Abstract
Pathogenic bacteria have developed extraordinary strategies for invading host cells. The highly conserved type III secretion system (T3SS) provides a regulated conduit between the bacterial and host cytoplasm for delivery of a specific set of bacterial effector proteins that serve to disrupt host signaling and metabolism for the benefit of the bacterium. Remarkably, the inner diameter of the T3SS apparatus requires that effector proteins pass through in at least a partially unfolded form. AvrPto, an effector protein of the plant pathogen Pseudomonas syringae, adopts a helical bundle fold of low stability (ΔGF→U = 2 kcal/mol at pH 7, 26.6 °C) and offers a model system for chaperone-independent secretion. P. syringae effector proteins encounter a pH gradient as they translocate from the bacterial cytoplasm (mildly acidic) into the host cell (neutral). Here, we demonstrate that AvrPto possesses a pH-sensitive folding switch controlled by conserved residue H87 that operates precisely in the pH range expected between the bacterial and host cytoplasm environments. These results provide a mechanism for how a bacterial effector protein employs an intrinsic pH sensor to unfold for translocation via the T3SS and refold once in the host cytoplasm and provide fundamental insights for developing strategies for delivery of engineered therapeutic proteins to target tissues.
Keywords: NMR, T3SS
Although the delivery of bacterial proteins into the cytoplasm of a host cell is a primary step in pathogen infection by Gram-negative bacteria, little is presently known regarding the detailed mechanisms of this delivery process. Gram-negative bacteria, such as the plant pathogen Pseudomonas syringae, use the broadly conserved T3SS to infect their hosts (1). This elaborate trafficking device, composed of multiple copies of >20 different proteins, provides a needle-like conduit for trafficking a suite of bacterial effector proteins (effectors), which act as agents of infection, into the host cell (Fig. 1A). Effectors vary widely, depending on the pathogen species and the targeted host, and the specific signals that target these proteins for translocation via the T3SS have been hypothesized but are not yet fully understood (2, 3). Many effectors counteract host defenses and have been shaped by the constant competition between the infecting pathogen and the resisting host cell (4). A critical gap in our understanding of T3SS-mediated infection lies in defining the properties of effectors that allow efficient T3SS transport.
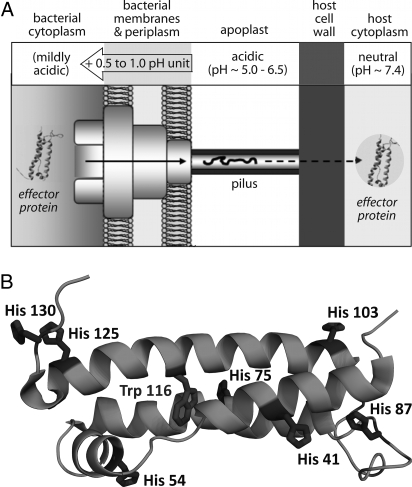
Fig. 1.
Variation of environmental pH for plant pathogen P. syringae effector proteins translocated via the T3SS and His residues in the structured core of AvrPto. (A) The T3SS supramolecular assembly spans across both bacterial membranes, the apoplast, and the host cell wall. Effector proteins first encounter the mildly acidic bacterial cytoplasm, must then at least partially unfold for translocation through the narrow pilus, and then must refold upon delivery to the neutral host cytoplasm. (B) Seven His residues are dispersed across the TrAvrPto structure; the lone Trp residue is buried at the center of the helix bundle.
Secretion through the T3SS is a complicated process that depends on several factors, including effector protein stability and folding kinetics. These invading proteins are trafficked along the hollow needle-like structure (pilus) of the T3SS, presumably through the pilus lumen (5). Because the inner diameter of the pilus is ≈2–3 nm (6, 7), most proteins should at least partially unfold to pass through unimpeded. The apparent requirement for unfolding suggests possible limits of effector protein stability or folding kinetics that must not be exceeded for efficient secretion. Studies of chimeras composed of effector YopE fused to various partners support the principle that high fold stability (8) or rapid folding (9) can indeed prevent secretion. Some effectors are known to have cognate secretion chaperones whose functional roles vary from influencing the temporal order of effector secretion to maintaining their cognate effector in partially unfolded form (10, 11). However, in P. syringae, only a few of its nearly 30 known effectors have been shown to be chaperone-dependent (12).
Bacteria live and function in a wide range of environments and possess regulatory mechanisms that allow them to rapidly alter gene expression, metabolism, motility, and secretion in response to environmental changes, such as pH. In the case of P. syringae, the bacterium launches its invasion upon arrival in the plant apoplast, the extracellular milieu surrounding the host cells, where the pH typically ranges from 5 to 6.5 (13). The intracellular bacterial pH is ≈0.5–1.0 pH unit higher, inferred from the T3SS requirement of a proton gradient, with internal cytoplasmic pH higher than external pH (14, 15). The cytoplasm of plant leaf cells is typically maintained at neutral pH (≈7.4). Hence, P. syringae effectors experience aqueous environments of different pH, ranging from mildly acidic (in the bacterial cytoplasm) to neutral (in the host cytoplasm) (Fig. 1A).
