling enzyme and the bacterial homologue of mitochondrial complex III, together with its quinone content suggest a scenario for the LP to HP transition in proteobacteria. The ensemble of data demonstrates that this transition necessarily involved the existence of ambivalent species containing both HP and LP chains.
Results
In purple bacterial photosynthesis, reducing equivalents produced by the light-induced charge separation within the photosynthetic reaction center (RC) are considered to be injected into the UQ pool, passed on to the cyt bc1 complex, and subsequently shuttled back by a soluble electron carrier protein toward the oxidizing side of the RC. In H. halophila, this electron carrier is a high-potential iron sulfur protein (HiPIP) (13). The redox potential of this HiPIP [Em = +165 mV (13)] turned out to be substantially lower than that of other purple bacterial HiPIPs (14) and instead falls within the range of potentials generally observed for electron donors to RCs in phototrophs using MK as pool quinone (15). Because no purple bacterium operating its photosynthetic chain on MK has been described thus far, this finding is surprising. We therefore set out to analyze the composition and redox potentials of the H. halophila photosynthetic chain in more detail.
Quinone Content of H. halophila.
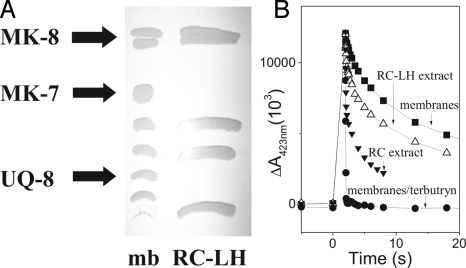
In the early work by Ventura et al. (10), MK-8 was reported to represent the major quinone species in membranes of all tested strains of H. halophila. We analyzed the content in quinones and their relative ratios for our strain by methanol/chloroform extraction and thin-layer chromatography (TLC) (Fig. 1A). UV/Vis spectroscopy identified 3 spots as redox active (see Material and Methods) compounds, the chemical nature of which was subsequently determined by mass spectrometry. Three quinone species were thus purified and their redox midpoint potentials determined. Bands not marked as quinones in Fig. 1A correspond to carotenoids and chlorophylls. We confirmed MK-8 as the major quinone representing 50–60% of the total quinone content, and UQ-8 as the sole ubiquinone species accounting for 35% (in strict anaerobiosis) to 45% (under microaerobiosis). The third quinone (≈5%) was identified to be methyl-MK-7 rather than MK-5 or MK-4 as reported in the past (9, 10). Cyclic voltammetry determined, for MK-8 and MK-7, a redox midpoint potential at Em,7.5 = −110 mV and at Em,7.5 = +70 mV for UQ-8. These data agree well with published Em values of MK and UQ determined at pH 7 [−74 mV and +112 mV, respectively (16)], taking a dependence on proton activity of −60 mV/pH into account (17).
Fig. 1.
Quinone content of membranes and RCs from H. halophila. (A) UV-fluorescence quenching spots detected on TLC plates loaded with methanol/chloroform extracts of membranes and RC–LH complexes. The indicated bands contained redox active compounds identified as MK-8, MK-7, and UQ-8 by mass spectroscopy and quantified by UV-redox difference spectroscopy. (B) Flash-induced absorbance changes (measured at 423 nm) of the special pair pigment in membranes in the presence (filled circles) and absence (filled squares) of the QB-site inhibitor terbutryn as well as in RC–LH complexes (open triangles) or in RC complexes purified as published in ref. 20 (closed triangles).
The fact that an individual quinone species is dominant, however, does not necessarily imply that it functionally represents the pool quinone. To assess whether MK or UQ constitutes the quinone pool in photosynthesis, we characterized the enzymes directly interacting with the pool, i.e., the photosynthetic RC and the cyt bc1 complex.
Photosynthetic RC: Em of the P/P+ Couple.
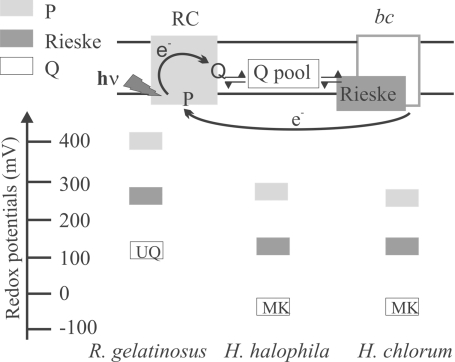
In purple bacterial photosynthetic RC, the absorption of a photon induces the ejection of an electron from the so-called special-pair pigment (P), situated close to the periplasmic side of the membrane, through a row of redox cofactors toward the quinone acceptors (see below) located in proximity of the cytoplasm. The redox midpoint potential of the P/P+ couple therefore is a key parameter indicative for the operating redox range of the whole chain. This Em value of P was determined by equilibrium optical redox titrations on isolated RCs and on membranes as well as through the redox potential dependence of the flash-induced absorption changes in membranes as described previously (13). All these experiments yielded the same result of Em7.5 = +270 ± 10 mV [see Fig. 2; data presented in supporting information (SI) Fig. S1]. Because we had observed that the Em of HiPIP in this halophilic species strongly depended on ionic strength (13), we also performed these experiments at physiological salt concentrations and obtained the same value for the Em of P (see Fig. S1). The measured potential is substantially lower than those observed thus far for special pair pigments of photosynthetic RCs, which are in the range of +400 mV < Em < +550 mV in species employing UQ or PQ as pool quinone (for a review, see ref. 11). Comparably low Em values were only reported for P/P+ from Chloroflexaceae, Chlorobiaceae and Heliobacteria (ref. 11; see Fig. 2), photosynthetic organisms that are phylogenetically very distant from purple bacteria and that employ electron transfer chains exclusively containing MK (2, 18, 19).
