Abstract
G protein-coupled receptors with seven transmembrane α-helices (GPCRs) comprise the largest receptor superfamily and are involved in detecting a wide variety of extracellular stimuli. The availability of high-resolution crystal structures of five prototypical GPCRs, bovine and squid rhodopsin, engineered A2A-adenosine, β1- and β2-adrenergic receptors, permits comparative analysis of features common to these and likely all GPCRs. We provide an analysis of the distribution of water molecules in the transmembrane region of these GPCR structures and find conserved contacts with microdomains demonstrated to be involved in receptor activation. Colocalization of water molecules associating with highly conserved and functionally important residues in several of these GPCR crystal structures supports the notion that these waters are likely to be as important to proper receptor function as the conserved residues. Moreover, in the absence of large conformational changes in rhodopsin after photoactivation, we propose that ordered waters contribute to the functional plasticity needed to transmit activation signals from the retinal-binding pocket to the cytoplasmic face of rhodopsin and that fundamental features of the mechanism of activation, involving these conserved waters, are shared by many if not all family A receptors.
Keywords: adrenergic receptor, allosteric modulator, ordered waters, transmembrane proteins, x-ray
G protein-coupled receptors (GPCRs) are membrane proteins that mediate signal transduction by recognizing a wide range of extracellular stimuli ranging from photons of light, biogenic amines and odorants, to small peptides that all induce receptor activation. Activated GPCRs signal through heterotrimeric G proteins to activate effector enzymes, resulting in rapid signal amplification. Given the new-found wealth of structural information available from determination of several GPCR crystal structures, our purpose here is to provide a structural analysis of components likely to participate in the allosteric communication between the ligand-binding site near the extracellular domain and the cytoplasmic site involved in G protein coupling and activation.
The first high-resolution crystal structure of a GPCR was that of bovine rhodopsin (1), an advance that facilitated the successful determination of other high-resolution bovine rhodopsin structures (2–5). One paradigm for GPCR activation involves large rigid body movements of up to 15 Å of the heptahelical transmembrane bundle upon receptor transition from an inactive to an active conformation (6). This notion has been challenged because large changes in these helices were not observed in the crystal structure of a late photointermediate of rhodopsin that retained the full-agonist, all-trans-retinylidene, and exhibited spectral features consistent with the metarhodopsin (Meta) II activated state (7). Newer studies using double electron–electron resonance (DEER) spectroscopy revised the predicted structural changes upon activation to be no more than 5 Å between helices (8). This reduced scale for the conformational changes associated with activation is within the observed differences in the helical arrangement of GPCRs with known structures (9) and is compatible with the crystallographic structure of photoactivated rhodopsin. These observations highlight the important question of how, in the absence of a large structural rearrangement, is the “activation signal” transferred from the ligand-binding pocket (or retinal isomerization site) near the N-terminal face of the receptor to the cytoplasmic face where G protein binding and activation occur.
In an attempt to address the question of what receptor components contribute to a shared mechanism of activation, we provide a detailed analysis of conserved features within available family A GPCR structures. In addition to ground-state bovine rhodopsin, these structures include inactive ligand free bovine opsin (10, 11), truncated squid rhodopsin (12), mutant human β2-adrenergic receptor T4-lysozyme fusion protein (13) and mutant β2-adrenergic receptor with Fab (14), mutant turkey β1-adrenergic receptor with antagonist bound (15), and mutant human A2A-adenosine T4-lysozyme fusion protein with antagonist bound (9). Comparative sequence analysis has revealed amino acid residues conserved between GPCR family A members (16). Sequence alignments of the putative seven-transmembrane-spanning helices common to all GPCRs reveal residues that are possibly linked to ligand binding, receptor activation, and G protein coupling.
Results
Conserved Features of Family A GPCRs.
A comparison of amino acid sequences of the crystallized constructs (omitting the T4 lysozyme amino acid sequence present in mutant β2-adrenergic receptor and A2A-adenosine receptors) revealed modest amino acid conservation ranging from 15% identity between bovine rhodopsin and mutant β2-adrenergic receptor to 58% sequence identity between mutant β1- and mutant β2-adrenergic receptor [supporting information (SI) Fig. S1]. Bovine rhodopsin and mutant β1-adrenergic receptor share 17% sequence identity, mutant A2A-adenosine receptor and bovine rhodopsin share 20% sequence identity, and squid rhodopsin shares 26% sequence identity with bovine rhodopsin. Multiple sequence alignments of the five receptors identified 24 conserved amino acid residues. The functional importance of these conserved residues is highlighted by the fact that 8 of the 24 rhodopsin mutations are associated with autosomal dominant retinitis pigmentosa (G106, C110, R135, Y136, P171, C187, P215, and P267) (17). Conserved segments are localized in the transmembrane domains, including the highly conserved E/DRY motif in the third transmembrane span, the WXPF/Y motif in the VI transmembrane span, and the NPXXY sequence localized at the end of transmembrane helix VII. Given that three of the five receptors compared here signal through different heterotrimeric G proteins, these residues are more likely associated with conserved structural and functional motifs that are associated with a common mechanism for activation as opposed to motifs that mediate G protein specificity.
