Abstract
Myosin VIIA is an unconventional myosin, responsible for human Usher syndrome type 1B, which causes hearing and visual loss. Here, we studied the molecular mechanism of regulation of myosin VIIA, which is currently unknown. Although it was originally thought that myosin VIIA is a dimeric myosin, our electron microscopic (EM) observations revealed that full-length Drosophila myosin VIIA (DM7A) is a monomer. Interestingly, the tail domain markedly inhibits the actin-activated ATPase activity of tailless DM7A at low Ca2+ but not high Ca2+. By examining various deletion constructs, we found that deletion of the distal IQ domain, the C-terminal region of the tail, and the N-terminal region of the tail abolishes the tail-induced inhibition of ATPase activity. Single-particle EM analysis of full-length DM7A at low Ca2+ suggests that the tail folds back on to the head, where it contacts both the motor core domain and the neck domain, forming an inhibited conformation. We concluded that unconventional myosin that may be present a monomer in the cell can be regulated by intramolecular interaction of the tail with the head.
Keywords: molecular motor, regulation, unconventional myosin, Usher syndrome type 1B, EM images
Myosin is a mechanoenzyme that was originally found in muscle to produce force by converting the chemical energy of ATP to a mechanical work. Although myosin was originally thought to be a force producer, recent studies have revealed that it can also function as a cargo transporter (1, 2). Of the 2 isoforms of myosin VII, most studies have been carried out on myosin VIIA because of its involvement in human auditory disease, Usher syndrome type 1B (3). In sensory hair cells, myosin VIIA is found in the pericuticular necklace and along the stereocilia and is thought to be involved in vesicle transport (4). Myosin VIIA is also present in the retina and may play a role in opsin transportation from the inner to the outer segment of photoreceptor cells (5). Myosin VIIA plays a role in phagocytosis of shed outer segments of photoreceptor cells (6, 7). It is therefore likely that it functions as a cargo transporter and force producer in distinct cellular components.
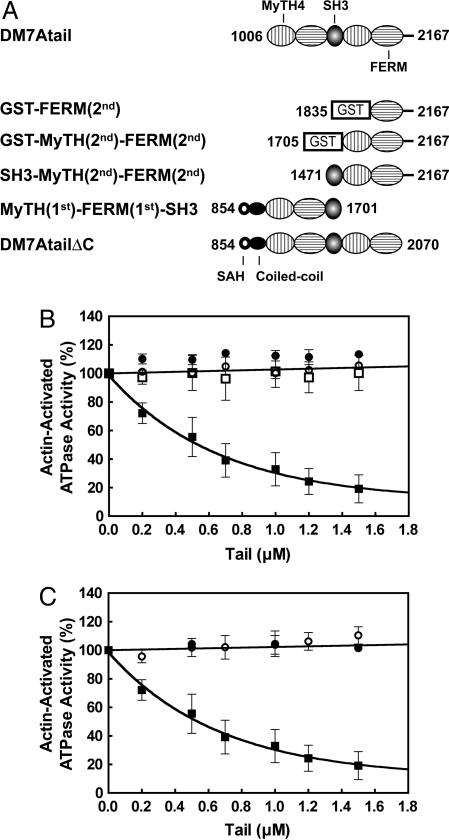
Myosin VIIA has a domain structure common to all myosin superfamily members. A conserved motor domain is present in its N-terminal region, which is followed by a neck domain consisting of 5 IQ motifs (isoleucine–glutamine motifs) that serve as light chain binding site (Fig. 1A). Similar to other myosin family members (8, 9), calmodulin copurifies with myosin VIIA (10) and is thought to function as the light chain. The tail consists of a proximal short α-helical region followed by a globular domain. A part of the α-helical region is homologous to the stable single α-helix (SAH) domain found in myosin X (11) and probably does not form a coiled-coil structure, but rather serves as an extension module enabling a long step size. The distal short coiled-coil region may serve for dimer formation, although there is no hard evidence that myosin VIIA forms a 2-headed structure. The globular tail domain contains 2 large repeats, each incorporating a MyTH4 (myosin tail homology 4) domain and a FERM (band 4.1-ezrin-radixin-moesin) domain (12).
Fig. 1.
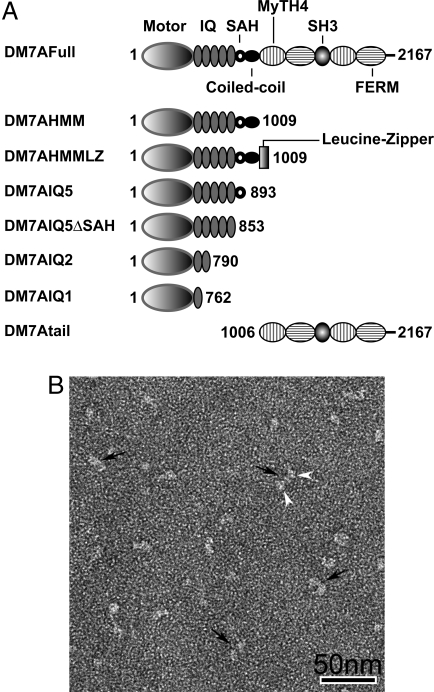
DM7A constructs and subunit structure of myosin VIIA. (A) Schematic diagram of the DM7A constructs. The amino acid numbers of the constructs are indicated. (B) General appearance of DM7AHMMLZ, showing dimeric conformation. White arrowheads point to 2-pear shaped heads of individual molecules (black arrows). (Scale bar: 50 nm.)