Interestingly, secretion of P. syringae effector AvrPto is strongly affected by pH. The P. syringae T3SS and its associated proteins are expressed when the bacteria are in the acidic apoplast in the host leaves (13, 16). Expression of these proteins can also be induced when P. syringae is cultured in a minimal hrp-inducing medium (17). Although AvrPto is expressed in the bacterium at external hrp medium pH (pHext) values of 6 or 7, it is secreted efficiently by the T3SS when pHext is 6 but not 7 (17). AvrPto has both virulence (disease-promoting) and avirulence (disease-preventing) roles in the host cell (18). Its avirulence role is mediated via a gene-for-gene interaction with plant kinase Pto, which elicits rapid host defense responses that facilitate highly localized cell death (18). AvrPto virulence function has recently been shown to involve direct targeting of host transmembrane receptor kinase BAK1 (and possibly others) to disrupt innate immune responses facilitated via the microbe-associated molecular patterns (MAMP) pathway (19). An additional AvrPto virulence mechanism appears to involve suppression of the microRNA pathway that is important in antibacterial basal defense in the host (20).
The structured region of AvrPto (referred to as TrAvrPto, herein composed of residues D29–I133) is essential for its avirulence activity, and for at least one of its virulence functions. Proper folding and subcellular localization of TrAvrPto have been shown to be required for elicitation of the hypersensitive response in host tissues, demonstrating the role of this region in avirulence (21). Similarly, the inability of AvrPto-S46P to bind to virulence target BAK1 (19) suggests that the same folding requirement applies for virulence, because the Pro Cδ would sterically clash with the carbonyl group of Q42 in the TrAvrPto structure (PDB ID 1R5E). Hence, for the example of AvrPto, the biological system requires the protein to be of low stability in the acidic bacterial cytoplasm and to refold once delivered into the neutral host cytoplasm, suggesting the presence of a pH folding switch at work in this protein.
The P. syringae effector AvrPto has no known secretion chaperone, and provides a useful model system to gain a detailed understanding of a pH-triggered folding switch that facilitates secretion of an effector via the T3SS. The NMR structure of TrAvrPto, in which the disordered N- and C-terminal tails of AvrPto are removed, reveals a fold composed of a 3-helix bundle with an orthogonal helix and an omega loop (Fig. 1B) (21). Importantly, NMR spectra for both AvrPto and TrAvrPto display 2 discrete populations, one corresponding to the folded conformation and one to a nonnative ensemble. Full-length AvrPto and TrAvrPto follow the same pH dependence of stability: Both are more stable at pH 7 than at pH 6 (21, 22). The folding kinetics and thermodynamics of TrAvrPto have been studied at pH 6.1, showing slow folding and unfolding rates (kUF = 1.8 s−1 and kFU = 0.33 s−1, respectively) and an unfolded population of 16% (22). In contrast, TrAvrPto is nearly completely folded at pH 7.0, with an unfolded population of only 2%. Hence, the AvrPto folded core possesses a pH-regulated folding switch that operates precisely in the pH range expected across the T3SS, i.e., between the bacterial and host cytoplasm (Fig. 1A).
In this study, we applied NMR, CD, and Trp fluorescence (Fl) spectroscopies to elucidate the mechanism of the pH-regulated folding switch that facilitates the acid denaturation of TrAvrPto. Although NMR provides atomic-level resolution of the pH dependence of TrAvrPto stability, CD and Fl demonstrate the concentration independence of the equilibrium and also characterize the thermal denaturation of this protein at different pH values. By using the atomic resolution of NMR, pKa values for each histidine side chain in native TrAvrPto were quantified. Most His residues display pKa values consistent with their solvent-exposed environment on the protein surface. However, buried H87 has an anomalously low pKa value. Thermodynamic modeling reveals the titration of H87 as the dominant force in the pH-regulated folding switch of TrAvrPto. Moreover, the H87Y mutation shifts the acid denaturation curve to significantly lower pH, confirming the essential role of H87 as a pH sensor for fold stability in the pH range of the biological pathogen–host system (pH ≈5–7). Intriguingly, H87 is conserved in homologous sequences for both AvrPto and distantly related AvrPtoB, suggesting broad functional importance. This detailed molecular mechanism used by P. syringae to deliver a subset of effector proteins into the host cell at the right time and the right place provides fundamental insights potentially applicable to the engineering of therapeutic proteins for bacterial delivery to target tissues.
Results and Discussion
NMR Reveals the Acid Denaturation of TrAvrPto.