Fig. 2.
Schematic comparison of the redox properties of selected cofactors in the RC and the cyt bc1 complex from H. halophila (this work) to the corresponding values in photosynthetic chains from other species based either on UQ- [exemplified by Rubrivivax gelatinosus (11, 36)] or MK- [represented by Heliobacillus chlorum (11, 23)] pools. The rectangles indicate the redox midpoint potentials of the respective cofactors on a redox scale.
Photosynthetic RC: Chemical Nature of QA and QB.
Purified purple bacterial RCs allow one to directly assess the type of pool quinone via the identification of the chemical nature of the quinone in the so-called QB site. Purple bacterial RCs contain 2 quinone-binding sites, QA and QB. QA is a tightly bound cofactor of the RC, whereas the quinone in the QB site is in dynamic equilibrium with the pool. Depending on the species, QA can be a MK- or a UQ-molecule. As discussed above, only UQ had been found thus far in purple bacterial quinone pools and hence also in QB. In 1984, Lefebvre et al. (20) reported that only MK could be observed after chemical extraction of purified RC. However, because purified RCs frequently are depleted in QB, the actual presence of QB has to be demonstrated before extraction of lipophilic components from RC samples. A convenient method to assay the occupation state of the QB-site consists of recording recombination kinetics after a single flash-induced charge separation (21). If QB is present and oxidized before the flash, that is, available as acceptor for the negative charge ejected from the excited P*, a slow (several seconds) back-reaction is observed. In the absence of QB, charge recombination is significantly faster due to back-reaction from the earlier electron acceptor QA. This situation can be induced in membranes by the addition of QB-site inhibitors such as terbutryn (21). Fig. 1B shows such back-reaction kinetics recorded on H. halophila membranes. When we measured flash-induced absorption changes on RC samples prepared according to ref. 20, we observed a dominant fast back-reaction resembling the terbutryn-inhibited case, demonstrating that these samples are strongly depleted in QB. The results reported in ref. 20 thus identify QA as MK, but do not inform on the identity of QB. We therefore decided to use a milder purification protocol yielding RC–light harvesting (RC–LH) complexes (see Materials and Methods). The back-reaction kinetics recorded on these RC–LH complexes predominantly showed the slow kinetics (Fig. 1B, triangles) with a small contribution of a fast phase resulting from a minor population of RC lacking quinones in the QB-site, thus demonstrating a high occupation level of the QB-site in RC–LH samples.
Lipophilic components have been extracted from this RC–LH and were loaded, together with those obtained from membrane fragments, on a TLC plate. Fig. 1A shows that whereas CH3-MK-7, MK-8, and UQ-8 are detectable in membranes, only MK-8 is found in RC–LH complexes, demonstrating that MK-8 occupies both the QA and the QB sites (see Fig. 2).
Cyt bc1 Complex: Em of the Rieske Cluster.
The physiological electron acceptor from the quinone pool in purple bacterial photosynthesis is the Rieske [2Fe-2S] center in the cyt bc1 complex. The Rieske cluster stands out from the bulk of other [Fe-S] centers by its unique EPR spectrum represented in Fig. S2 and, for example, featuring a gy value at ≈1.9. This peculiar spectrum is due to the replacement of 2 cysteine ligands present in ordinary clusters by histidines (for review see ref. 7). The presence of a Rieske cluster in a specific protein is readily recognized by means of 2 distinctive binding motifs in its amino acid sequence. The EPR spectrum of this center is often straightforwardly observed in membrane fragments, permitting the determination of its Em value in nonpurified samples. The Em of the Rieske cluster has therefore in the past been exploited to recognize species operating on MK- or HPQ-chains (7).
In H. halophila, however, our attempts to titrate the Rieske cluster directly in membrane fragments by EPR were hampered by the abundance of other [Fe-S] clusters with signals at gy = 1.92. We therefore developed an enrichment protocol for the cyt bc1 complex. The obtained fractions do not yet correspond to a biochemically pure complex but were sufficiently depleted in other [Fe-S] centers to allow EPR titrations of the Rieske cluster, yielding an Em,7.5 at +110 ± 10 mV (Fig. S2 Inset, filled squares; evaluated at gy = 1.905).