Motor activity was first demonstrated in rat myosin VIIA to be a plus-end-directed myosin whose ATPase activity was activated >10-fold by actin (13). It was shown recently that Drosophila myosin VIIA is a high-duty ratio motor, in which the head spends most of its cross-bridge cycle time in strong-actin-binding form and is thus suitable for “processive” movement (14). Consistent with this finding, the tail-truncated myosin VIIA can move processively on actin when constructed with a leucine zipper module to force dimerization (15).
One of the most important aspects of myosin physiological function is how it is regulated. The regulation of unconventional myosin is best understood for myosin Va, where activity of myosin Va is closely correlated with a major conformational transition, which is regulated by Ca2+ binding to the calmodulin light chain (16–18). Nothing is known about myosin VIIA regulation. A critical issue is whether the molecule is dimeric, because this may affect both its regulation and processive movement (cf. ref. 19).
Here, we have studied the regulation mechanism of Drosophila myosin VIIA. We found that the full-length Drosophila myosin VIIA (DM7A) molecule is predominantly a monomer and that the globular tail domain interacts with the head and IQ domains to form a folded compact structure, which correlates with inhibition of the actin-activated ATP hydrolysis cycle.
Results
Expression and Purification of DM7A Constructs.
We produced a series of truncated DM7A constructs to study the regulation mechanism (Fig. 1A). The isolated DM7A heavy chain was copurified with calmodulin light chain (Fig. S1). The apparent molecular mass of each constructs determined from its mobility in SDS/PAGE was 250, 116, 120, 103, 98, 90, 87, and 120 kDa for DM7AFull, DM7AHMM, DM7AHMMLZ, DM7AIQ5, DM7AIQ5ΔSAH DM7AIQ2, DM7AIQ1 and DM7Atail, respectively. These values agree well with the calculated molecular masses of these constructs.
The Tail Domain Inhibits the ATPase Activity of DM7AHMM.
Because myosin VIIA contains a predicted coiled-coil domain in the proximal tail region, it has been thought to form a 2-headed molecule like myosin Va (19). We examined whether DM7A formed a stable 2-headed structure by chemical cross-linking. Fig. S2 shows SDS/PAGE of the cross-linked products. DM7AHMM containing the entire predicted coiled-coil failed to produce a cross-linked heavy-chain dimer, suggesting that DM7AHMM is predominantly monomeric. However, the DM7AHMMLZ construct, in which a leucine zipper motif was introduced at the C-terminal end, yielded a high molecular mass band corresponding to the heavy-chain dimer of DM7AHMMLZ. The result suggests that the coiled-coil domain of myosin VIIA does not hold the heavy chains together to form a dimer unlike myosin Va. Negative-staining electron microscopy (EM) revealed that DM7AHMM formed mostly single-headed structures, had a pear-like appearance, and was ≈12–15 nm in length (see Fig. 6 A and E). The structure of the dimer with the C-terminal leucine zipper, however, showed 2-pear shaped heads connected to each other (Fig. 1B), consistent with the SDS/PAGE analysis.
Fig. 6.
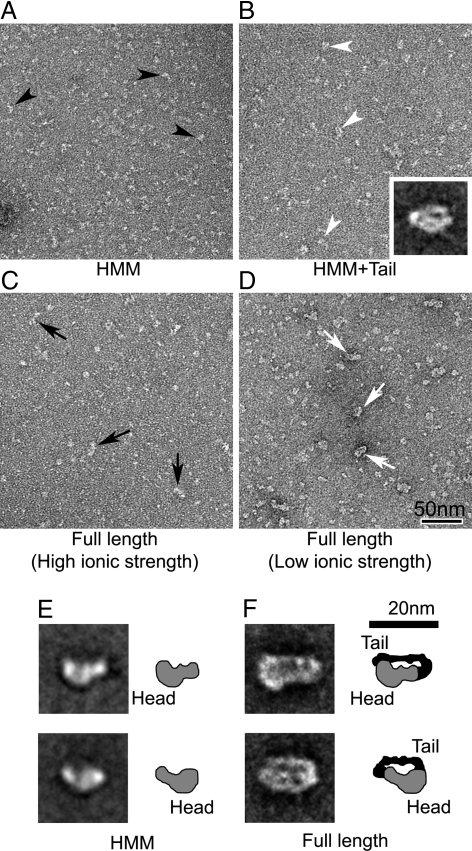
EM of DM7AHMM and DM7AFull in EGTA. (A and B) Negatively-stained fields of DM7AHMM in the absence and presence of the tail domain. Black and white arrowheads indicate individual HMM molecules and molecules interacting with the tail domain, respectively. (B Inset) Averaged image of head–tail interacting molecules (containing 37 individual images). (C and D) Fields of DM7AFull in 300 mM NaCl and 50 mM NaAcetate solution. Black and white arrows indicate the appearance of full-length molecules in the high and low ionic strength conditions, appearing narrower and wider, respectively. (E and F) Averaged images of HMM and full-length molecules (compare A and D, respectively). Averaged images in E show 2 different views (thought to represent molecules face-up and face-down on the carbon substrate), containing 104 images (Upper) and 80 images (Lower). Averaged images of full-length molecules in F, containing 21 images in each, were selected based on separation between the head and the tail domains in the closely-packed structures. Diagrams in E and F suggest possible interpretation of densities of the head (gray) and the tail domain (black), identified from the averages.