The pH dependence of the folded population of TrAvrPto was measured at 26.6 °C as a function of pH by using 2D NMR. The 15N–1H fast-HSQC (fHSQC) experiment (23) was used to monitor the populations of folded and unfolded TrAvrPto at different pH values. This experiment resolves individual scalar-coupled 15N–1H bonds within the protein as separate peaks whose position in frequency space reflects the average local chemical environment of each group. NMR has the additional advantage of sensitivity to time scale of conformational exchange processes, where slow exchange on the NMR time scale yields distinct sets of peaks with population-weighted volumes, whereas fast exchange yields a single averaged set of peaks with population-weighted positions. The presence of 2 discrete populations of peaks in the 2D 15N–1H NMR spectrum corresponding to folded and unfolded TrAvrPto states in slow exchange (21) allows quantitative analysis of the folding equilibrium as a function of pH.
Thirteen N–H groups give rise to well-resolved folded (F) and unfolded (U) peaks in the 15N–1H fHSQC spectrum: the backbone amide groups of A47, G48, A61, S64, T76, T91, G92, S94, G95, G99, A112, and G128 and the Trp side-chain indole group, W116 ε1. A representative set of peaks corresponding to G95 illustrate the pH-dependent change in equilibrium F and U populations (Fig. 2A). The average folded population (pF) derived from NMR peak volumes for these 13 residues shows clear acid denaturation as the pH is lowered from 7 to 4 (Fig. 2B). Importantly, the NMR-derived curves are conserved for all 13 of these structurally dispersed groups, as reflected by the error bars for each pF data point (Fig. 2B). Within the native conformation, these residues represent diverse environments, e.g., α-helix (e.g., T76, A112), solvent-exposed (e.g., G95), and buried (e.g., W116 ε1), indicating that the observed acid denaturation reflects global unfolding (Fig. 1B).
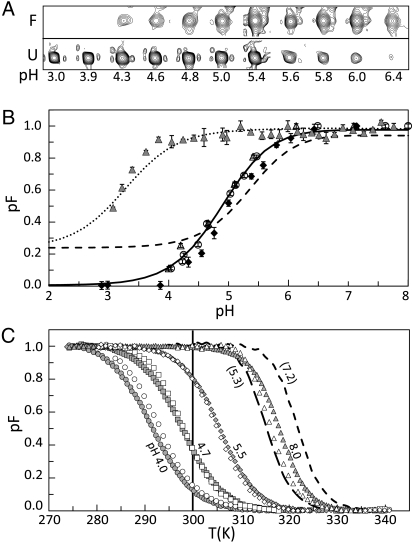
Fig. 2.
Acid and thermal denaturation of WT TrAvrPto (WT) and H87Y-TrAvrPto (H87Y). (A) NMR peaks for a representative residue (Gly 95) showing the folded and unfolded peaks at the same contour level at different pH values. (B) Population of the folded state (pF) versus pH at 26.6 °C for the WT from NMR (filled diamonds), CD (open circles) Fl (open triangles) experiments and for H87Y from Fl (filled triangles) experiment. Solid lines, fitted curve for WT; dashed line, fitted by using WT pKa data without including COOH titration; dotted line, curve fitted to H87Y Fl data by using all WT pKa values except H87Y. (C) Thermal denaturation data for WT and H87Y. WT Fl at pH 4.0 (open circles), 4.7 (open squares), 5.4 (open diamonds), and 7.1 (open triangles). WT CD at pH 4.0 (filled circles), 4.6 (filled squares), 5.5 (filled diamonds), and 8.0 (filled triangles). H87Y Fl at pH: 5.3 (long-dashed line), 7.2 (short-dashed line). Vertical line marks 26.6 °C.
To ensure that the observed pH-dependent stability of TrAvrPto is not concentration dependent, a 15N–1H fHSQC spectrum was obtained at 80 μM TrAvrPto concentration at pH 5.0 (chosen to provide similar folded and unfolded populations). The pF values obtained are within error of those observed at 0.4 mM [supporting information (SI) Fig. S1 and Table S1]. These results demonstrate that the same TrAvrPto stability is obtained at millimolar and micromolar concentrations in a highly pH-sensitive region (i.e., in the steepest region of the denaturation curve). This indicates that the higher concentration of the NMR samples does not impart artifacts in TrAvrPto stability and supports previous NMR work that ruled out the presence of dimers and higher-order aggregates at millimolar concentrations (22).
Thermal Stability of TrAvrPto.
