Tailed bacterial viruses (bacteriophages) are ubiquitously distributed in nature and are likely the most abundant organisms on the biosphere (1). Spending most of their time outside of the host, a bacterial cell, in often hostile external environments, they come to “life” upon encountering the receptor molecules on the host cell surface. The virus consists of a head (capsid) into which the DNA (genome) is packaged and a tail that delivers the genome into the bacterium. The capsid is pressurized because of packing of highly negatively-charged, relatively rigid dsDNA to near-crystalline density (≈500 μg/mL). The internal capsid pressure, ≈6 MPa or >10 times that of bottled champagne (2), provides a driving force for delivery of viral genome into host cell. One of the longstanding questions in phage biology has been how these viruses contain the DNA pressure and trigger release only upon recognition of a specific host cell. In this issue of PNAS, a study by Lhuillier et al. (3) describes the pseudoatomic structure of a DNA gate from the Bacillus subtilis bacteriophage SPP1, which “zips” the capsid after the genome is packaged and unzips it when the virus is ready to infect the host. It is a compelling story, which began with the first in vitro virus assembly experiments described by Edgar and Wood >40 years ago (4) and is applicable not only to phages but also to large eukaryotic viruses such as herpes viruses.
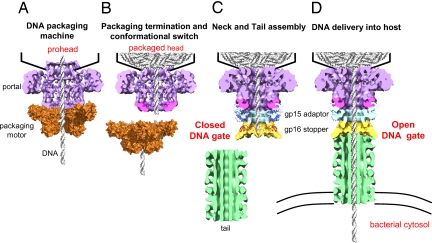
In the assembly of a typical tailed bactreiophage, a capsid shell of precise dimensions is assembled, often with a single protein subunit, around a protein scaffold. A mushroom-shaped dodecameric portal ring (Fig. 1) acts as an initiator of head assembly and remains at the special 5-fold vertex of the otherwise isometric capsid, facilitating all subsequent transactions: DNA packaging, tail attachment, and DNA delivery. The scaffold is removed, creating an empty space for housing the viral genome. The packaging proteins cut the concatemeric viral DNA and dock at the protruding end of the portal, inserting the DNA end into the ≈3.5-nm portal channel. The ATP-fueled packaging machine thus assembled powers translocation of the DNA into the capsid (5). After packaging 1 viral genome that is equivalent to 1 “headful,” the motor dissociates and cuts again, separating itself from the DNA-full head. The “neck” proteins now assemble on the portal, sealing the packaged genome and providing a platform for tail attachment. When the virus infects a host cell, the seal is broken and the tail becomes an open channel connecting the virus capsid to the cytosol, facilitating the delivery of DNA into host cell. The efficiency of delivery in some phages reaches the theoretical limit, with each virion productively infecting a bacterium.
Fig. 1.
A model for DNA gating in SPP1 and tailed bacteriophages. (A) The nascent prohead portal binds to the packaging proteins and a DNA packaging machine is assembled. (B) After packaging 1 viral genome (headful), a conformational change (pink) causes termination of packaging and dissociation of the motor. The last packaged DNA is restrained by the portal channel. (C) The neck proteins (adaptor and stopper) assemble, forming a closed DNA gate. Another conformational change widens the portal channel. The DNA descends and lands on the stopper, primed for delivery. The tail assembles on the neck. (D) Interaction with the host receptor triggers conformational changes that transmit a signal to the stopper, unzipping the DNA gate. The DNA is released into the bacterium through the tail tube.
The SPP1 head–tail connector consists of 3 proteins that assemble as dodecamers: portal (gp6), adaptor (gp15), and stopper (gp16), the latter 2 being the neck proteins. The connector is at the heart of the late assembly steps the virus must execute with exquisite precision to generate an infectious virion. A number of intriguing questions arise and their examination gives a glimpse of how viruses solved this problem.
How is the assembly sequence determined? Both the packaging proteins and the neck proteins are expressed in the same timeframe and bind to the portal, but in a strict sequence. The packaging proteins must bind first to assemble the DNA packaging machine (Fig. 1A), otherwise, i.e., binding of neck proteins first would result in abortive assembly. The portal conformation in the nascent prohead is found to be different from that in phage (6), implying that the prohead portal interacts with the packaging proteins but not the neck proteins. However, in vitro, the purified phage P22 portal (gp10) and neck (gp4) proteins do interact, forming a complex of expected stoichiometry (7). Perhaps, the isolated portal is conformationally less restrained and the very high protein concentrations in the binding reaction in the absence of the packaging proteins allow for gp10-gp4 binding.
How is the transition from packaging to neck assembly coordinated? A powerful packaging motor overcomes the repulsive forces and bending energies that oppose confinement of DNA inside the virus capsid. The internal pressure builds as the capsid fills and when it reaches headful, the motor nearly stalls (8) and dissociates from the capsid. Previous studies (2, 9), e.g., certain siz mutations in the SPP1 portal either overfill or underfill the head, suggest that head filling is accompanied by a portal conformational change that signals the packaging motor to dissociate (Fig. 1B), i.e., the portal may no longer be competent to interact with the packaging proteins. Studies with several phages show that such DNA-full heads can be isolated from neck mutant extracts and complemented in vitro with neck and tail proteins to produce infectious virions (10). These results imply that the portal channel probably acts as a valve to restrain the packaged DNA. Otherwise, the internal pressure would have pushed at least some of the DNA out, and extrusion of even a short piece of DNA would have resulted in abortive assembly.