To verify that the observed center indeed arises from the Rieske cluster of the cyt bc1 complex, we titrated the sample in the presence of the QO-site inhibitor stigmatellin that increases the Em of the Rieske cluster specifically in this complex (22). Presence of 20 μM stigmatellin resulted in the appearance of 2 separate waves, one with the Em of the uninhibited center and a second one with an Em increased by almost 100 mV (Fig. S2 Inset, crosses). Upon increasing the concentration of stigmatellin to 100 μM, the wave characteristic for the uninhibited enzyme disappeared and the g = 1.9 center titrated homogeneously at +190 mV (Fig. S2 Inset, open circles). These results suggest that all EPR-detectable [Fe-S] center in this sample indeed arises from the cyt bc1 complexes' Rieske cluster.
The Em-value of +110 mV of the H. halophila Rieske center is thus some 150 mV below the values typically determined in HPQH2-oxidizing species but close to those obtained for MKH2-oxidizing organisms (6, 7). This value reaches the lowest midpoint potential reported for cyt bc (recently renamed Rieske/cytb) complexes' Rieske centers so far. The high concentration of inhibitor required to obtain saturation of the QO-site indicates that the affinity of the QO-site for stigmatellin is significantly lower than for typical bc1 complexes. A strongly decreased affinity for stigmatellin has also been reported previously for the MKH2-oxidizing Rieske/cytb complex from Heliobacterium chlorum (23), corroborating the identification of MK as the pool quinone in H. halophila.
Genomic Analysis of Photosynthetic Enzymes in H. halophila.
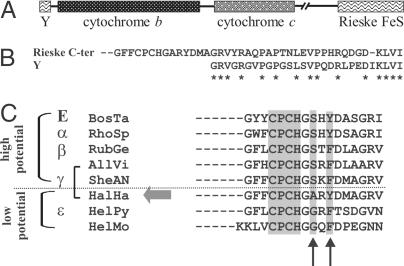
The genome of H. halophila SL1 has recently been sequenced. Genes coding for the photosynthetic RC's L, M, and tetraheme subunits were found in canonical order within the photosynthetic gene cluster. The organization of the genes coding for the cyt bc1 complex, however, is suspicious. With the sole exception of the cyanobacteria, all species containing a Rieske/cytb complex feature a conserved diad of genes with the Rieske gene directly upstream of that coding for cyt b (24). In striking contrast, no gene coding for a Rieske protein is detected close to those coding for cyt b (fbcB) and cyt c1 (fbcC) in the H. halophila genome (see Fig. 3A). The one and only gene in this genome showing significant similarity to Rieske proteins is situated in a completely different genomic region. However, we noticed that directly upstream of the fbcB gene, i.e., where a “well-behaved” Rieske-gene was to be expected, a sequence stretch is present with an astonishing similarity to the C terminus of the Rieske gene (Fig. 3 A and B). This stretch most probably represents the relics of a lost fbcF gene, situated in its canonical position upstream of fbcB (see Discussion).
Fig. 3.
Genomic organization and sequence characteristics of the cyt bc1 complex in H. halophila. (A) Genomic arrangement of the genes coding for the 3 principal subunits of the cyt. bc1 complex. “Y” denotes a sequence stretch that probably represents a fragment of a gene of the Rieske protein. (B) Sequence comparison of Y to the C-terminal end of the Rieske gene located distal from the cytb/cytc1-diad. (C) Comparison of the Rieske protein's sequence range containing the second cluster-binding motif (shaded) and the 2 amino acid residues (shaded, arrows) considered to strongly influence the redox potential of the cluster. H. halophila is highlighted by a large shaded arrow, “α”–“ε” denote subgroups of the proteobacterial phylum and “E” stands for eukaryotic mitochondria. The sequences (accession numbers and parent species) used in the alignment are given in Table S1.
As discussed in a previous article (25), Rieske proteins are composed structurally of 2 domains, one of which essentially is represented by the N-terminal half of the sequence, and the second one corresponds to the [2Fe-2S] cluster-binding domain. The first domain is only weakly conserved and requires structural information to be reliably aligned. The cluster-binding domain, however, is strongly conserved over representatives from all 3 domains of life. In particular, 2 motifs involved in the cluster binding are highly similar. Site-directed mutagenesis studies (reviewed in ref. 7) have demonstrated that the Em value of the Rieske cluster is critically influenced by 2 residues close to the second cluster-binding motif (Fig. 3C). In so-called “HP Rieske” proteins, a serine and a tyrosine residue situated 2 and 4 positions downstream of this motif (Fig. 3C) engage in hydrogen bonds with the [Fe-S] cluster and thereby increase its Em value. The absence of these 2 hydrogen bonds in a typical “LP Rieske” cluster results in Em values lowered by ≈150 mV. The serine residue has been shown to have a much stronger impact than the tyrosine, resulting by itself in a redox shift of ≈100 mV. In contrast to the characterized LP Rieske proteins, all known α/β/γ-proteobacterial sequences except that from H. halophila conserve the serine residue and thus probably represent HP- Rieske proteins. The absence of this residue suggests that the H. halophila protein features a significantly lower Em value than for other proteobacteria, in line with our electrochemical titration (Fig. S2 Inset). It is noteworthy that no other ORF featuring the characteristic sequence motifs of a Rieske center was found in the H. halophila genome. The association of the physicochemical parameters determined in our work on the [2Fe-2S] center to the product of the Rieske gene is thus unambiguous and clearly indicates that the bc-Rieske is of the MKH2-oxidizing type.