Activated states for members of the GPCR superfamily can be defined in part as states having the capacity to associate with and activate G protein. Bovine rhodopsin, squid rhodopsin, A2A-adenosine, and β1- and β2-adrenergic receptors differ in their basal levels of activity. Indeed, the β2-adrenergic receptor has been shown to couple to G proteins independent of agonist binding (18) compared with rhodopsin, which exhibits virtually no basal activity (19). Additionally, for rhodopsin, hydrolysis of chromophore leading to the formation of opsin results in a state of the receptor that exhibits a very low level of G protein-activating capacity, with rates approximately ≈250 times lower than that of Meta II (20). In general, residues associated with ligand binding cluster near the extracellular portion (intradiscal surface/extracellular side) of the helical bundle in family A GPCRs, and residues associated with receptor constitutive activity or folding form a switch region localized near the ligand-binding pocket and extracellular loop 2 (E2). In contrast, residues that mediate G protein coupling are found nearest the cytoplasmic face of the receptor. Interactions between the heterotrimeric G proteins and activated receptors involve the C-terminal portion of helix III, residues in the cytoplasmic loop 3, and the C-terminal tail of the GPCR and the α-subunit of the heterotrimeric G protein (21, 22). The structure of bovine opsin with a high-affinity synthetic peptide mimetic of Gtα bound provides further insight into some of the contacts that may mediate G protein binding to receptor; however, we do not consider this to be an active state of the receptor given that there is electron density in the ligand-binding pocket consistent with the presence of hydrolyzed chromophore, which has been demonstrated to reduce greatly the capacity of opsin to activate G protein (11, 20). This form is functionally similar to the ground states of other GPCRs that exhibit low levels of basal activity.
Conserved Polar Interactions with Water.
The importance of water for proper protein function is implicit because water is a fundamental component supporting and sustaining nearly all biological activity (23). Many lines of evidence have indicated the presence of water in rhodopsin, and receptor hydration has been shown to play a critical role in the formation of its active state, i.e., Meta II. Wald et al. (24) showed that dried films of this GPCR are capable of forming Meta I, a precursor to the active form of this receptor, but not activated Meta II. This observation can be explained by water functioning as an allosteric mediator in rhodopsin, such that proper structural transition from Meta I to Meta II requires the presence of water in the helical bundle of this receptor. Indeed, crystallographic studies have demonstrated that water functions as an allosteric mediator in dimeric hemoglobin (25). The study of water molecules has become a fixture of modern crystallography, as for example, structures at 2 Å resolution are likely to have 1 water per residue built into the crystallographic model (26). Integral membrane proteins are no exception because a recent survey of buried water molecules found in crystal structures of helical transmembrane proteins revealed a high number of interactions between water and main chain oxygens and nitrogens of the α-helices.
Although our comparison of six of the high-resolution structures from five different rhodopsin-like GPCR superfamily members evidenced significant conservation for the major structure elements (Fig. 1), we were surprised to find ordered waters that colocalized in the transmembrane regions after structural superpositioning of the crystallographic models. These ordered waters were found at 15 locations throughout the helical bundle of five different GPCR structures (Fig. 2A). Because water occupancy can be partial, and thus specific water molecules of structural importance can be missed in the examination of a single crystallographic structure, a value of this study is the identification of waters that are consistently observed in a number of structures, with allowance of positional variation that is to be expected for the different members of the same family. In the following description of the 15 water clusters present in the GPCR structures, residues are numbered according to the Ballesteros and Weinstein (27) numbering scheme (i.e., D2.50), where the first number denotes the transmembrane helix and the most conserved residue in a transmembrane helix is assigned a value of 50 (TM no.x.50), additional rhodopsin residues are numbered when appropriate. The bound waters were observed either in unique locations, i.e., waters 1, 5, and 14, or in clusters, i.e., waters 2, 3, 4, 6, 7, 8, 9, 10, 11, 12, 13, and 15, where two or more receptor structures had bound waters in close proximity.
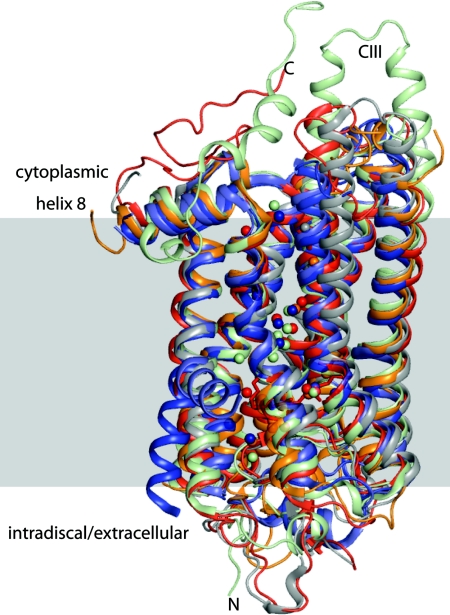
Fig. 1.
Structural superpositioning diagram of high-resolution crystal structures of bovine rhodopsin (red), squid rhodopsin (wheat), mutant β1-adrenergic receptor (light blue), mutant β2-adrenergic receptor (navy blue), and bovine opsin (gray) demonstrating a high level of overall structural similarity. Also shown are those water molecules that colocalize in the transmembrane helices.
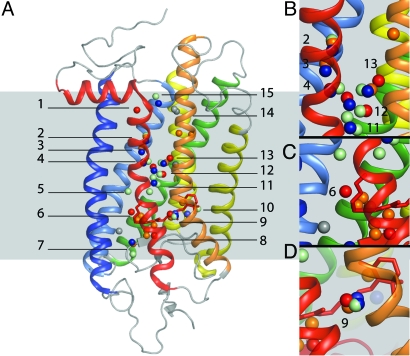
Fig. 2.
Water molecules observed in the crystal structures of family A GPCRs. Structural superpositioning of the Cα chains reveals that many of the ordered waters found in available high-resolution GPCR crystal structures are colocalized. (A) Positions of the waters (shown as spheres) from the structures of bovine rhodopsin (red), squid rhodopsin (wheat), bovine opsin (light gray), bovine opsin with peptide (dark gray), mutant β1-adrenergic receptor (light blue), mutant β2-adrenergic receptor (navy blue), and A2A-adenosine receptor (orange) after superpositioning each GPCR structure with that of bovine rhodopsin. Waters are localized to 15 regions within the helical bundles of these GPCRs, shown here in the context of the structure of rhodopsin. (B) Water cluster conserved between squid rhodopsin, mutant β2-adrenergic receptor and bovine rhodopsin. Waters 2, 3, and 4 notably make contacts with D2.50 and Y7.53 and are within hydrogen-bonding distance of highly conserved residues in H2, H6, and H7. Waters 11, 12, and 13 are part of an extended network of waters that may constitute the communication pathway from regions adjacent to the ligand-binding pocket to the cytoplasmic face of the receptors. (C) Water 6, unique to bovine rhodopsin, interacts with the Schiff base counterion E113; this water potentially plays a role in Schiff base stabilization and proton transfer during photoactivation and interconversion between MI and MII. (D) Water cluster 9, found in 5 of the 6 GPCR structures near the highly conserved WxP6.50F/Y motif.