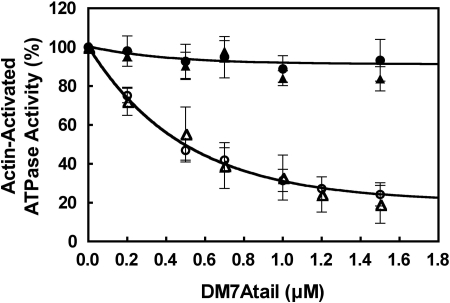
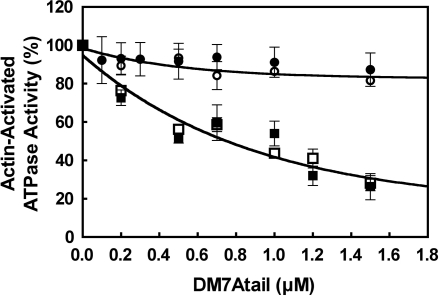
It is known that the tail domain significantly inhibits the actin-activated ATPase activity of myosin Va (19), a stable 2-headed molecule. We therefore examined whether the tail could inhibit the ATPase activity of the 2-headed construct of myosin VIIA (DM7AHMMLZ). The tail domain significantly inhibited the actin-activated ATPase activity of DM7AHMMLZ in EGTA (Fig. 2). The inhibition of ATPase activity was dose-dependent, and the apparent affinity between DM7AHMMLZ and the tail domain (0.50 μM) was obtained. This inhibition was found only in EGTA and not in the presence of 100 μM Ca2+. Next, we examined whether or not dimer formation was critical for inhibition. The ATPase activity of DM7AHMM without the C-terminal leucine zipper was again significantly inhibited by the tail domain in EGTA (Fig. 2). The apparent affinity between the tail domain and DM7AHMM was 0.67 μM, virtually identical to that for DM7AHMMLZ. These results indicate that dimer formation is not critical for the tail-induced inhibition of myosin VIIA.
Fig. 2.
Inhibition of the actin-activated ATPase activity of DM7AHMM by exogenous tail domain. Effect of exogenous tail domain (DM7Atail) on the actin-activated ATPase activity of DM7AHMMLZ and DM7AHMM is shown. ○, DM7AHMMLZ (1 mM EGTA); ●, DM7AHMMLZ (pCa4); △, DM7AHMM (1 mM EGTA); ▲, DM7AHMM (pCa4). Assay conditions are as described in Materials and Methods. Values are mean with SE from 4 independent experiments. One hundred percent activities in EGTA and pCa4 conditions were 1.4 and 1.1 s−1, respectively.
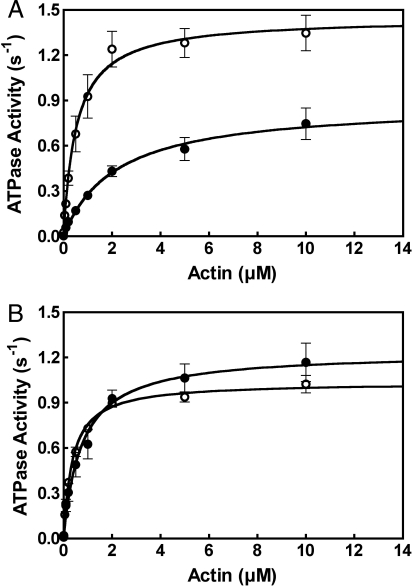
Consistent with the tail inhibition of DM7AHMM, the actin-activated ATPase activity of the full-length myosin VIIA (DM7AFull), containing its own endogenous tail, showed significantly lower ATPase activity than DM7AHMM in EGTA (Fig. 3A). However, the ATPase activities of DM7AFull and DM7AHMM in Ca2+ were virtually the same (Fig. 3B).
Fig. 3.
Decreased actin-activated ATPase activity of DM7AFull in EGTA but not in pCa4. The actin-activated ATPase activities of DM7AFull and DM7AHMM were measured as a function of actin concentration in EGTA (A) and pCa4 (B), respectively. ○, DM7AHMM; ●, DM7AFull. Values are mean with SE from 4 independent experiments. Assay conditions are as described in Materials and Methods.
The above results suggest that there is interaction of the tail domain with the head/neck domain of DM7AFull, which is abolished by Ca2+. Because calmodulin (CaM) is the light chain of DM7A, associated at the neck, we examined the pCa dependence on the tail-induced inhibition. As shown in Fig. S3A, the ATPase activity was inhibited at pCa6 or higher and abolished below pCa5. It has been shown that high Ca2+ dissociates CaM from myosin Va and myosin I. We therefore examined whether the dissociation of CaM from the DM7A heavy chain was responsible for the Ca2+-dependent regulation of actin-activated ATPase activity. DM7AHMM was incubated in the presence of 1 mM EGTA-CaCl2 buffer system, then subjected to a pull-down assay. As shown in Fig. S3B, the bound CaM of DM7AHMM (4.0 ± 0.5 mol/mol heavy chain) did not change, whereas the bound CaM of myosin Va HMM was significantly reduced, consistent with the previous report (20). These results suggest that the dissociation of the CaM is not responsible for disruption of the tail-induced inhibition. We suggest that the binding of Ca2+ induces a conformational change in the bound CaM, which influences the interaction between the tail and the head/neck of myosin VIIA. It should be noted that one of 5 IQ domains may not be occupied by CaM.