How do the neck proteins assemble into a DNA gate? Lhuillier et al. (3) for the first time provide a structural basis for the assembly of neck proteins into a DNA gate that seals the packaged capsid. They solved the structures of both gp15 and gp16 by NMR and fitted these structures and the previously-determined X-ray structure of the portal into the 10-Å cryoEM density map of the connector. The pseudoatomic structure thus generated sheds light not only on the dynamics of assembly but also on the mechanism of capsid closure and opening. Both of the neck proteins consist of large unstructured/flexible regions (20–30% of total sequence), as was found in the homologous proteins gpW and gpFII from phage λ (11, 12), which prevent premature olgomerization. The portal conformational change after DNA packaging exposes an interaction site for the largely α-helical gp15. Binding to portal causes a large structural rearrangement in gp15, 15-Å motion of N-terminal helices relative to C-terminal helices through a flexible hinge. This process allows intercalation of helices from the adjacent gp15 subunits between the N and C segments. Assembly on portal and oligomerization occur hand in hand, and rapidly, producing a dodecameric adaptor with a central channel that is contiguous with the portal channel. The highly basic N-terminal surface of gp15 is now exposed, which attracts the acidic region of gp16. gp16 binding places a large unstructured loop toward the center of the channel. Assembly of 12 such gp16 molecules places the 12 loops into the center, filling up the channel space. The psuedoatomic structure shows that the loops become structured in the assembled state, forming a 12-stranded parallel β-sheet stopper.
The efficiency of delivery in some phages reaches the theoretical limit.
How is the DNA gate primed for genome delivery? Another portal conformational change must occur after neck assembly (13), widening the portal channel and causing the DNA to descend until it lands on the stopper (Fig. 1C). Thus, the DNA, likely the last packaged DNA, spans the entire connector channel. Indeed, a 60-bp segment of the last packaged DNA was shown to be associated with the tail when the phage was disrupted with denaturing agents (14). Lhuillier et al. (3) introduced 2 cysteine substitutions into each β-strand of the stopper β-sheet. Consistent with their predictions, the phage in which the β-strands were cross-linked by disulphide bridges failed to eject DNA upon exposure to the host receptor YueB780. The same phage ejected the DNA when the cross-links were removed by treatment with DTT. These results suggest that the DNA is stopped at the gate; if it were to descend further down into the tail tube, cross-linking would not have much effect on ejection. In other words, the connector not only seals the packaged viral genome but also primes it for delivery. During infection, specific interactions between the tip of the tail and the host receptor transmit a signal to the stopper, unzipping the β-strands and opening the gate (Fig. 1D).
This study (3) closed certain critical gaps in our understanding of late assembly events and helped define a probable pathway for infectious virus assembly (Fig. 1). Of particular interest is the control of sequence, kinetics, and fidelity of virus assembly by folding the unstructured regions of interacting molecules. This design seems to have special relevance to plugging holes in the virus for DNA containment, as was also evident in the assembly of gpD, the phage λ outer capsid protein that cements the holes between capsomers (15). Future studies should elucidate the interacting residues and the dynamics of proposed conformational changes, especially in the portal, and the mechanisms of signal transmission from host surface to head–tail connector and unzipping of the DNA gate. Finally, it is conceivable that the DNA gate may assume particular importance in nanomedicine research if the packaged viral capsids were to be used as vehicles to deliver DNA therapeutics.
Acknowledgments.
My research has been funded by National Science Foundation Grant MCB-423528 and National Institutes of Health Grant AI056443.
Footnotes
The author declares no conflict of interest.
See companion article on page 8507.
References
- 1.Hendrix RW. Bacteriophage: Evolution of the majority. Theor Popul Biol. 2002;61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- 2.Lander GC, et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 3.Lhuillier S, et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci USA. 2009;106:8507–8512. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgar RS, Wood WB. Morphogenesis of bacteriophage T4 in extracts of mutant-infected cells. Proc Natl Acad Sci USA. 1966;55:498–505. doi: 10.1073/pnas.55.3.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:642–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 6.Orlova EV, et al. Structure of a viral DNA gatekeeper at 10-Å resolution by cryoelectron microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olia AS, et al. Binding-induced stabilization and assembly of the phage P22 tail accessory factor gp4. J Mol Biol. 2006;363:558–576. doi: 10.1016/j.jmb.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Smith DE, et al. The bacteriophage ϕ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 9.Tavares P, et al. Identification of a gene in Bacillus subtilis bacteriophage SPPl determining the amount of packaged DNA. J Mol Biol. 1992;225:81–92. doi: 10.1016/0022-2836(92)91027-m. [DOI] [PubMed] [Google Scholar]
- 10.Bode VC, Gillin FD. The arrangement of DNA in lambda phage heads. I. Biological consequences of micrococcal nuclease attack on a portion of the chromosome exposed in tailless heads. J Mol Biol. 1971;62:493–502. doi: 10.1016/0022-2836(71)90150-1. [DOI] [PubMed] [Google Scholar]
- 11.Maxwell KL, et al. The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold. J Mol Biol. 2001;308:9–14. doi: 10.1006/jmbi.2001.4582. [DOI] [PubMed] [Google Scholar]
- 12.Maxwell KL, et al. The solution structure of the bacteriophage lambda head-tail joining protein, gpFII. J Mol Biol. 2002;318:1395–1404. doi: 10.1016/s0022-2836(02)00276-0. [DOI] [PubMed] [Google Scholar]
- 13.Zheng H, et al. A conformational switch in bacteriophage P22 portal protein primes genome injection. Mol Cell. 2008;29:1–8. doi: 10.1016/j.molcel.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tavares P, et al. Sequential headful packaging and fate of the cleaved DNA ends in bacteriophage SPP1. J Mol Biol. 1996;264:954–967. doi: 10.1006/jmbi.1996.0689. [DOI] [PubMed] [Google Scholar]
- 15.Lander GC, et al. Bacteriophage lambda stabilization by auxiliary protein gpD: Timing, location, and mechanism of attachment determined by cryo-EM. Structure (London) 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]