The initial crystal structure of rhodopsin revealed the presence of a single ordered water buried within the transmembrane domain (1, 3) whereas 12 ordered waters were localized in and around the transmembrane domain in the highest resolution X-ray crystal structure of ground-state rhodopsin [Protein Data Bank (PDB) ID code 1u19] (4), and the structural and functional importance of some of these waters has been addressed recently (28). A comparison of five ground-state rhodopsin crystal structures at resolutions of 2.8 Å or better revealed waters or hydroxyl groups that colocalized with 8 waters modeled in the 2.2-Å structure (PDB ID code 1u19). Thus, for rhodopsin, we focused our comparisons on these 8 waters. Five of the 8 water molecules seen in multiple rhodopsin structures contacted residues linked to human autosomal dominant retinitis pigmentosa, and ordered waters overall hydrogen-bonded with 7 residues associated with this blinding disease (G902.57w6, E1814.70w8, C1874.76w8, G1884.77w8, S1864.75w8, C2646.47w9, and S2987.45w12).
The structure of squid rhodopsin revealed 13 waters of which 3 colocalized with waters in bovine rhodopsin and 3 colocalized with waters in the mutant β2-adrenergic receptor (Fig. 1). The A2A-adenosine receptor structure includes 15 ordered waters, 7 of which were in a solvent-accessible cavity and 8 were positioned within the transmembrane domain. Four waters in the transmembrane domain of the A2A-adenosine receptor colocalized with waters in bovine, squid, and the mutant β2-adrenergic receptor. The mutant β2-adrenergic structures evidenced 9 ordered waters that colocalized either with waters in bovine rhodopsin or squid rhodopsin; 2 of these ordered waters were noted within the helical bundle of the mutant β1-adrenergic receptor. There were 5 ordered waters in the structure of the opsin (PDB ID code 3cap) and 3 in the structure of opsin with Gt peptide analog bound (PDB ID code 3dqb), but these waters did not colocalize with waters observed in other rhodopsin crystal structures (10, 11). Water contacts for GPCR structures are summarized in Figs. 2 and 3 and Table S1.
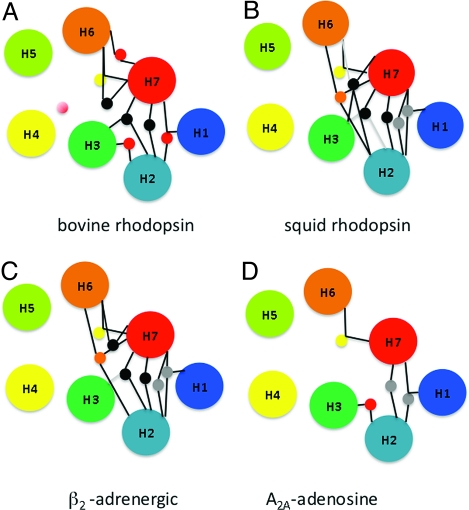
Fig. 3.
Conservation of hydrogen-bonding networks. Colocalized waters present in the crystal structures of bovine rhodopsin (A), squid rhodopsin (B), mutant β2-adrenergic receptor (C), and A2A-adenosine receptor (D) make contact with highly conserved amino acid residues. The lines connecting waters and helices indicate putative hydrogen bonds. For water clusters, yellow circles indicate conserved networks shared by all receptors, gray circles indicate interactions shared among squid rhodopsin, mutant β2-adrenergic, and A2A-adrenergic receptor, black circles indicate conserved networks shared among bovine rhodopsin, squid rhodopsin, and mutant β2-adrenergic receptor, red circles indicate conserved networks shared between bovine rhodopsin and mutant A2A-adenosine receptor, and finally, orange circles indicate conserved networks shared between squid rhodopsin and mutant β2-adrenergic receptor.
In this section we summarize the interactions made by several of the waters in the 15 water clusters. By employing the CONTACT module of the CCP4 program suite (29), we assigned hydrogen bonding or polar interactions when a water molecule was found to be <3.8 Å away from a nitrogen or oxygen in either the side chain or Cα backbone of the receptor (29).
Water clusters 1, 2, 3, 12, and 13 all make contact with the NPXXY motif found in helix VII. Water 1 is unique to rhodopsin and made contacts with T621.57, N732.40 (40% conservation in family A GPCRs), and Y3067.53 (92% conservation in family A GPCRs) of the NPXXY motif. Mutagenesis data indicate that residues contacting water 1 are functionally important in rhodopsin (30). Interestingly, the Y3067.53 to Ala mutation evidenced an increased stability of MII formation that did not correlate with an increase in G protein activation (31). Water cluster 2 consisted of 3 waters (1 from the mutant β2-adrenergic receptor, 1 from the mutant A2A-adenosine receptor, and 1 from squid rhodopsin) that made contacts with I/V2.42, A2.47, and T/Y7.53 connecting helices II and VII. Functional studies of the β2-adrenergic receptor revealed that the Y7.53 to Ala mutation exhibited a reduction in phosphorylation and receptor internalization and reduced G protein coupling (32, 33). Mutagenesis data for both rhodopsin and the β2-adrenergic receptor suggest that Y7.53 of the NPXXY motif contributes to both G protein activation and desensitization processes. Water cluster 3 contained 3 waters, 1 from the mutant β2-adrenergic receptor, 1 from the mutant A2A-adenosine receptor, and 1 from squid rhodopsin. Conserved contacts with side chains of residues N1.50, D2.50, and N7.49 mediate linkage among helices I, II, and VII similar to contacts maintained by water 1 in bovine rhodopsin. Mutation of D2.50 to Asn was reported to reduce epinephrine-induced cAMP accumulation; this was interpreted as a constitutively inactivating mutation and demonstrates the functional importance of D2.50 (34).