To examine the temperature dependence of TrAvrPto stability and to confirm the acid denaturation of TrAvrPto at low protein concentration (80 μM), thermal denaturation of the protein was monitored by CD and Fl at different pH values (Fig. 2B and Fig. S2). CD provides a measure of secondary structure composition (24), whereas Fl reports on the level of solvent exposure of Trp residues (25). The single TrAvrPto Trp is buried in the core of the 3-helix bundle (Fig. 1B) and provides a probe of tertiary structure. The temperature-dependent CD and Fl signals were converted to pF vs. temperature (T) (see Materials and Methods), and the resulting curves show that, for pH values in the range of ≈4–6, small changes in T yield large changes in pF (and hence, unfolded protein) (Fig. 2C). These results suggest that translocation of AvrPto via the T3SS should be highly sensitive to temperature, with more efficient translocation at higher temperatures corresponding to higher unfolded AvrPto populations. Translocation assays performed in plants equilibrated to 24 °C show definitive delivery of AvrPto to the host-cell cytoplasm (26). However, in culture medium at pH 6, P. syringae efficiently secretes AvrPto at 20 °C but not at 30 °C (17). Clearly, additional factors must be involved in governing the efficiency of AvrPto secretion at higher temperatures. Studies of the temperature dependence of AvrPto translocation in planta will enable exploration of how the thermal stability of the folded core of AvrPto determined here might influence the delivery of AvrPto into the host cytoplasm.
The CD- and Fl-derived thermal denaturation curves enable the pF value at any temperature to be determined and thereby provide independent measures of the acid denaturation of TrAvrPto at 26.6 °C (the temperature of NMR experiments). Both CD- and Fl-derived pF vs. pH data agree well with the NMR-derived data (Fig. 2B), again confirming that the acid denaturation of TrAvrPto does not depend on concentration, and verifying the pH dependence of fold stability.
H87 Has an Anomalously Low pKa Value in Native TrAvrPto.
In general, the pH dependence of protein stability is subject to the relative ability of the F and U conformations to accommodate each ionization state of a titratable group (27). These capacities are reflected by the differences between individual pKa values for individual ionizable groups in the folded (pKaF) and unfolded (pKaU) forms of a protein. For any titratable group, when pKaF < pKaU, a decrease in pH will favor the unfolded form in which the protonated group is more energetically favored, and the titrating group provides a driving force for acid denaturation. The sensitivity of the TrAvrPto pF vs. pH curve in the pH 5–7 range suggests a role of histidine and motivates the determination of pKa values for each of the 7 His residues in this protein.
The resolution of NMR peaks in the 15N–1H HMQC spectrum allows the tautomeric state and pKa value of individual His side chains to be characterized. This HMQC experiment was optimized to observe 2-bond scalar-coupled 15N–C–1H groups to probe the local environments of the 2 15N nuclei in His side chains, 15Nε2 and 15Nδ1 (28). The pH titration of a His side chain at physiological pH is between the positively charged state (His+) where both 15N are protonated, and the 2 neutral (His0) tautomers, one protonated at 15Nε2 (Nε2–H His0) and the other at 15Nδ1 (Nδ1–H His0). Therefore, even in the absence of protein folding, the HMQC data are affected by 2 separate reactions: the pH titration between His+ and His0 and tautomer exchange between Nε2–H His0 and Nδ1–H His0.
All 7 His residues in TrAvrPto titrate in a manner consistent with fast proton exchange on the NMR time scale and are predominantly in the more energetically favored Nε2–H tautomer (28) at pH 7.0 (Fig. S3A). Although H130 displays a single tautomer state, the other His residues exhibit fast (H41, H54, H75, and H125) or intermediate (H87 and H103) tautomer exchange (Fig. S3B) (28–30).
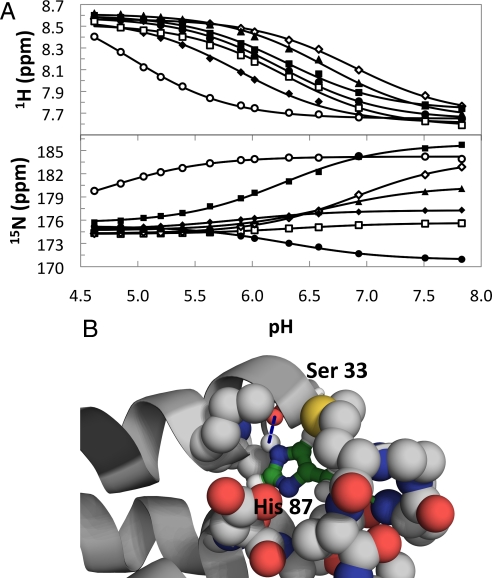
The pKa values for each His side chain in folded TrAvrPto were quantified based on peak shifts during pH titration (Fig. 3A). Although peaks for 15Nε2 and 15Nδ1 of each side chain are resolved, tautomer exchange means that the fitted pKa values are averaged over both groups. Three of the 7 His residues in folded TrAvrPto display pKa values (pKaF) consistent with their solvent-exposed locations on the protein surface (pKa,H41F = 6.32 ± 0.04, pKa,H103F = 6.28 ± 0.02, pKa,H130F = 6.28 ± 0.03), whereas 2 display somewhat elevated values (pKa,H75F = 6.89 ± 0.01, pKa,H125F = 6.65 ± 0.01) and one a somewhat depressed value (pKa,H54F = 5.88 ± 0.01). The most striking outlier is H87, which displays a dramatically low pKa,H87F value (pKa,H87F = 4.8 ± 0.1) relative to a typical solvent-exposed His side chain (pKa ≈ 6.4). The H87 side chain is buried (desolvated), and its Nε2-H is a hydrogen-bond donor to the S33 carbonyl group (Fig. 3B). The desolvation and the formation of this tertiary contact rationalize both the low pKaF value and the intermediate tautomer exchange observed for H87 (28–30). The low pKa value determined for H87, a full 1.5 pH units lower than an average solvent exposed histidine in an unfolded protein, should provide a major driving force for acid denaturation.