To determine whether myosin VIIA inhibition was likely to be through ionic interaction, we tested the effect of ionic strength on the tail-induced inhibition. The inhibition showed significant dependence on salt concentration, being more profound <100 mM KCl and absent at 300 mM KCl (Fig. S4). These results suggest that the inhibition depends on ionic interaction between the tail domain and the head/neck domain of DM7A. This property is similar to myosin Va, in which inhibition is achieved by interaction between basic residues in the tail and acidic residue in the head (21).
Determination of the Regions Critical for Tail Inhibition of DM7AHMM Motor Activity.
To determine the sites critical for the interaction of DM7AHMM with the tail, and thus the inhibition of ATPase activity, we produced a series of truncation mutants of DM7A (see Fig. 1A). The actin-activated ATPase activities of DM7AIQ1 and DM7AIQ2 were not inhibited by the tail domain in contrast to DM7AHMM (Fig. 4). To further clarify the elements responsible, we produced DM7A/ΔCC (DM7AIQ5) and DM7A/ΔCC/ΔSAH (DM7AIQ5ΔSAH) constructs, in which the coiled-coil and SAH are eliminated, respectively, and examined the effect of the tail domain on their ATPase activity. As shown in Fig. 4, both constructs were significantly inhibited by the tail domain with similar potency to DM7AHMM. These results indicated that the distal IQ domains are critical for tail inhibition.
Fig. 4.
Determination of the domain in DM7AHMM critical for the tail-induced inhibition of the actin-activated ATPase activity. Effect of exogenous tail domain on the actin-activated ATPase activity of various truncated DM7A constructs under EGTA conditions is shown. ○, DM7AIQ2; ●, DM7AIQ1; □, DM7AIQ5ΔSAH; ■, DM7AIQ5. Values are mean with SE from 4 independent experiments. One hundred percent represents as follows: DM7AIQ2, 1.8 s−1; DM7AIQ1, 1.7 s−1; DM7AIQ5ΔSAH, 1.3 s−1; DM7AIQ5, 1.5 s−1.
We also attempted to determine the regions in the tail critical for the inhibition. A series of deletion mutants of the tail domain was produced as shown in Fig. 5A. The deletion of MyTH(1st) and FERM(1st) from the entire tail domain abolished its inhibitory activity (Fig. 5B). However, deletion of the C-terminal end (DM7AtailΔC) also abolished the inhibitory activity (Fig. 5C). These results suggest that both regions of the tail, i.e., the N-terminal domain and the C-terminal end, are required for inhibition and both are important for the binding of the tail domain to the motor/IQ domains of DM7A.
Fig. 5.
Determination of the regions in the tail domain critical for the inhibition of the actin-activated ATPase activity of DM7AHMM. (A) Schematic representation of the various tail domain constructs. (B) Effect of tail domain constructs, in which the N-terminal domains are truncated to various extent on the actin-activated ATPase activity of DM7AHMM under EGTA conditions is shown. ○, GST-MyTH(2nd)-FERM(2nd); ●, SH3-MyTH(2nd)-FERM(2nd); □, GST-FERM(2nd); ■, DM7Atail. (C) Effect of tail domain constructs, in which the C-terminal domains are truncated to various extent on the actin-activated ATPase activity of DM7AHMM under EGTA conditions is shown. ○, MyTH(1st)-FERM(1st)-SH3; ●, DM7AtailΔC; ■, DM7Atail. One hundred percent represents 1.4 s−1.
Visualization of the Interaction Between the Head/Neck and the Tail Domain in the Inhibited Form of Single-Headed DM7A.
To clarify the structural basis of the tail inhibition mechanism of DM7A, we used negative-staining EM. As described earlier, DM7AHMM molecules had a pear-like shape with a length of 12–15 nm and width 5–7 nm (Fig. 6 A and E). In the presence of exogenous tail domains (≈130 kDa), they showed additional densities around the head (visible in averaged image, Fig. 6B; cf. Fig. 6E), which increased the width to ≈10 nm, although they appeared similar in length. This result suggests that the tail domain contributes mainly to the additional density on the head, presumably because of its binding at this location. To confirm this observation, full-length molecules (DM7AFull) were examined. Fig. 6 C and D shows the appearances of DM7AFull in high and low ionic strengths. As shown in Fig. 6 C and D, DM7AFull images show that the full-length myosin VIIA is monomeric. The images were quite different from those of the forced dimer of DM7AHMM, which showed a dumbbell shape caused by the 2 heads. This finding is supported by a cross-linking experiment of DM7AHMM (Fig. S2), in which DM7AHMM but not the forced dimer of DM7AHMM failed to produce an interheavy chain cross-linking product.
The main difference at the lower ionic strength was a consistent increase in the number of wider molecules (≈10 nm) (white arrows in Fig. 6 D and F) compared with those at high ionic strength (black arrows in Fig. 6C). Single-particle image processing determined the structural characteristics of these wider full-length molecules, providing suggestive evidence for intramolecular interaction between the tail domain and the head (Fig. 6F).