Water cluster 12 includes 4 waters, 2 from bovine rhodopsin, 1 from mutant β2-adrenergic receptor, and 1 from squid rhodopsin. These waters extend the network of connections between helix II and VII for the mutant β2-adrenergic receptor and squid rhodopsin with water–water contacts bridging the helix contacts. The hydrogen-bonding network mediated by water cluster 12 in bovine rhodopsin is among helices II, III, VI, and VII. Contacts are seen between water 12a (atom number 2020 in PDB ID code 1u19) and residues M2576.4, Y3017.48, and S2987.45 linking the WXPF/Y switch region with the NPXXY motif. The second water in cluster 12 present in the rhodopsin crystal structure, 12b (atom number 2030 in PDB ID code 1u19), makes contacts with D832.50, G1203.36, S2987.45, and N3027.49, connecting highly conserved residues in transmembrane helices I and VII. The linkage between D832.50 and N3027.49 in the NPXXY motif near cytoplasmic helix 8 has been shown to change upon photoactivation (35). This linkage is mediated in part by the ordered waters in cluster 12 with conserved contacts between water and residue D832.50 and S/N7.45 present in all three structures. FTIR studies of bovine rhodopsin revealed that water that contributes to an interactive network between D832.50, G1203.35, and E1223.37, and water 12b in rhodopsin is likely to be the water observed in these FTIR studies (36).
Water 13 is found in bovine and squid rhodopsin, the opsin structure with peptide-bound, mutant β2-adrenergic receptor, and mutant A2A-adenosine receptor. In squid rhodopsin, mutant β2-adrenergic and mutant A2A-adenosine receptors, water 13 is part of a network of waters stabilizing interactions, primarily between helix VI and helix VII and secondarily via water–water bonding to helix II. Water 13 has a conserved interaction with residue N3027.49 of the NPXXY motif, again likely to stabilize the inactive state of the receptors.
Cluster 4 was composed of waters from bovine rhodopsin, squid rhodopsin, and the mutant β2-adrenergic receptor (Fig. 2 A and B). Water 4 mediates contacts between helices II and VII of bovine rhodopsin through residues D832.50 (94% conservation in family A GPCRs) and S2987.45, V3007.47, and N3027.49 (75% conservation in family A GPCRs). Mutation of D832.50 favored the formation of Meta II (37, 38); FTIR studies additionally suggest that D832.50 is protonated in both rhodopsin and Meta II (39). Thus, water is likely defining the functional role of this residue rather than an intrinsic property such as protonation state. Residue D832.50 appears to make contact with waters in this cluster and may function as a nucleation sight for an extended hydrogen-bonding network evident in bovine rhodopsin, squid rhodopsin, and the mutant β2-adrenergic receptor. Water 4 was found to be within 3.8 Å of waters in clusters 12 and 13 in the rhodopsin structure (Fig. 2B), possibly constituting part of a water wire similar to the water organization in bacteriorhodopsin responsible for proton transfer occurring during the photo cycle of this evolutionarily distant but related chromophore-binding, integral membrane protein.
Water 6, noted in bovine rhodopsin and the A2A-adenosine receptor, was localized near the ligand-binding pocket of bovine rhodopsin and made contacts with E1133.28, G902.57, and F912.58. In the A2A-adenosine receptor, water 6 made contacts with I3.28, F3.31, and V3.32. Importantly, ligand interaction with the conserved negatively charged Asp at position 3.32 in biogenic amine receptors and an analogous hydrophobic interaction was observed between ligand and residue 3.32 in the A2A-adenosine receptor structure (9). Functional importance of G902.57 was demonstrated when mutation to Asp in rhodopsin reportedly led to constitutive activation of Gt (16). Interestingly, the G902.57 to Asp mutation in human rhodopsin has been reported to cause congenital night blindness, highlighting the importance of this region for proper receptor function (40). Extensive site-directed mutagenesis studies indicated that residue E1133.28 is the counterion for the Schiff base linkage and that it functions as the proton acceptor during the Schiff base deprotonation that occurs during photoactivation.
Strikingly, water 9 was found in five of the six other GPCR structures (Fig. 2D). Water cluster 9 made conserved main-chain contacts with residues at positions C2646.47 (74% conservation), Y2686.51 (36% conservation), and P2917.38 in bovine rhodopsin, squid rhodopsin, mutant A2A-adenosine, mutant β1- and mutant β2-adrenergic receptors. Water cluster 9 was found adjacent to the ligand-binding pocket and near the conserved WxP6.50F/Y motif; main-chain hydrogen bonding likely stabilizes the Pro kink and allows W6.48 to be held away from the chromophore-binding pocket (Figs. S2 and S3). Inspection of opsin structures lacking this water reveals that W2656.48 is localized toward the empty chromophore-binding pocket and away from the position where water 9 is found in the other five structures (Figs. S2 and S3). Ligand-binding studies have revealed that residue 6.47 in constitutively active mutant β2-adrenergic receptor constructs is more accessible to sulfhydryl-reactive reagents, implicating a rearrangement in helix VI after the receptor attains an active conformation (41). Mutation of Y2686.51 to phenylalanine in rhodopsin effected spectral changes and an increased rate of Schiff base hydrolysis after photoactivation (42).