Fig. 3.
Determination of histidine pKa values reveals buried H87 as an outlier. (A) 15Nε2 and 1Hε1 chemical shift changes as a function of pH yield pKa values for individual His residues: H41 (open squares), H54 (filled diamonds), H75 (open diamonds), H87 (open circles), H103 (filled squares), H125 (filled triangles), and H130 (filled circles). Lines depict best-fit curves using Eq. S8. (B) The H87 pH-regulated folding switch features a long-range tertiary hydrogen bond (H87 Nε2···S33 C O, dashed line) and significant desolvation of the H87 side chain. Protonation of H87 disfavors this local environment.
O, dashed line) and significant desolvation of the H87 side chain. Protonation of H87 disfavors this local environment.
His-87 Drives the Acid Denaturation of TrAvrPto.
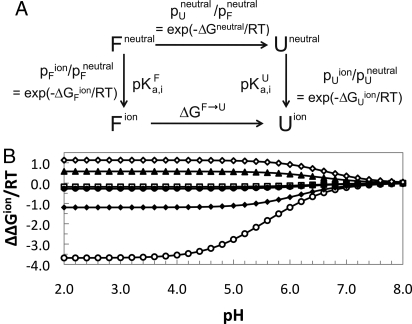
Fitting of the 29 data points gathered from the 3 different spectroscopic techniques to the 2-state, multititration model of Yang and Honig (Fig. 4A) (27) demonstrates that H87 has a major influence on the acid denaturation of TrAvrPto. In the simplest case, we consider the impact of the measured histidine pKa values for the folded state, assuming all His residues in the unfolded state have a pKaU = 6.4 (typical for solvent-exposed His side chains), and allowing the pH-independent component of the free energy, ΔGneutral, to be optimized by the fit (see Materials and Methods). The fitted curve adequately models the higher pH region of the acid denaturation data, but does not predict the complete denaturation at low pH (Fig. 2B). This suggests contribution from a not-yet-accounted-for ionizable group titrating in the pH range of 2–5 (e.g., Asp or Glu side chain). A COOH titration event was added with 2 additional parameters (pKa,COOHU and pKa,COOHF) that were optimized without restriction. This approach yielded a good fit of the entire acid denaturation profile (Fig. 2B). The resulting fitted titration parameters (pKa,COOHU = 4.8 ± 0.05 and pKa,COOHF = 2.7 ± 0.4) are physically reasonable for a COOH group involved in a salt bridge in the folded form. The fitted ΔGneutral (−0.74 ± 0.5 kcal/mol), although not as well determined, indicates that in the hypothetical neutral state, the unfolded form is favored (Fig. 4A), and the charged carboxylate group is critical for stability of the folded state. The ΔΔGion contribution of the COOH group (2.9 kcal/mol at pH 6.5 and above) is too high for a single solvent-exposed salt bridge (31) and suggests that multiple COOH groups are involved in stabilization of the folded protein. Thus, the observed pH dependence of TrAvrPto stability can be modeled well by titration of its 7 His residues and 1 or more of its 4 Glu and 9 Asp residues. This modeling demonstrates that natively buried H87 is the major contributor to the pH-dependent component of free energy (ΔΔGion) in the pH range of 5–7.5 (Fig. 4B), the expected range of pH encountered in the biological system (Fig. 1A).
Fig. 4.
Multititration 2-state model. (A) This model separates out the pH-dependent components (ΔΔGion = ΔGUion − ΔGFion) of the denaturation free energy (ΔGF→U) and relates the populations of the F and U states to the pKa of individual ionizable residues (i) in each state i via ΔGjion (see Materials and Methods, Eq. 2). (B) Corresponding contribution (based on Eq. 2) of each His residue [H41 (open squares), H54 (filled diamonds), H75 (open diamonds), H87 (open circles), H103 (filled squares, overlapped with H130 data), H125 (filled triangles), H130 (filled circles)] to the ΔΔGion of the protein as a function of pH.