Discussion
All photosynthetic species in α-, β- and γ-proteobacteria have thus far been considered to operate HP bioenergetic chains based on UQ as pool quinone. A well-studied case within the γ-subgroup is provided by the photosynthetic cycle of the Chromatiacea Allochromatium vinosum. In this species, UQ is the predominant quinone in membranes (26, 27). The sequence of the Rieske protein (see Fig. 3C), the nature of the RC's secondary electron acceptor QB (26), and the redox potential of other cofactors in the RC (11) all confirm the HP nature of this electron-transport system.
Surprisingly, the Halorhodospira species H. halophila, another photosynthetic member of the γ-proteobacteria containing both UQ and MK, clearly operates a LP photosynthetic cycle based on MK (Fig. 2). To assay whether this is a common trait of the Halorhodospira/Ectothiorhodospira subclade of the γ-proteobacteria, we have analyzed published data on other closely related organisms. Results reported on Ectothiorhodospira haloalkaliphila (formerly E. mobilis) BN 9903 indicate that this species contains UQ as predominant quinone in membranes (10) and as secondary electron acceptor QB in its RC (28) as well as HP cyt in its cyt bc1 complex (29) demonstrating that E. haloalkaliphila uses a UQ-pool in its photosynthetic cycle. The clade of the Halorhodospiraceae/Ectothiorhodospiraceae therefore is heterogeneous with some subclasses working on LP and others on HP bioenergetic chains when grown phototrophically.
Scenario for the Evolutionary Transition from Low- to High-Potential Bioenergetic Chains in Proteobacteria.
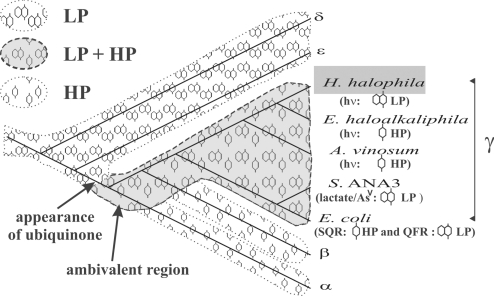
The proteobacterial phylum is heterogeneous both with respect to the occurrence of MK and/or UQ and with respect to the range of redox potentials covered by their respective bioenergetic chains. ε- and δ-proteobacteria contain only MK (2) and correspondingly operate LP energy-conserving chains exclusively (6, 8). MK must therefore be supposed to represent the ancestral type of quinone in proteobacteria. Given the observation that in other places of the phylogenetic tree, chemically very different HPQ have appeared (see Introduction), it seems extremely unlikely to us that the UQ molecule arose independently in α-, in β- and in γ-proteobacteria. This suggests that UQ was already present in the common ancestor of the α- and the β-/γ- subgroups (β- and γ-proteobacteria are sister-clades, see Fig. 4).
Fig. 4.
Distribution of MK- and UQ-based chains on a proteobacterial phylogenetic tree. This tree (reconstructed by the neighbor-joining method) is based on 16S r-RNA genes. The shaded area indicates extant and extinct lineages where both MK and UQ functioned alternatively. The probable evolutionary appearance of the UQ molecule after the divergence of the δ- and ε-subgroups and before the α/β/γ-radiation is indicated. The α, β, δ, and ε species used for the tree reconstruction are, Rhodobacter sphaeroides, Rubrivivax gelatinosus, EbS7, and Helicobacter pylori, respectively. Identification of HP and/or LP chains are based on our work for H. halophila, on refs. 10, 28, and 29 for E. haloalkaliphila, on refs. 11, 26, and 27 for A. vinosum, on ref. 30 for Shewanella ANA-3, and on ref. 12 for E. coli.
Because no LP bioenergetic chains have been reported for α- and β-proteobacteria thus far, we consider for the time being these 2 lineages as having abandoned energy-conserving chains based on MK pools (Fig. 4). The situation is different for the γ-subgroup. E. coli is able to switch between LP and HP chains (12). Biochemical and genetic studies demonstrated that the nonphotosynthetic γ-proteobacterium Shewanella ANA-3 performs bioenergetic electron transport from lactate to arsenate based on a MK-pool (30). The genomic analysis of this strain, however, shows the presence of 3 O2-reductases (31) indicative of an optional aerobic lifestyle. The sequence of the Rieske protein in its cyt. bc1 complex (Fig. 3C) furthermore indicated the HP nature of the chain this enzyme is embedded in. It therefore appears probable to us that Shewanella is also able to switch from LP to HP chains. As discussed above, the Ectothiorhodospira/Halorhodospira-clade contains species that drive photosynthetic chains based either on MK- or on UQ-pools. Besides MK-pool-based photosynthesis, H. halophila probably contains UQ-pool-based bioenergetic chains bypassing the cyt. bc1 complex. Genomic analysis of the species shows the absence of heme-copper oxidases but the presence of a quinol-oxidizing bd oxidase (Hhal_2420 and Hhal_2419) (31). This oxidase may support growth under microaerobic conditions. It is noteworthy that the relative content of UQ has been shown to increase under these growth conditions (from 35% to 45%). H. halophila therefore probably represents one more case of a strain able to use both UQ- and MK-pools in distinct bioenergetic chains. Whether these chains are sequestered from each other in a given cell by an as-yet-unknown mechanism or whether individual cells exclusively use 1 of the 2 types of quinones, remains to be determined for the respective species.