In squid rhodopsin and mutant β2-adrenergic receptor structures, waters from clusters 2, 3, 4, 11, 12, and 13 are part of extended hydrogen-bonded water networks, often referred to as water wires (Fig. S4). These extended water networks may represent part of a pathway connecting a proton donor site on the cytoplasmic face of the receptor with an acceptor site buried within the helical bundle. Water wires permit a more rapid and controlled transmission of protons than diffusion (43). The apparent absence of these networks in other structures may simply be the result of their lack of sufficient electron density.
Inspection of the waters bound in the five different GPCR structures reveals that more than half of the observed interactions occurred between amino acid side chains and water. This high level of side-chain interactions likely reflects an evolutionary conservation of structural and functional motifs because these waters interact with highly conserved residues. These ordered waters colocalize and mediate contacts between highly conserved residues stabilizing structurally and functionally important helix–helix interactive networks. Seven of the 8 waters present in the bovine rhodopsin structure colocalized with waters in structures of squid rhodopsin, mutant β2-adrenergic, and mutant A2A-adenosine receptors. Waters connecting helices II and VII, VI and VII, and I, II, and VII were shared among bovine rhodopsin, squid rhodopsin, and the mutant β2-adrenergic receptor (Fig. 3). Waters mediating connections between helices I, II, and VII as well as helices II and VII were common to bovine, squid rhodopsin, and mutant A2A-adenosine receptors. Along with the extended water wire, squid rhodopsin and mutant β2-adrenergic receptor both showed the presence of water mediating an interhelical interaction among helices II, VI, and VII. In bovine rhodopsin, mutations of residues that make contact with waters 9 and 12 are both associated with retinal disease. Given that water 9 contacts residues that line the ligand-binding pocket and participate in conformational changes during receptor activation, it seems likely that this water plays more than just a static structural role. Although ordered waters bound in different GPCRs may either be structurally or functionally important, it is difficult to distinguish between these two roles for any of the waters found in the available structures.
Discussion
Changes upon Receptor Activation.
In the absence of high-resolution crystal structures of activated states of any GPCR, we must infer changes that coincide with receptor activation from the existing biochemical and biophysical data. Many lines of evidence support the notion that there are conformational alterations in the helical bundle of GPCRs after activation. However, the nature and magnitude of these conformational changes during receptor activation are the subject of much debate. Formation of the active state of rhodopsin has been shown to be sensitive to the hydration state of this receptor (24, 44), implicating the importance of water in the activation process. Early biochemical studies indicated that activation of rhodopsin involved the movement of two protons (45); however, the role of the embedded waters in the proton sensitivity is unclear.
In addition to release of the ionic lock, changes in the interhelical interactive network involving the highly conserved NPXXY motif linking residues of helices VI and VII occur after receptor activation (31), and changes in this network are likely mediated in part by the observed embedded bound waters (5, 28). Along with bovine rhodopsin, β2-adrenergic receptor (46–48) and 5HT1 receptor (49) activation was shown to involve rotation of helix VI.
Early studies of rhodopsin detected mobility changes of helix III near the cytoplasmic face after light excitation, as evidenced by increased spin label mobility and chemical reactivity of Cys-140 upon receptor activation (50). In the photoactivated crystal structure of rhodopsin, there is increased disorder in several of the subunits in the region containing residue E1223.37 that correlate with mechanistic components of receptor activation. Radiolytic footprinting studies of rhodopsin revealed that M862.53 near E1223.37, close to structural waters in the transmembrane bundle, became more mobile in the photoactivated state, possibly reflecting changes the location of the internal waters, strongly suggesting that the increased disorder observed in the crystal structure is indeed a result of photoactivation. Solid-state NMR spectroscopic measurements indicated that little to no movement of helix III occurred after photoactivation (51).
The highly conserved E/DRY motif forming the “ionic lock” at the cytoplasmic end of helix III is thought to hold the receptor in the inactive state by R1353.50 forming a salt bridge with E2476.32 in helix VI that holds helices III and VI together. It had been shown that disruption of the ionic lock in the β2-adrenergic receptor leads to receptor activation, indicating that the ionic lock motif is functionally conserved. Recent work has demonstrated the critical role of E1343.49 and R1353.50 of the conserved ERY motif in rhodopsin activation (52). However, the mechanism linking protonation/deprotonation of the ionic lock to protonation/deprotonation of the Schiff base linkage is still unknown. In the case of rhodopsin, with the exception of K296 covalently modified with chromophore and residues E1223.37 on helix III and H2115.46 on helix V, which are involved in a salt bridge, all other charged residues in the transmembrane domain make contacts with waters. Are bound waters within the helical bundle facilitating those changes responsible for the observed pH-sensitive interconversion between inactive Meta I and active Meta II? It is tempting to speculate that these events are mediated by waters acting as allosteric effectors and by bound waters transferring protons via water wires.
Changes in Water Organization and Hydration After Activation.
Several lines of experimental evidence support the idea of changes in the state of the bound waters after photoactivation of rhodopsin. Changes in the hydrogen-bonding network after photoactivation are evident from studies using NMR, resonance Raman, and FTIR spectroscopy; however, little is know regarding changes in water-mediated contacts with respect to other members of the GPCR superfamily. Deng et al. (53) used continuous-flow resonance Raman spectroscopy to demonstrate that the Schiff base can be deuterated in unactivated rhodopsin. This study suggests the presence of 1 or 2 waters near or hydrogen-bonded to the Schiff base linkage, consistent with waters 6, 8, and 9 found in proximity to the chromophore. The mechanism of deuterium uptake is unlikely because of bulk solvent penetration and exchange if the methodologies developed to measure water interactions by FTIR are considered; these require dehydration of rhodopsin films and subsequent rehydration to drive water into the helical bundle. Proton transfer through amino acid side chains and water is likely to explain deuteration of the Schiff base linkage. Rath et al. (54) have shown that films of unactivated rhodopsin undergo very slow deuterium exchange when rehydrated in the presence of D2O and that the exchange at amide II bonds increased after photoactivation, consistent with increased disorder in the polypeptide chain after activation.