Averaged images (showing a molecular width of ≈10 nm) suggest that the tail domain folds backward to the head (compare with densities and diagrams of DM7AHMM head in Fig. 6E), such that it contacts both the motor and neck domains (compare averages in Fig. 6 B and F). It should be noted that averaged images of DM7AFull showed many views of closely-packed structures (see Fig. S5). Among them, averaged images shown in Fig. 6F were selected that best identify structural information of motor/neck domain and the tail domain from the DM7AFull. Taken together with the results that the actin-activated ATPase activities of DM7AHMM is inhibited by the addition of the tail domain (Fig. 2), this structure supports a model in which inhibition results from folding back of the tail on to the head of this single-headed myosin.
Discussion
Single-Headed Structure of DM7A.
The present study revealed that DM7A is a single-headed myosin, based on cross-linking experiment (Fig. S2) and EM observation (Fig. 6). It has previously been thought that myosin VII is 2-headed because of the presence of a coiled-coil domain on the C-terminal side of the IQ domain (neck domain) (12, 13, 22–24). It was suggested previously that rat myosin VIIA is a dimer based on gel filtration and native gel electrophoresis experiments (13). However, because of the potential influence of molecular shape and surface charges in these experiments, the data could also be consistent with a monomer. It is also possible that the structure could differ between species. It was recently reported that myosin X, having a predicted coiled-coil domain is predominantly monomeric, although ≈10% of dimer molecules are observed (11). It was also shown that a part of the predicted coiled-coil of myosin X forms a rigid stable SAH, which may serve as a part of the lever arm (11). The predicted coiled-coil of myosin VI also does not form stable coiled-coil and may also function as part of a lever arm (25). The predicted coiled-coil of DM7A is short (33 residues) and the proximal portion of the predicted coiled-coil is homologous to the sequence motif found in SAH (11). Therefore, the present finding is consistent with the structural analysis of the predicted coiled-coil domain of myosin X and myosin VI and shows that the short coiled-coil domain of myosin VIIA is not sufficient to form a stable dimer, although it might in part support formation of a 2-headed structure in cells in conjunction with the binding of specific targeting molecules (26–30).
Mechanism of the Tail Domain-Induced Inhibition of the ATPase Activity.
A critical finding is that the tail domain of myosin VIIA functions as an intramolecular inhibitor. We found that the tail domain of DM7A significantly inhibits the actin-activated ATPase activity of the tailless DM7A, DM7AHMM in low Ca2+ (Fig. 2). Consistently, the actin-activated ATPase activity of the DM7AFull is significantly lower than DM7AHMM in low Ca2+. These results suggest that the tail domain interacts with the head/neck domain, thus inhibiting the ATPase activity. By examining the various truncated constructs of DM7A, we identified the distal IQ domains critical for the tail-induced inhibition (Fig. 4). Furthermore, we found by making a series of truncated tail domains that both the N-terminal domain and C-terminal tip of the tail are required for inhibitory activity of the tail. The results suggest that the tail domain has the 2 head/neck bound subdomains and that these bind to the distal IQ domain and the motor domain. EM analysis provided a structural basis for the tail-induced inhibition. We found that the DM7AFull molecules show a compact, apparently folded conformation at low ionic strength (Fig. 6F), whereas they appear unfolded in high ionic strength (Fig. 6C). The observed salt-dependent structural change correlates with the tail-dependent inhibition that is abolished at high ionic strength (Fig. S4). Single-particle analysis of the compact conformation revealed that the tail domain interacts with the head/neck domain. Image analysis suggested that the tail domain can bind to the head/neck domain at 2 sites, one at the motor domain top and the other at the distal IQ domains. This structural analysis agrees with the biochemical functional analysis. Based on these findings, we propose the following model for the regulation of myosin VIIA. In physiological ionic conditions, myosin VIIA in low Ca2+ forms an inhibited conformation, in which the 2 distinct regions of the tail domain bind to the head at the motor core domain and the distal IQ domains, respectively, thus functioning as a “latch” to prevent free movement of the myosin head and decreasing the ATP hydrolysis cycle rate.
Previously, we and other groups have found that the 2-headed myosin Va forms an inhibited conformation, in which the globular tail domain folds back to form a triangular conformation (16–18). Furthermore, exogenous globular tail domain potently inhibits ATPase activity of myosin Va HMM and induces formation of the triangular inhibited conformation (19). Because deletion of the C-teminal end of the first long coiled-coil of myosin Va eliminates the inhibition, we proposed a model that the tail binds to the C-terminal end of the first coiled-coil and to the motor domain, thus functioning as a latch preventing the swinging of the neck (19). The inhibition of DM7A by the tail resembles to the tail-dependent inhibition of myosin Va, in which the tail binds to the head. However, there are several differences. First, myosin Va is a 2-headed myosin and a single-headed construct containing the entire IQ domain with a short coiled-coil domain is not inhibited by the tail (19). These findings together with the present results suggest that the tail binding elements and the mechanism of tail-induced inhibition of DM7A is different from that of myosin Va. Nevertheless, it is likely that the binding of the tail at the distal IQ domain and the motor core domain inhibits the lever-arm swing motion. We proposed previously that this prevention of lever-arm movement by binding of the tail at the motor domain decreases the Pi off-rate of myosin Va in the inhibited state (8, 21). The inhibition of acto-DM7AHMM ATPase activity by the tail domain is more pronounced at low actin than at high actin concentration, suggesting that the inhibition is partially reversed by increasing actin concentration. This is different from the tail inhibition of myosin Va, in which high actin concentration does not attenuate the inhibition (8) and suggests that the tail binds at or near the actin binding sites of myosin VIIA, although we cannot exclude the possibility that the reverse inhibition by actin is an allosteric effect.