The capacity to switch between LP and HP bioenergetic chains therefore appears to be a characteristic found all over the γ-proteobacterial subgroup rather than being a specificity of E. coli and related Enterobacteria (Fig. 4). This suggests that the LP-to-HP transition, having occurred some 2 billion years ago, probably proceeded via ambivalent organisms, able to use both types of bioenergetic chains and that extant species from the γ-proteobacterial subgroup still feature these ambivalent characteristics (Fig. 4).
Evolutionary Driving Forces Favoring the Presence of 2 Distinct Sets of Electron-Transfer Enzymes in Bioenergetic Chains Based on MK- and UQ-Pools.
The setup of extant electron transfer chains, suggests that the alternative use of MK and UQ as pool quinone necessarily entails the redox adaptation of surrounding electron transfer components and in particular the cyt bc1 complex, as outlined above. The need to up-shift the redox potentials of cofactors in this enzyme in response to the presence of UQ is energetically obvious because the higher potential UQ would be a very poor electron donor to the redox cofactors of an “MK-type” Rieske/cytb complex (Fig. 2). By contrast, the reasons why a “UQ-type” complex should not work with MKs are less obvious. From an energetic point of view, the increased difference between the low potential of MK and the cofactors of a UQ-adapted bc complex would result in a more favorable oxidation of MKH2 to semiquinone at QO, the rate-limiting step in the Q-cycle (32). One might therefore expect that MK-containing organisms simply “take advantage” of this additional driving force and get along with a single, UQ-type bc1 complex. However, it has recently been shown that conservation of the Q-cycle driving force is critical to preventing deleterious superoxide production at the QO site (32–35). Substitution of the LP rhodoquinol (RQH2) for UQH2 in mitochondrial cyt. bc1 complex resulted in dramatic increases in superoxide production. Work on a series of alternative quinol substrates (33) and changing the redox potential of the [2Fe-2S] cluster (32) demonstrated that the increased superoxide production is attributable to changes in the driving force for oxidation of QH2 to its semiquinone. According to these results, a MK pool functioning with a UQ-type cyt bc1 complex would thus very likely have extremely deleterious effects on the parent organisms. In other words, the potential of the Rieske cluster needs to be tuned to that of the quinol substrate to avoid the cyt bc1 complex turning into a powerful source of reactive oxygen species.
How Do Ambivalent Species Cope with the Problem of Global Redox Tuning?
In E. coli and in the majority of related Enterobacteria, the radical solution of doing away with a cyt bc1 complex was obviously arrived at. However, because the cyt bc1 complex is an important energy-coupling segment of the chain, its lack will substantially lower the energetic efficiency of the system, and we surmise that only their abundant source of food permits Enterobacteria to afford this waste in energy.
An alternative to loss of the cyt bc1 complex would be to duplicate and redox-optimize this enzyme or at least its crucial redox subunit, the Rieske protein. Double and even multiple occurrence of genes coding for Rieske proteins is observed in several prokaryotic phyla (24). In one case, it has been demonstrated that the products of the duplicate genes are indeed assembled into the cyt bc1 complex and yield a functional enzyme (36). However, thus far, none of these “second” Rieske proteins seems to have potentials different from that of the “canonical” Rieske protein encoded within the fbc-operon (36). In the γ-proteobacterium Acidithiobacillus ferrooxidans, even 2 complete cyt bc1 complexes were found, but they appear to be optimized to fit 2 distinct metabolic pathways rather than to operate at differing redox potentials (37). These cases demonstrate that duplication of the Rieske gene and even of the complete cyt bc operon can and does occur in proteobacteria.
It is therefore straightforward to assume that the bioenergetically ambivalent ancestor of α/β/γ-proteobacteria used 2 distinct copies of the cyt bc complex, distinguishing themselves at least in the Rieske protein. They were (as were probably other bioenergetic enzymes) optimized to operate in the context of MK- and UQ-pools, respectively. All of the studied species from these subgroups obviously have lost the LP cyt bc complex, whereas H. halophila has abandoned the HP version of the enzyme. Further species resembling H. halophila to this respect certainly exist, and we tend to believe that even ambivalent γ-proteobacteria expressing HP or LP versions of the cyt bc1 complex as a function of growth conditions and resulting nature of the quinone pool are still to be discovered.
It furthermore seems likely to us that all lineages undergoing the LP-to-HP transition in response to the increasing oxidation level of the environment had to go through intermediate species possessing ambivalent redox capacities. A thorough investigation of basally branching species of cyanobacteria and sulfolobales may help to better understand this critical evolutionary transition that allowed life to cope with the deleterious effects of rising levels of O2 2.5 billion years ago.
Materials and Methods
Growth of H. halophila.