The contribution of H87 to the pH-dependent folding switch was tested by an H87Y mutation. Fluorescence data of the pH titration of the H87Y mutant shows a shift of the acid denaturation to a significantly lower pH (Fig. 2B). Modeling this data by using the same pKa values (excluding H87) of the WT TrAvrPto and allowing ΔGneutral to vary fit the H87Y fluorescence data well (Fig. 2B). The fitted ΔGneutral (−0.68 ± 0.01 kcal/mol) indicates that the H87Y mutation has shifted the Fneutral → Uneutral equilibrium toward Fneutral. This result is supported by an increase in melting temperature for the H87Y mutant relative to WT at pH 5.3 and 7.2 (Fig. 2C). These results provide remarkable validation of the 2-state multititration model and confirm the essential role of H87 in tuning the stability of TrAvrPto in the pH range of 5–7.
TrAvrPto is not the only protein in which its acid denaturation is controlled by the protonation state of a buried histidine. Other examples in which specific His residues are identified as key factors in the pH stability of a protein include the villin headpiece domain (30), and sperm whale apoglobin (29). With typical pKaF values in the vicinity of the pH of biological environments, His side chains are indeed used in nature to impart pH-sensing mechanisms within protein-folding equilibria.
H87 Is Conserved for AvrPto and AvrPtoB.
The conservation of an amino acid across multiple homologous proteins generally implies functional importance. Based on structural considerations, TrAvrPto has been aligned with an 80-residue segment of AvrPtoB (AvrPtoB121–200) despite low sequence identity (14%) (32). One distinct ortholog of TrAvrPto and 7 unique sequences of AvrPtoB121–200 orthologs were identified and aligned by using standard methods as described (SI Text). Final alignment was produced taking into account the structure-based TrAvrPto–AvrPtoB121–200 alignment (Fig. S3C) (32). Strikingly, H87 is the only His residue absolutely conserved within these sequences. The conservation of H87 implies that a His is required in this position in the protein. Given that H87 has no known catalytic role, the importance of this buried residue is expected to be structural. Importantly, most found homologs (with the exception of Ralstonia solanacearum) are Pseudomonas effector proteins secreted via the T3SS and would encounter similar pH environments upon translocation. Altogether, the stability, titration, structural, and simulation data presented here consistently point to the buried H87 as a pH-regulated folding switch for AvrPto. The conservation of this His among several related T3SS effector proteins suggests that it is a broadly used mechanism for sensing and responding to changes in pH.
The pH-sensitive stability of AvrPto characterized here, and its putative role in facilitating translocation of AvrPto via the T3SS, is strikingly reminiscent of a completely different pH-sensitive translocation system used by anthrax (33), botulinum, and diphtheria toxins (34). These toxins infect host cells via receptor-mediated endocytosis, and deliver specific proteins to the host cytoplasm from the acidic environment of the endosome. Elegant biophysical studies of the anthrax system, in which 2 large toxin proteins (lethal factor, LF, and edema factor, EF) are translocated from the endosome to the cytosol through a narrow (15 Å in diameter) integral membrane pore formed by 7 subunits of toxin protein PA63, show that acid denaturation of LF and EF is an initial step in their translocation (35). Their acid-induced unfolding is attributed to desolvated His residues in the native structure of each protein that are expected to have depressed pKa values and, consequently, to drive acid denaturation in the pH range of the endosome. This remarkable parallel with the behavior of AvrPto and its translocation via the T3SS illustrates how the fundamental pH sensitivity of His residues has been used in disparate translocation systems to generate finely tuned pH-regulated folding switches in unrelated protein folds to facilitate their delivery into the cytoplasm of a host cell.
Conclusions
The work presented here reveals important insights into how an effector protein can be efficiently unfolded in the bacterial cytoplasm and secreted by the T3SS but still refold into its functional form once inside the host cell cytoplasm. Under mildly acidic conditions, mimicking the pH when P. syringae is in the host plant tissue, TrAvrPto exists in a slowly exchanging equilibrium between folded and unfolded states. In the neutral pH environment of the host-cell cytoplasm, TrAvrPto is at its most stable, which ensures that most of the injected protein is in the folded and functional conformation. The observed loss of folded conformation as the pH is lowered from 7 to 5 is regulated predominantly by the ionization of the H87 side chain, a residue conserved for AvrPto and AvrPtoB homologs.
The dynamic behavior of AvrPto is complex. Three separate chemical exchange reactions have been observed and characterized: tautomer exchange, pH titration, and folding. Together, these processes yield a description of a pH-regulated thermodynamic equilibrium of AvrPto that correlates beautifully with the requirements of the biological system in which it operates, providing a detailed mechanism by which this protein senses and alters its conformation in response to changes in pH. The H87Y mutant confirms the critical role of H87 in fine tuning the pH dependence of AvrPto stability. The question that remains to be answered is whether the H87-mediated pH folding switch of AvrPto is directly coupled to the efficiency of AvrPto translocation via the P. syringae T3SS.