Vertebrate smooth muscle or nonmuscle myosin II, whose motor activity is regulated by phosphorylation of the regulatory light chain in the neck forms an inhibited conformation, in which the motor domain is bent back toward the tail to form a folded structure (31). Recent structural analysis has revealed that the 2 heads show an asymmetric arrangement in this conformation, in which the tip of one head (the blocked head) binds to the converter/essential light chain region of the other (free head) (32–35). It has been proposed that the interhead interaction is essential for the inhibition of the motor activity, whereas the proposed model for the inhibition of myosin VIIA does not require head–head interaction. However, it is plausible that inhibition of the converter/lever-arm movement is a general mechanism for the inhibition of myosin motors.
Mechanism for Ca2+-Dependent Regulation of DM7A.
Our result indicated that the tail-induced inhibition is abolished in the presence of micromolar concentration of Ca2+ (pCa6–pCa5), presumably because of the binding of Ca2+ to CaM, which is associated with the neck domain. It has been shown that CaM light chains can be dissociated from the heavy chain of myosin Va HMM (20, 36) at high Ca2+ (pCa4) and this causes the inhibition of the ATPase activity, although it is rescued by the presence of exogenous CaM (20). We found that the Ca2+-dependent reversal of inhibition does not involve the dissociation of bound CaM from DM7A heavy chain. The result suggests that the reversal of inhibition by Ca2+ is caused by a Ca2+-induced conformational change in bound CaM. Because the distal IQ domain region is one of the binding sites of the tail, it is likely that the Ca2+-induced conformational change of bound CaM at this distal IQ domain attenuates the tail binding, thus abolishing the inhibition of the ATPase activity. This is quite different from the regulation of myosin Va, in which the tail does not bind to the distal IQ domain (21).
Cargo Molecules and Regulation.
We previously showed that a cargo protein of myosin Va, melanophilin, which binds to the tail domain, activates the actin-activated ATPase activity of myosin Va, presumably by disrupting the inhibited conformation (37). Several myosin VIIA binding proteins have been reported (26–30). These proteins associate with the tail domain of myosin VIIA, and therefore, it is plausible that they compete with the head/neck domain for tail binding, thus releasing the inhibition. It is expected that the motor function of myosin VIIA is required only when it binds to its docking partners. Presumably myosin VIIA is inhibited until it binds to the docking partners, thus saving ATP consumption in cells. Another important issue is whether or not myosin VIIA forms a dimer in cells. It was shown that DM7A-forced dimer can move processively in vitro (15), and it was suggested that myosin VIIA is involved in melonosome transportation in retinal pigmental epithelia (27). The present study indicates that DM7A does not form a stable dimer; however, it is plausible that the binding of docking partners in cells may induce the dimer formation, and thus functions as a cargo transporter. It requires further study to clarify the cargo molecule-dependent regulation of myosin VIIA.
Materials and Methods
Materials.
See SI Text.
Cloning and Expression of DM7A Protein.
DM7A cDNA was obtained from Drosophila total RNA by using reverse transcriptase-coupled PCR. The cDNA fragment encoding DM7AFull was subcloned into modified pFastBacHT baculovirus transfer vector containing a FLAG tag sequence at the 5′ end. C-terminal-truncated DM7A constructs (DM7AHMM, DM7AHMMLZ, DM7AIQ5, DM7AIQ5ΔSAH, DM7AIQ2, and DM7AIQ1) were produced by introducing a stop codon at various sites of DM7AFull (Fig. 1A). DM7AHMMLZ was produced by fusing DM7AHMM with the GCN4 leucine zipper.
The cDNA coding the DM7Atail was constructed by PCR and subcloned into modified pFastBacHT baculovirus transfer vector containing a FLAG tag sequence. C-terminal-truncated DM7Atail constructs [MyTH(1st)-FERM(1st)-SH3 and DM7AtailΔC] were produced by introducing a stop codon at various sites of DM7Atail (Fig. 5A). N-terminal-truncated DM7Atail [SH3-MyTH(2nd)-FERM(2nd)] was constructed by PCR and subcloned into pFastBacHT transfer vector. The cDNAs coding the Pro1705–Asn2167 [GST-MyHH(2nd)-FERM(2nd)] and Gln1835–Asn2167 [GST-FERM(2nd)] were subcloned into modified pET30c vector (Invitrogen) containing a GST protein sequence.
DM7A constructs were expressed in Sf9 cells, except for GST-fused constructs (38). The proteins were purified by anti-FLAG resin (19), except for MyTH(1st)-FERM(1st)-SH3 and DM7AtailΔC that were purified by Ni-NTA-agarose affinity chromatography (16). GST-fused constructs were expressed in Escherichia coli and purified by Glutathione Sepharose 4B chromatography as described (39). The purified DM7A samples were dialyzed against 30 mM Hepes·KOH (pH 7.5), 300 mM KCl, 2 mM MgCl2, 0.2 mM EGTA, 1 mM DTT, and 10% sucrose. The concentration of DM7A samples were determined as described (16).
Steady-State ATPase Assay.