H. halophila SL1(DSM244) was grown photosynthetically and anaerobically at 35 °C in ATCC 1448 medium in 1-L bottles.
Preparation of Photosynthetic RCs and RC–LH Complexes.
Cells were suspended in 50 mM 3-[morpholino]propanesulfonic acid (Mops) at pH 7.5 (buffer A) and broken by 1 passage through a French pressure cell. Unbroken cells were eliminated by low-speed centrifugation. Ultracentrifugation separated the “total soluble fraction” in the supernatant from the “membrane-fragments fraction” in the pellet. RCs were initially purified by using LDAO as described by Lefebvre et al. (20). This preparation turned out (see Results) to be devoid of QB. We therefore extracted RC–LH complexes using LDAO as described in ref. 20 but submitted the crude LDAO extract directly to ultracentrifugation without dilution of LDAO. The supernatant was then loaded on a 30–10% sucrose gradient equilibrated in 0.01% dodecyl maltoside (DM) in buffer A. After ultracentrifugation overnight, RC–LH complexes were collected in the 25% sucrose fraction and dialyzed against buffer A/0.01% DM.
Preparation of cyt bc1 Complexes.
Membrane fragments were obtained as described above. Cyt bc1 complexes were extracted for 1 h at 4 °C by using DM. Membrane fragments were resuspended at 8 mg/mL proteins in Mops/glycerol 20%/500 mM NaCl. DM was added to a final concentration of 6.6 mg/mL. The sample was then ultracentrifuged for 3 h at 200,000 × g and at 4 °C. The supernatant was dialyzed against Mops 50 mM/glycerol 20%/0.01% DM (buffer A) and subsequently ultracentrifuged as above. This supernatant was adsorbed onto a DEAE biogelA column equilibrated in buffer A. The column was washed with buffer A/0.1 M NaCl and eluted with a linear gradient from 0.1 to 0.5 M NaCl. Fractions containing cyt bc1 complex eluting at ≈0.37 M NaCl were pooled, concentrated 5 times by using PM 30,000 ultrafiltration membranes yielding 2 μM of cyt. bc1 complex and finally dialyzed against buffer A/50 mM NaCl.
Analysis of Quinone Content.
Quinones were extracted from membrane fragments or RC–LH samples. One volume of sample was mixed with 1 volume of chloroform and 2 volumes of methanol to extract hydrophobic components. Subsequently, 2 volumes of chloroform and 1 volume of water were added. This mixture was centrifuged for 10 min at 15,000 × g, and the bottom phase was sampled, evaporated under a stream of argon in the dark, resuspended in ethanol, and finally loaded on a silica gel TLC plate containing a fluorescent indicator. To identify quinones in RC–LH samples, home-purified MK-8 and UQ-8 were loaded as standards. The plates were developed in darkness with toluene. Fluorescence-quenching spots were detected under UV light, scraped off the plate, and resuspended in ethanol. Quinones were identified by UV/Vis difference spectroscopy and their exact chemical nature was determined by mass spectroscopy. Borohydride was used as reductant.
Redox Titrations.
Membrane samples or purified samples were redox titrated at 15 °C as described by Dutton (38) in 50 mM Mops (pH 7.5) (with 0.01% DM in the case of cyt bc1 complexes and 0.1% LDAO in the case of RC) in the presence or absence of 130 g/L NaCl, in the presence of the following redox mediators at 100 μM for EPR- or 15 μM for optical titrations: 1,4 p-benzoquinone, 2,5-dimethyl-p-benzoquinone, 2-hydroxy 1,2-naphthoquinone, 1,4-naphthoquinone, dimethyl-1,4-naphthoquinone, 2,5-dihydroxy-p-benzoquinone, dihydroxy-1,4-naphthoquinone, anthraquinone-2-sulfonate. Reductive and oxidative titrations were carried out by using sodium dithionite and ferricyanide, respectively. The RC used for titrations was prepared according to ref. 20.
Optical spectra were recorded on a Carry 5E spectrophotometer. EPR spectra were recorded on a Bruker ESP 300 X-band spectrometer fitted with an Oxford Instruments liquid-helium cryostat and temperature control system.
Light-Induced Absorption Changes.
Membrane fragments were diluted in 50 mM Mops (pH 7.5) and RC preparations in 50 mM Mops (pH 7.5)/0.01% DM. Xenon flash-induced absorption changes were measured at 423 nm by a laboratory-built spectrophotometer (7). Duration of actinic flash was ≈10 μs.
Amino Acid Sequence Analysis.
Database searches were performed by using BLASTP (39) on the amino acid sequence database at the National Center for Biotechnology (NCBI), Washington, DC. Amino acid sequences were aligned by CLUSTALX (40) using the Blossom matrix and refined based on the available structures as detailed in ref. 25.
Supplementary Material
Acknowledgments.
D.M.K.'s contribution to this work was supported by the Centre National de la Recherche Scientifique through a 4 months' sabbatical fellowship in 2006.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813173106/DCSupplemental.