Changes in the amount of water present in the helical bundle have been measured by several different methods. Monitoring the effects of active osmolytes and ethanol on the Meta I/Meta II equilibrium demonstrated that hydrogen-bonded water is released from rhodopsin upon transition from Meta I to Meta II (55, 56). FTIR studies have revealed changes in the interaction of internal water molecules in rhodopsin during photoactivation that involved loss of bound water after formation of bathorhodopsin (57). Mutagenesis studies indicating required freedom of motion for residues in the NPXXY motif for G protein activation are consistent with the release of water that ablates structural constraints holding the receptor in the “off” state. Considering the demonstrated requirement for waters in forming Meta II and the effects of osmolytes, these results suggest that a change in active water occurs that is required for Meta II formation.
Solid-state NMR spectroscopy has indicated that interactions between G1213.36 and W2656.48 are lost after rhodopsin activation. Additionally, inspection of the crystal structures of opsin lacking water 9 that interacts with residues adjacent to W2656.48 revealed that the loss of this water coincides with the structural reorganization that occurs near W2656.48 in the opsin (Figs. S2 and S3). Other small changes in the interaction between M862.53 and G1203.35 also occur after photoactivaton (51).
In recent studies employing the radiolytic footprinting method developed by Chance et al. (58), we have demonstrated that waters bound within the transmembrane domain of rhodopsin can be activated and used to monitor the dynamics of certain regions within the transmembrane regions of this receptor, including studies of photoactivated states. We also have monitored the communication between bulk water and structural waters in ground, photoactivated, and opsin states. Hydroxyl radical labeling of rhodopsin in native disk membranes or affinity-purified receptor in the presence of rapidly mixed 50% O18 labeled water resulted in minimal exchange of water from the bulk solvent to the core for any of these three states. These results strongly suggest that the helical bundle of rhodopsin does not communicate with bulk solvent at the cytoplasmic face of the receptor on the time scale of activation. Local changes in structural constraints, including reorganization of structural waters, appear to mediate the conformational changes that occur in rhodopsin after photoactivation and mediate allosteric communication. These data are consistent with changes in side-chain interactions detected by solid-state NMR spectroscopy (51) because the rate and extent of M862.53 hydroxylation were increased in Meta II relative to unactivated rhodopsin. Water may leave the core of the protein before Meta II formation, and some bound waters may be required for Meta II formation, but bulk water uptake is not required for receptor activation. Major changes in hydration and hydrogen bonding after photoactivation are likely to be limited to movement of waters already bound within the receptor in its ground state, including the conserved waters identified here. Constitutively activating mutations of the opioid receptor are located in groups similar in distribution to the water clusters described here (59) and the regions of rhodopsin that are labeled by hydroxyl radicals. Constraints holding rhodopsin, and by extension all family A GPCRs, in ground-state conformations are likely to be mediated by waters bridging contacts between highly conserved motifs (28), and changes in water contacts may facilitate communication from the ligand-binding pocket to the cytoplasmic face where G protein binding and activation occur.
Conclusions
Inspection of available high-resolution crystal structures provides insights into common structural features that may be linked to conserved functions shared by all GPCRs. Colocalization of waters found in several different GPCR crystal structures and the putative interaction of such waters with highly conserved and functionally important residues support the notion that these waters are functionally important and are as likely as their conserved residue partners to be essential to the mechanism of activation. Ordered waters interact with residues that are important in disease, receptor activation, and signaling. In the absence of ligand, as in the case of bovine opsin, fewer ordered waters are bound to the receptor. This likely reflects an important role for release of tightly bound waters upon receptor activation, even though the extent of water reorganization relative to crystallographically identified ordered waters is unknown. Improved understanding of the role of these water molecules should shed light on conserved features involved in receptor stabilization, ligand binding, and activation. The presence of ordered, bound waters in many of the available structures suggests that these waters mediate the structural plasticity linked to the ligand-induced shift in conformational equilibrium. Conserved ordered waters likely impose structural constraints favoring inactive states of these receptors. Reorganization of bound water may well be necessary for receptor activation, as shown for bovine rhodopsin and likely reflected by the structure of opsin. Local conformational changes have been reported in rhodopsin after photoactivation, and the regions where such changes occur are close to those regions where ordered waters are observed in the structure of ground-state rhodopsin. Water likely participates and may mediate the structural transition from inactive to active conformations of family A GPCRs, if not all receptors in this superfamily.
Methods
Multiple sequence alignments of the GPCRs analyzed were made employing ClustalW (60). Analysis of water contacts in structures was done by using the CONTACT module of the CCP4 program suite (29). The following crystal structures were examined; 1u19, 1f88, 1gzm, 1hzx, 1l9h, 3cap, 3dqb, 2rh1, 2vt4, 2z73, 3eml, and interactions between water and protein residues were assigned if nitrogen and/or oxygen atoms of the protein were 3.8 Å or less from the oxygen of water. All crystallographic models in figures were rendered employing PyMOL.
Supplementary Material
Acknowledgments.
We thank Dr. Marta Filizola, Dr. Marcin Golczak, Dr. David T. Lodowski, Philip Kisier, and Dr. Leslie T. Webster, Jr., for critical comments on the manuscript. This work was supported by National Institutes of Health Grants EY09339, GM079191, and EB01979. T.E.A was supported by grant T32EY007157.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903545106/DCSupplemental.