The studies presented herein firmly validate a model in which the pH dependence of stability is governed by differences between the pKaU and pKaF values for all titrating groups in a protein (27). Given this model, it should be possible to introduce pH-regulation of stability into any protein fold that can accommodate 1 or more desolvated His residues. Indeed, conservation of H87 in AvrPto and AvrPtoB homologues suggest that nature has accomplished this in the context of a 3-helix bundle fold. These principles should be straightforward to apply to the engineering of therapeutic proteins, either for their delivery via a translocation system that requires unfolding or for regulation of their function by pH.
Materials and Methods
Sample Preparation.
Proteins were expressed in Escherichia coli and affinity purified using standard protocols as described in detail in SI Text. The H87Y TrAvrPto mutant was obtained by modifying the original expression vector (22) as described (SI Text). All samples used for spectroscopic measurements were dialyzed into McIlvaine's citric acid-phosphate buffer (36) prepared at the desired pH and diluted to a 230 μM ionic strength as described in detail (SI Text).
CD and Fl Spectroscopy.
CD and Fl experiments were performed on 80 μM TrAvrPto samples by using standard protocols as described in detail in SI Text. Thermal denaturation of TrAvrPto was quantified by monitoring signal at 220 nm (CD) or 343 nm (Fl) as a function of temperature (Fig. 2C and Fig. S2) over the range 1–80 °C (CD) or 7–80 °C (Fl) from pH 4.0 to 8.0 for a total of 12 (CD) and 7 (Fl) pH samples. Standard methods were used to convert the temperature dependence of signals to pF values and their associated uncertainties as described in SI Text. Additionally, the acid denaturation of TrAvrPto-H87Y was measured by Fl as described (SI Text).
NMR Spectroscopy.
All NMR spectra were collected at 26.6 °C on a Varian Inova 600-MHz spectrometer with a {H,C,N} z axis gradient probe. Data were processed by using NMRPipe and NMRDraw (37), and peak volumes were determined by using Sparky (38). Integration settings assumed a Gaussian line shape with allowed adjustment of peak positions and line widths.
The pH dependence of TrAvrPto stability was measured by using fully relaxed 15N–1H fHSQC NMR spectra (23) of 0.3–0.6 mM TrAvrPto samples at pH 2.88–7.14 as described (SI Text). The folded population for each NH was calculated from the folded and unfolded peak volumes (VF and VU):
where AU and AF are factors that correct for different average relaxation rates in the unfolded (R2H,Uavg) and folded (R2H,Favg) states during INEPT transfer (Fig. S4), determined as described (SI Text). The reported pF and uncertainty at each pH (Fig. 2B) are the average and standard deviation over the individual pF values obtained by using Eq. 1 for all NHs used in the analysis. Concentration dependence of pF was investigated by comparing fully relaxed fHSQC spectra obtained for a 420 μM TrAvrPto sample and for the same sample diluted to 80 μM (Fig. S1 and Table S1).
Histidine pKa values were quantified by using a series of 15N–1H HMQC spectra acquired at pH 4.62–7.83 (Fig. 3A and Fig. S3B) by fitting the resulting chemical shifts vs. pH to the standard titration equation as described (SI Text). The 15N and 1H chemical shifts were fitted separately, with the reported pKa values reflecting the average and standard deviation for each His residue.
Fitting to the 2-State Multititration Model for TrAvrPto pH Dependence of Stability.
The pF values obtained from NMR, Fl, and CD were globally fit to a 2-state multititration model (27) (Fig. 4A). In this model, the denaturation free energy (ΔGF→U = ΔGneutral + ΔΔGion) is divided into pH-dependent (ΔΔGion) and pH-independent (ΔGneutral) components, where ΔGneutral is the denaturation free energy of the protein when all its ionizable groups are in the neutral state, and ΔΔGion = ΔGUion − ΔGFion, where ΔGjion (j = U or F) is the difference in free energy between the ionized and neutral forms of state j. Neglecting interactions between charged groups in each state, ΔGjion is given by ref. 27:
where N is the number of ionizable groups, γi is −1 or +1 for an acidic or basic group, respectively, and pKa,ij is the pKa of the ith ionizable group in state j. Because ΔGjion depends only on the pKa,ij values and pH, the above model expresses the pH dependence of ΔGF→U (and hence, of pF) for a given set of pKa,ij values (Fig. 4A). Notably, ΔΔGion depends on differences between pKa values in the U and F states; titrating groups with elevated or depressed pKaF relative to pKaU will dominate the pH dependence of stability. Populations were calculated from ΔGneutral, ΔGUion and ΔGFion (Fig. 4A), and the total folded population was calculated as pFobs = pFneutral + pFion, the sum of neutral and ionized folded populations. By using the experimentally determined pKaF values for the 7 histidines, assuming pKaU for all, and including an unknown COOH group (fitted parameters pKa,COOHF and pKa,COOHU the fit to the experimental pF vs. pH data for WT TrAvrPto was optimized by using least-squares analysis to extract pKa,COOHF, pKa,COOHU, and ΔGneutral. The errors in the fitted parameters were estimated by a Monte Carlo analysis as described (SI Text). The H87Y pF vs. pH experimental data were fitted by eliminating the contribution of H87 from the calculation of ΔΔGion, but otherwise by using the same pKa values as for the WT best fit and varying ΔGneutral to optimize the fit. The fitting routines were implemented in MATLAB 2007a, and the final figures were generated in Microsoft Excel 2007.