References
- 1.Mitchell P. Vectorial chemiosmotic processes. Annu Rev Biochem. 1977;46:996–1005. doi: 10.1146/annurev.bi.46.070177.005024. [DOI] [PubMed] [Google Scholar]
- 2.Collins MD, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiraishi A, Hoshino Y. Distribution of rhodoquinone in Rhodospirillaceae and its taxonomic implication. J Gen Appl Microbiol. 1984;30:435–448. [Google Scholar]
- 4.Schäfer G, Moll R, Schmidt CL. Respiratory enzymes from Sulfolobus acidocaldarius. Methods Enzymol. 2001;331:369–410. doi: 10.1016/s0076-6879(01)31071-6. [DOI] [PubMed] [Google Scholar]
- 5.Nitschke W, Kramer DM, Riedel A, Liebl U. From naphtho- to benzoquinones -(r)evolutionary reorganizations of electron transfer chains. In: Mathis P, editor. Photosynthesis: From Light to Biosphere. Vol I. Dordrecht, The Netherlands: Kluwer Academic; 1995. pp. 945–950. [Google Scholar]
- 6.Schütz M, et al. Early evolution of cytochrome bc complexes. J Mol Biol. 2000;300:663–675. doi: 10.1006/jmbi.2000.3915. [DOI] [PubMed] [Google Scholar]
- 7.Schoepp B, Brugna M, Lebrun E, Nitschke W. Iron-sulfur centers involved in photosynthetic light reactions. Adv Inorg Chem. 1999;47:335–360. [Google Scholar]
- 8.Brugna M. France: University of Aix-Marseille; 2000. Les complexes de type bc: Aspects fonctionnels et évolution moléculaire. PhD Thesis. [Google Scholar]
- 9.Imhoff JF. Quinones of phototrophic purple bacteria. FEMS Microbiol Lett. 1984;25:85–89. [Google Scholar]
- 10.Ventura S, Giovannetti L, Gori A, Viti C, Materassi R. Total DNA restriction pattern and quinone composition of members of the family Ectothiorhodospiraceae. System Appl Microbiol. 1993;16:405–410. [Google Scholar]
- 11.Nitschke W, Dracheva SM. Reaction center associated cytochromes. In: Blankenship RE, Madigan MT, Bauer CE, editors. Anoxygenic Photosynthetic Bacteria. Dordrecht, The Netherlands: Kluwer Academic; 1995. pp. 775–805. [Google Scholar]
- 12.Jones RW, Garland PB. The function of ubiquinone and menaquinone in the respiratory chain of Escherichia coli. In: Trumpower BL, editor. Functions of quinones in energy conserving systems. New York, USA: Academic Press; 1982. pp. 465–476. [Google Scholar]
- 13.Lieutaud C, Alric J, Bauzan M, Nitschke W, Schoepp-Cothenet B. Study of the high-potential iron sulfur protein in Halorhodospira halophila confirms that it is distinct from cytochrome c as electron carrier. Proc Natl Acad Sci USA. 2005;102:3260–3265. doi: 10.1073/pnas.0407768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menin L, et al. Role of HiPIP as electron donor to the RC bound cytochrome in photosynthetic purple bacteria. Photosynth Res. 1998;55:343–348. [Google Scholar]
- 15.Meyer TE, Donohue TJ. Cytochromes, iron-sulfur, and copper proteins mediating electron transfer from the cyt bc1 complex to photosynthetic reaction complexes. In: Blankenship RE, Madigan MT, Bauer CE, editors. Anoxygenic Photosynthetic Bacteria. Dordrecht, The Netherlands: Kluwer Academic; 1995. pp. 725–745. [Google Scholar]
- 16.Kröger A, Unden G. The function of menaquinone in bacterial electron transport. In: Lenaz G, editor. Coenzyme Q. London: Wiley; 1985. pp. 285–300. [Google Scholar]
- 17.Takamiya KI, Dutton PL. Ubiquinone in Rhodopseudomonas sphaeroides. Some thermodynamic properties. Biochim Biophys Acta. 1979;546:1–16. doi: 10.1016/0005-2728(79)90166-x. [DOI] [PubMed] [Google Scholar]
- 18.Hale MB, Blankenship RE, Fuller RC. Menaquinone is the sole quinone in the facultatively aerobic green photosynthetic bacterium. Chloroflexus aurantiacus. Biochim Biophys Acta. 1983;723:376–382. [Google Scholar]
- 19.Hiraishi A. Occurrence of menaquinone as the sole isoprenoid quinone in the photosynthetic bacterium Heliobacterium chlorum. Arch Microbiol. 1989;151:378–379. [Google Scholar]
- 20.Lefebvre S, Picorel R, Cloutier Y, Gingras G. Photoreaction center of Ectothiorhodospira sp. Pigment, heme, quinone, and polypeptide composition. Biochemistry. 1984;23:5279–5288. [Google Scholar]
- 21.Ginet N, Lavergne J. Equilibrium and kinetic parameters for the binding of inhibitors to the QB pocket in bacterial chromatophores: Dependence on the state of QA. Biochemistry. 2001;40:1812–1823. doi: 10.1021/bi001686a. [DOI] [PubMed] [Google Scholar]
- 22.von Jagow G, Ohnishi T. The chromone inhibitor stigmatellin—Binding to the UQ oxidation center at the C-side of the mitochondrial membrane. FEBS Lett. 1985;17:311–315. doi: 10.1016/0014-5793(85)80929-7. [DOI] [PubMed] [Google Scholar]