References
- 1.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 3.Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada T, et al. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 5.Okada T, et al. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: A perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 7.Salom D, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci USA. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaakola VP, et al. The 2.6 ångström crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 11.Scheerer P, et al. Crystal structure of opsin in its G protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 12.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 13.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen SG, et al. Crystal structure of the human β2-adrenergic G protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 15.Warne T, et al. Structure of a β1-adrenergic G protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madabushi S, et al. Evolutionary trace of G protein-coupled receptors reveals clusters of residues that determine global and class-specific functions. J Biol Chem. 2004;279:8126–8132. doi: 10.1074/jbc.M312671200. [DOI] [PubMed] [Google Scholar]
- 17.Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobilka B, Schertler GF. New G protein-coupled receptor crystal structures: Insights and limitations. Trends Pharmacol Sci. 2008;29:79–83. doi: 10.1016/j.tips.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Melia TJ, Jr, Cowan CW, Angleson JK, Wensel TG. A comparison of the efficiency of G protein activation by ligand-free and light-activated forms of rhodopsin. Biophys J. 1997;73:3182–3191. doi: 10.1016/S0006-3495(97)78344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jager S, Palczewski K, Hofmann KP. Opsin/all-trans–retinal complex activates transducin by different mechanisms than photolyzed rhodopsin. Biochemistry. 1996;35:2901–2908. doi: 10.1021/bi9524068. [DOI] [PubMed] [Google Scholar]
- 21.Kobilka BK, et al. Chimeric α2-,β2-adrenergic receptors: delineation of domains involved in effector coupling and ligand binding specificity. Science. 1988;240:1310–1316. doi: 10.1126/science.2836950. [DOI] [PubMed] [Google Scholar]
- 22.Angel TE, Kraft PC, Dratz EA. Metarhodopsin-II stabilization by cross-linked Gtα C-terminal peptides and implications for the mechanism of GPCR-G protein coupling. Vision Res. 2006;46:4547–4555. doi: 10.1016/j.visres.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Ball P. Water as an active constituent in cell biology. Chem Rev. 2008;108:74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- 24.Wald G, Durell J, St George CC. The light reaction in the bleaching of rhodopsin. Science. 1950;111:179–181. doi: 10.1126/science.111.2877.179. [DOI] [PubMed] [Google Scholar]
- 25.Royer WE, Jr, Pardanani A, Gibson QH, Peterson ES, Friedman JM. Ordered water molecules as key allosteric mediators in a cooperative dimeric hemoglobin. Proc Natl Acad Sci USA. 1996;93:14526–14531. doi: 10.1073/pnas.93.25.14526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carugo O, Bordo D. How many water molecules can be detected by protein crystallography? Acta Crystallogr D. 1999;55:479–483. doi: 10.1107/s0907444998012086. [DOI] [PubMed] [Google Scholar]
- 27.Ballesteros J, Weinstein H. Integrated methods for the construction of three-dimensional models of structure–function relations in G protein-coupled receptors. Methods Neurosci. 1985;25:366–428. [Google Scholar]
- 28.Pardo L, Deupi X, Dolker N, Lopez-Rodriguez ML, Campillo M. The role of internal water molecules in the structure and function of the rhodopsin family of G protein-coupled receptors. Chembiochem. 2007;8:19–24. doi: 10.1002/cbic.200600429. [DOI] [PubMed] [Google Scholar]
- 29.Anonymous. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Shi W, Osawa S, Dickerson CD, Weiss ER. Rhodopsin mutants discriminate sites important for the activation of rhodopsin kinase and Gt. J Biol Chem. 1995;270:2112–2119. doi: 10.1074/jbc.270.5.2112. [DOI] [PubMed] [Google Scholar]
- 31.Fritze O, et al. Role of the conserved NPxxY(x) 5,6F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci USA. 2003;100:2290–2295. doi: 10.1073/pnas.0435715100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferguson SS, et al. Role of phosphorylation in agonist-promoted β2-adrenergic receptor sequestration: Rescue of a sequestration-defective mutant receptor by β-ARK1. J Biol Chem. 1995;270:24782–24789. doi: 10.1074/jbc.270.42.24782. [DOI] [PubMed] [Google Scholar]
- 33.Menard L, et al. Members of the G protein-coupled receptor kinase family that phosphorylate the β2-adrenergic receptor facilitate sequestration. Biochemistry. 1996;35:4155–4160. doi: 10.1021/bi952961+. [DOI] [PubMed] [Google Scholar]
- 34.Liapakis G, Chan WC, Papadokostaki M, Javitch JA. Synergistic contributions of the functional groups of epinephrine to its affinity and efficacy at the β2-adrenergic receptor. Mol Pharmacol. 2004;65:1181–1190. doi: 10.1124/mol.65.5.1181. [DOI] [PubMed] [Google Scholar]
- 35.Lehmann N, Alexiev U, Fahmy K. Linkage between the intramembrane H-bond network around aspartic acid 83 and the cytosolic environment of helix 8 in photoactivated rhodopsin. J Mol Biol. 2007;366:1129–1141. doi: 10.1016/j.jmb.2006.11.098. [DOI] [PubMed] [Google Scholar]
- 36.Nagata T, Terakita A, Kandori H, Shichida Y, Maeda A. The hydrogen-bonding network of water molecules and the peptide backbone in the region connecting Asp83, Gly120, and Glu113 in bovine rhodopsin. Biochemistry. 1998;37:17216–17222. doi: 10.1021/bi9810149. [DOI] [PubMed] [Google Scholar]