The steady-state ATPase activity was measured in the buffer containing 20 μM actin, 30 mM Hepes·KOH (pH 7.5), 50 mM KCl, 2 mM MgCl2, 1 mM DTT, 15 μg/mL calmodulin, and 1 mM EGTA or 0.1 mM CaCl2 in the presence of an ATP regeneration system (20 units/mL pyruvate kinase and 2 mM phosphoenolpyruvate) at 25 °C. Five microliters of 1–2 μM DM7A was added to the reaction buffer described above to final concentration of 0.05–0.1 μM and volume of 100 μL. Reaction solution was preincubated at 25 °C for 10 min before adding ATP to start the reaction. The ATPase activity was stopped at various times between 5 and 60 min, and the activity was obtained from the slope of the time course of liberated pyruvate. The liberated pyruvate was determined as described (16).
Negative Staining and Single-Particle Image Processing.
For negative staining, purified DM7AHMM, DM7AHMMLZ, or DM7AFull [700 nM stock protein in high salt (300 mM KCl)] was mixed with 21 μM CaM (ratio of 1:30 to enhance CaM binding) and then 10-fold diluted with 50 mM Na acetate (300 mM NaCl for high-salt observations), 1 mM EGTA, 2 mM MgCl2, 10 mM Mops, and 200 μM ATP, pH 7.5. After dilution, 5 μL of the final mixture was applied to a carbon-coated grid that had been glow-discharged (Harrick Plasma) for 3 min in air, and the grid was immediately (≈5 s) negatively-stained by using 1% uranyl acetate (35). The same procedure was used for the HMM + Tail specimens made by mixing the above final DM7AHMM mixture (70 nM HMM myosin and 2.1 μM CaM) with ≈700 nM exogenous tail fragments. Grids were examined in a Philips CM120 electron microscope (FEI) operated at 80 kV. Images were recorded on a 2K×2K F224HD slow scan CCD camera (TVIPS) at a magnification of 65,000 (0.37 nm per pixel). Single-particle image processing was carried out with SPIDER (Health Research Inc.), and averaged images were produced by alignment and classification of windowed particles (80 × 80 pixels) from micrographs: 1,164 particles (DM7AHMM), 668 particles (DM7AHMM + Tail), and 595 particles (DM7AFull). Representative averages showing the clearest view of the structures were selected from 20–30 total class averages.
Glutar-Aldehyde Cross-Linking.
See SI Text.
Pull-Down Assay.
See SI Text.
Supplementary Material
Note Added in Proof:
While this paper was under review, another report was published on the regulation of Drosophila myosin VIIA (Yang et al., 2009 (40)). The EM images showed that the full-length myosin VIIA is monomeric and has a compact structure similar to Fig. S5, but not Fig. 6F. It was concluded that the C-terminal end of the tail was tightly bent back against the motor domain, inhibiting ATPase activity, although the images did not directly visualize the binding between the tail and the motor domain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812930106/DCSupplemental.
References
- 1.Brown ME, Bridgman PC. Myosin function in nervous and sensory systems. J Neurobiol. 2004;58:118–130. doi: 10.1002/neu.10285. [DOI] [PubMed] [Google Scholar]
- 2.Mallik R, Gross SP. Molecular motors: Strategies to get along. Curr Biol. 2004;14:R971–R982. doi: 10.1016/j.cub.2004.10.046. [DOI] [PubMed] [Google Scholar]
- 3.Weil D, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 4.Hasson T, et al. Unconventional myosins in inner-ear sensory epithelia. J Cell Biol. 1997;137:1287–1307. doi: 10.1083/jcb.137.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, Udovichenko IP, Brown SD, Steel KP, Williams DS. Myosin VIIa participates in opsin transport through the photoreceptor cilium. J Neurosci. 1999;19:6267–6274. doi: 10.1523/JNEUROSCI.19-15-06267.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Ondek B, Williams DS. Mutant myosin VIIa causes defective melanosome distribution in the RPE of shaker-1 mice. Nat Genet. 1998;19:117–118. doi: 10.1038/470. [DOI] [PubMed] [Google Scholar]
- 7.Gibbs D, Kitamoto J, Williams DS. Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc Natl Acad Sci USA. 2003;100:6481–6486. doi: 10.1073/pnas.1130432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato O, Li XD, Ikebe M. Myosin Va becomes a low duty ratio motor in the inhibited form. J Biol Chem. 2007;282:13228–13239. doi: 10.1074/jbc.M610766200. [DOI] [PubMed] [Google Scholar]
- 9.Homma K, Ikebe M. Myosin X is a high duty ratio motor. J Biol Chem. 2005;280:29381–29391. doi: 10.1074/jbc.M504779200. [DOI] [PubMed] [Google Scholar]
- 10.Todorov PT, Hardisty RE, Brown SD. Myosin VIIA is specifically associated with calmodulin and microtubule-associated protein-2B (MAP-2B) Biochem J. 2001;354:267–274. doi: 10.1042/0264-6021:3540267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight PJ, et al. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J Biol Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 12.Chen ZY, et al. Molecular cloning and domain structure of human myosin-VIIa, the gene product defective in Usher syndrome 1B. Genomics. 1996;36:440–448. doi: 10.1006/geno.1996.0489. [DOI] [PubMed] [Google Scholar]