- 23.Liebl U, Rutherford AW, Nitschke W. Evidence for a unique Rieske iron-sulphur centerin Heliobacterium chlorum. FEBS Lett. 1990;261(2):427–430. [Google Scholar]
- 24.Kramer DM, Nitschke W, Cooley JW. The cytochrome bc1 and related bc complexes, the Rieske/cytochrome b complex as the functional core of a central electron/proton transfer complex. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT, editors. The Purple Phototrophic Bacteria. Dordrecht, The Nederlands: Springler; 2009. pp. 451–473. [Google Scholar]
- 25.Lebrun E, et al. The Rieske protein: A case study on the pitfalls of multiple sequence alignments and phylogenetic reconstruction. Mol Biol Evol. 2006;23(6):1180–1191. doi: 10.1093/molbev/msk010. [DOI] [PubMed] [Google Scholar]
- 26.Halsey YD, Parson WW. Identification of ubiquinone as the secondary electron acceptor in the photosynthetic apparatus of Chromatium vinosum. Biochim Biophys Acta. 1974;347:404–416. doi: 10.1016/0005-2728(74)90079-6. [DOI] [PubMed] [Google Scholar]
- 27.Parson WW. In: The Photosynthetic Bacteria. Clayton RK, Siström WF, editors. New York: Plenum; 1978. pp. 455–469. [Google Scholar]
- 28.Leguijt T, Hellingwerf KJ. Characterization of reaction center/antenna complexes from bacterio-chloroplyll a containing Ectothiorhodospiraceae. Biochim Biophys Acta. 1991;1057:353–360. [Google Scholar]
- 29.Leguijt T, Engels PW, Crielaard W, Albracht SPJ, Hellingwerf KJ. Abundance, subunit composition, redox properties and catalytic activity of the cytochrome bc1 complex from alkaliphilic and halophilic, photosynthetic members of the family Ectothiorhodospiraceae. J Bact. 1993;175:1629–1636. doi: 10.1128/jb.175.6.1629-1636.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy JN, Saltikov CW. The cymA gene, encoding a tetraheme c-type cytochrome, is required for arsenate respiration in Schewanella species. J Bacteriol. 2007;189(6):2283–2290. doi: 10.1128/JB.01698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brochier-Armanet C, Talla E, Gribaldo S. The multiple evolutionary histories of dioxygen reductases: Implications for the origin and evolution of aerobic respiration. Mol Biol Evol. 2009;26(2):285–297. doi: 10.1093/molbev/msn246. [DOI] [PubMed] [Google Scholar]
- 32.Forquer I, Covian R, Bowman MK, Trumpower B, Kramer DM. Similar transition states mediate the Q-cycle and superoxide production by the cytochrome bc1 complex. J Biol Chem. 2006;281:38459–38465. doi: 10.1074/jbc.M605119200. [DOI] [PubMed] [Google Scholar]
- 33.Cape JL, et al. The respiratory substrate rhodoquinol induces Q-cycle bypass reactions in the yeast cytochrome bc1 Complex. J Biol Chem. 2005;280:34654–34660. doi: 10.1074/jbc.M507616200. [DOI] [PubMed] [Google Scholar]
- 34.Cape JL, Bowman MK, Kramer DM. Understanding the cytochrome bc complexes by what they don't do. The Q-cycle at 30. Trends Plants Sci. 2006;11:46–55. doi: 10.1016/j.tplants.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Cape JL, Bowman MK, Kramer DM. A semiquinone intermediate generated at the QO site of the cytochrome bc1 complex. Importance for the Q-cycle and superoxide production. Proc Natl Acad Sci USA. 2007;104:7887–7892. doi: 10.1073/pnas.0702621104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouchane S, Nitschke W, Bianco P, Verméglio A, Astier C. Multiple Rieske genes in prokaryotes: Exchangeable Rieske subunits in the cytochrome bc1-complex of Rubrivivax gelatinosus. Mol Microbiol. 2005;57:261–275. doi: 10.1111/j.1365-2958.2005.04685.x. [DOI] [PubMed] [Google Scholar]
- 37.Brasseur G, Levican G, Bonnefoy V, Holmes D, Jedlicki E, Lemesle-Meunier D. Apparent redundancy of electron transfer pathways via bc(1) complexes and terminal oxidases in the extremophilic chemolithoautotrophic. Acidithiobacillus ferrooxidans. Biochim Biophys Acta. 2004;1656:114–126. doi: 10.1016/j.bbabio.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Dutton PL. Oxidation-reduction potential dependence of the interaction of cytochromes, bacteriochlorophyll and carotenoids at 11°K in chromatophores of Chromatium D and Rhodopseudomonas gelatinosa. Biochim Biophys Acta. 1971;226:63–80. doi: 10.1016/0005-2728(71)90178-2. [DOI] [PubMed] [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipmann DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Higgins DG, Sharp PM. Fast and selective multiple sequence alignments on a microcomputer. CABIOS. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.