- 37.Weitz CJ, Nathans J. Rhodopsin activation: Effects on the metarhodopsin I–metarhodopsin II equilibrium of neutralization or introduction of charged amino acids within putative transmembrane segments. Biochemistry. 1993;32:14176–14182. doi: 10.1021/bi00214a016. [DOI] [PubMed] [Google Scholar]
- 38.DeCaluwe GL, Bovee-Geurts PH, Rath P, Rothschild KJ, de Grip WJ. Effect of carboxyl mutations on functional properties of bovine rhodopsin. Biophys Chem. 1995;56:79–87. doi: 10.1016/0301-4622(95)00018-s. [DOI] [PubMed] [Google Scholar]
- 39.Fahmy K, et al. Protonation states of membrane-embedded carboxylic acid groups in rhodopsin and metarhodopsin II: A Fourier-transform infrared spectroscopy study of site-directed mutants. Proc Natl Acad Sci USA. 1993;90:10206–10210. doi: 10.1073/pnas.90.21.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zvyaga TA, Fahmy K, Siebert F, Sakmar TP. Characterization of the mutant visual pigment responsible for congenital night blindness: A biochemical and Fourier-transform infrared spectroscopy study. Biochemistry. 1996;35:7536–7545. doi: 10.1021/bi960391n. [DOI] [PubMed] [Google Scholar]
- 41.Javitch JA, Fu D, Liapakis G, Chen J. Constitutive activation of the β2-adrenergic receptor alters the orientation of its sixth membrane-spanning segment. J Biol Chem. 1997;272:18546–18549. doi: 10.1074/jbc.272.30.18546. [DOI] [PubMed] [Google Scholar]
- 42.Ridge KD, Bhattacharya S, Nakayama TA, Khorana HG. Light-stable rhodopsin. II. An opsin mutant (Trp-265–Phe) and a retinal analog with a nonisomerizable 11-cis configuration form a photostable chromophore. J Biol Chem. 1992;267:6770–6775. [PubMed] [Google Scholar]
- 43.Chaplin M. Do we underestimate the importance of water in cell biology? Nat Rev Mol Cell Biol. 2006;7:861–866. doi: 10.1038/nrm2021. [DOI] [PubMed] [Google Scholar]
- 44.Nishimura S, Sasaki J, Kandori H, Lugtenburg J, Maeda A. Structural changes in the lumirhodopsin-to-metarhodopsin I conversion of air-dried bovine rhodopsin. Biochemistry. 1995;34:16758–16763. doi: 10.1021/bi00051a025. [DOI] [PubMed] [Google Scholar]
- 45.Arnis S, Hofmann KP. Two different forms of metarhodopsin II: Schiff base deprotonation precedes proton uptake and signaling state. Proc Natl Acad Sci USA. 1993;90:7849–7853. doi: 10.1073/pnas.90.16.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gether U, et al. Agonists induce conformational changes in transmembrane domains III and VI of the β2-adrenoceptor. EMBO J. 1997;16:6737–6747. doi: 10.1093/emboj/16.22.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jensen AD, et al. Agonist-induced conformational changes at the cytoplasmic side of transmembrane segment 6 in the β2-adrenergic receptor mapped by site-selective fluorescent labeling. J Biol Chem. 2001;276:9279–9290. doi: 10.1074/jbc.M004871200. [DOI] [PubMed] [Google Scholar]
- 48.Sheikh SP, et al. Similar structures and shared switch mechanisms of the β2-adrenoceptor and the parathyroid hormone receptor: Zn(II) bridges between helices III and VI block activation. J Biol Chem. 1999;274:17033–17041. doi: 10.1074/jbc.274.24.17033. [DOI] [PubMed] [Google Scholar]
- 49.Shapiro DA, Kristiansen K, Weiner DM, Kroeze WK, Roth BL. Evidence for a model of agonist-induced activation of 5-hydroxytryptamine 2A serotonin receptors that involves the disruption of a strong ionic interaction between helices 3 and 6. J Biol Chem. 2002;277:11441–11449. doi: 10.1074/jbc.M111675200. [DOI] [PubMed] [Google Scholar]
- 50.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 51.Crocker E, et al. Location of Trp265 in metarhodopsin II: Implications for the activation mechanism of the visual receptor rhodopsin. J Mol Biol. 2006;357:163–172. doi: 10.1016/j.jmb.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 52.Vogel R, et al. Functional role of the “ionic lock”: An interhelical hydrogen-bond network in family A heptahelical receptors. J Mol Biol. 2008;380:648–655. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 53.Deng H, Huang L, Callender R, Ebrey T. Evidence for a bound water molecule next to the retinal Schiff base in bacteriorhodopsin and rhodopsin: A resonance Raman study of the Schiff base hydrogen/deuterium exchange. Biophys J. 1994;66:1129–1136. doi: 10.1016/S0006-3495(94)80893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rath P, DeGrip WJ, Rothschild KJ. Photoactivation of rhodopsin causes an increased hydrogen–deuterium exchange of buried peptide groups. Biophys J. 1998;74:192–198. doi: 10.1016/S0006-3495(98)77779-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitchell DC, Litman BJ. Effect of protein hydration on receptor conformation: Decreased levels of bound water promote metarhodopsin II formation. Biochemistry. 1999;38:7617–7623. doi: 10.1021/bi990634m. [DOI] [PubMed] [Google Scholar]
- 56.Mitchell DC, Litman BJ. Effect of ethanol and osmotic stress on receptor conformation: Reduced water activity amplifies the effect of ethanol on metarhodopsin II formation. J Biol Chem. 2000;275:5355–5360. doi: 10.1074/jbc.275.8.5355. [DOI] [PubMed] [Google Scholar]
- 57.Furutani Y, Shichida Y, Kandori H. Structural changes of water molecules during the photoactivation processes in bovine rhodopsin. Biochemistry. 2003;42:9619–9625. doi: 10.1021/bi034592k. [DOI] [PubMed] [Google Scholar]
- 58.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 59.Decaillot FM, et al. Opioid receptor random mutagenesis reveals a mechanism for G protein-coupled receptor activation. Nat Struct Biol. 2003;10:629–636. doi: 10.1038/nsb950. [DOI] [PubMed] [Google Scholar]
- 60.Larkin MA, et al. ClustalW and ClustalX version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.