- 13.Inoue A, Ikebe M. Characterization of the motor activity of mammalian myosin VIIA. J Biol Chem. 2003;278:5478–5487. doi: 10.1074/jbc.M210489200. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe S, Ikebe R, Ikebe M. Drosophila myosin VIIA is a high duty ratio motor with a unique kinetic mechanism. J Biol Chem. 2006;281:7151–7160. doi: 10.1074/jbc.M511592200. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, et al. Dimerized Drosophila myosin VIIa: A processive motor. Proc Natl Acad Sci USA. 2006;103:5746–5751. doi: 10.1073/pnas.0509935103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li XD, Mabuchi K, Ikebe R, Ikebe M. Ca2+-induced activation of ATPase activity of myosin Va is accompanied with a large conformational change. Biochem Biophys Res Commun. 2004;315:538–545. doi: 10.1016/j.bbrc.2004.01.084. [DOI] [PubMed] [Google Scholar]
- 17.Wang F, et al. Regulated conformation of myosin V. J Biol Chem. 2004;279:2333–2336. doi: 10.1074/jbc.C300488200. [DOI] [PubMed] [Google Scholar]
- 18.Krementsov DN, Krementsova EB, Trybus KM. Myosin V: Regulation by calcium, calmodulin, and the tail domain. J Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li XD, Jung HS, Mabuchi K, Craig R, Ikebe M. The globular tail domain of myosin Va functions as an inhibitor of the myosin Va motor. J Biol Chem. 2006;281:21789–21798. doi: 10.1074/jbc.M602957200. [DOI] [PubMed] [Google Scholar]
- 20.Homma K, Saito J, Ikebe R, Ikebe M. Ca2+-dependent regulation of the motor activity of myosin V. J Biol Chem. 2000;275:34766–34771. doi: 10.1074/jbc.M003132200. [DOI] [PubMed] [Google Scholar]
- 21.Li XD, et al. The globular tail domain puts on the brake to stop the ATPase cycle of myosin Va. Proc Natl Acad Sci USA. 2008;105:1140–1145. doi: 10.1073/pnas.0709741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mooseker MS, Cheney RE. Unconventional myosins. Annu Rev Cell Dev Biol. 1995;11:633–675. doi: 10.1146/annurev.cb.11.110195.003221. [DOI] [PubMed] [Google Scholar]
- 23.Hasson T, Mooseker MS. Vertebrate unconventional myosins. J Biol Chem. 1996;271:16431–16434. doi: 10.1074/jbc.271.28.16431. [DOI] [PubMed] [Google Scholar]
- 24.Sellers JR. Myosins: A diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 25.Spink BJ, Sivaramakrishnan S, Lipfert J, Doniach S, Spudich JA. Long single α-helical tail domains bridge the gap between structure and function of myosin VI. Nat Struct Mol Biol. 2008;15:591–597. doi: 10.1038/nsmb.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kussel-Andermann P, et al. Vezatin, a novel transmembrane protein, bridges myosin VIIA to the cadherin-catenins complex. EMBO J. 2000;19:6020–6029. doi: 10.1093/emboj/19.22.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Amraoui A, et al. MyRIP, a novel Rab effector, enables myosin VIIa recruitment to retinal melanosomes. EMBO Rep. 2002;3:463–470. doi: 10.1093/embo-reports/kvf090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuda M, Kuroda TS. Slac2-c (synaptotagmin-like protein homologue lacking C2 domains-c), a novel linker protein that interacts with Rab27, myosin Va/VIIa, and actin. J Biol Chem. 2002;277:43096–43103. doi: 10.1074/jbc.M203862200. [DOI] [PubMed] [Google Scholar]
- 29.Etournay R, et al. PHR1, an integral membrane protein of the inner ear sensory cells, directly interacts with myosin 1c and myosin VIIa. J Cell Sci. 2005;118:2891–2899. doi: 10.1242/jcs.02424. [DOI] [PubMed] [Google Scholar]
- 30.Etournay R, et al. Shroom2, a myosin-VIIa- and actin-binding protein, directly interacts with ZO-1 at tight junctions. J Cell Sci. 2007;120:2838–2850. doi: 10.1242/jcs.002568. [DOI] [PubMed] [Google Scholar]
- 31.Ikebe M. Regulation of the function of mammalian myosin and its conformational change. Biochem Biophys Res Commun. 2008;369:157–164. doi: 10.1016/j.bbrc.2008.01.057. [DOI] [PubMed] [Google Scholar]
- 32.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodhead JL, et al. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 34.Burgess SA, et al. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Jung HS, Komatsu S, Ikebe M, Craig R. Head–head and head–tail interaction: A general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trybus KM, Krementsova E, Freyzon Y. Kinetic characterization of a monomeric unconventional myosin V construct. J Biol Chem. 1999;274:27448–27456. doi: 10.1074/jbc.274.39.27448. [DOI] [PubMed] [Google Scholar]
- 37.Li XD, Ikebe R, Ikebe M. Activation of myosin Va function by melanophilin, a specific docking partner of myosin Va. J Biol Chem. 2005;280:17815–17822. doi: 10.1074/jbc.M413295200. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe S, Umeki N, Ikebe R, Ikebe M. Impacts of Usher syndrome type IB mutations on human myosin VIIa motor function. Biochemistry. 2008;47:9505–9513. doi: 10.1021/bi8007142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koga Y, Ikebe M. A novel regulatory mechanism of myosin light chain phosphorylation via binding of 14-3-3 to myosin phosphatase. Mol Biol Cell. 2008;19:1062–1071. doi: 10.1091/mbc.E07-07-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y, et al. A FERM domain autoregulates Drosophila myosin 7a activity. Proc Natl Acad Sci USA. 2009;106:4189–4194. doi: 10.1073/pnas.0808682106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.