Supplementary Material
Acknowledgments.
We thank Dr. Cynthia Kinsland of the Cornell University Protein Facility (Ithaca, NY), Dr. Brian Zoltowski, Dr. Lea Vacca Michel, and Prof. Brian Crane for assistance with fluorescence and CD measurements and Dr. Jens Nielsen of University College, Dublin, Ireland, for helpful discussions. This work was supported by National Science Foundation Grant MCB-0641582 and National Institutes of Health Molecular Biophysics Training Grant T32 GM08267.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809138106/DCSupplemental.
References
- 1.Galan JE, Collmer A. Type III secretion machines: Bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd SA, Norman M, Rosqvist R, Wolf-Watz H. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol Microbiol. 2001;39:520–531. doi: 10.1046/j.1365-2958.2001.02271.x. [DOI] [PubMed] [Google Scholar]
- 3.Anderson DM, Fouts DE, Collmer A, Schneewind O. Reciprocal secretion of proteins by the bacterial type III machines of plant and animal pathogens suggests universal recognition of mRNA targeting signals. Proc Natl Acad Sci USA. 1999;96:12839–12843. doi: 10.1073/pnas.96.22.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfano JR, Collmer A. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu Rev Phytopathol. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- 5.Johnson S, Deane JE, Lea SM. The type III needle and the damage done. Curr Opin Struct Biol. 2005;15:700–707. doi: 10.1016/j.sbi.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Blocker A, et al. Structure and composition of the Shigella flexneri“needle complex”, a part of its type III secreton. Mol Microbiol. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- 7.Cordes FS, et al. Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem. 2003;278:17103–17107. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- 8.Lee VT, Schneewind O. Yop fusions to tightly folded protein domains and their effects on Yersinia enterocolitica type III secretion. J Bacteriol. 2002;184:3740–3745. doi: 10.1128/JB.184.13.3740-3745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorg JA, Miller NC, Marketon MM, Schneewind O. Rejection of impassable substrates by Yersinia type III secretion machines. J Bacteriol. 2005;187:7090–7102. doi: 10.1128/JB.187.20.7090-7102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh P. Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev. 2004;68:771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stebbins CE, Galan JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- 12.Buttner D, Bonas U. Who comes first? How plant pathogenic bacteria orchestrate type III secretion. Curr Opin Microbiol. 2006;9:193–200. doi: 10.1016/j.mib.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Grignon C, Sentenac H. pH and ionic conditions in the apoplast. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:102–128. [Google Scholar]
- 14.Minamino T, Imae Y, Oosawa F, Kobayashi Y, Oosawa K. Effect of intracellular pH on rotational speed of bacterial flagellar motors. J Bacteriol. 2003;185:1190–1194. doi: 10.1128/JB.185.4.1190-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilharm G, et al. Yersinia enterocolitica type III secretion depends on the proton motive force but not on the flagellar motor components MotA and MotB. Infect Immun. 2004;72:4004–4009. doi: 10.1128/IAI.72.7.4004-4009.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Q, Thilmony R, Zwiesler-Vollick J, He SY. Type III protein secretion in Pseudomonas syringae. Microbes Infect. 2003;5:301–310. doi: 10.1016/s1286-4579(03)00032-7. [DOI] [PubMed] [Google Scholar]
- 17.van Dijk K, et al. The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature- and pH-sensitive manner. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedley KF, Martin GB. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol. 2003;41:215–243. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- 19.Shan L, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wulf J, Pascuzzi PE, Fahmy A, Martin GB, Nicholson LK. The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis. Structure (London) 2004;12:1257–1268. doi: 10.1016/j.str.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 22.Dawson JE, Nicholson LK. Folding kinetics and thermodynamics of Pseudomonas syringae effector protein AvrPto provide insight into translocation via the type III secretion system. Protein Sci. 2008;17:1109–1119. doi: 10.1110/ps.034223.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulder FAA, Spronk CAEM, Slijper M, Kaptein R, Boelens R. Improved HSQC experiments for the observation of exchange broadened signals. J Biomol NMR. 1996;8:223–228. doi: 10.1007/BF00211169. [DOI] [PubMed] [Google Scholar]
- 24.Martin SR, Schilstra MJ. Circular dichroism and its application to the study of biomolecules. Methods Cell Biol. 2008;84:263–293. doi: 10.1016/S0091-679X(07)84010-6. [DOI] [PubMed] [Google Scholar]
- 25.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 26.Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J Bacteriol. 2004;186:543–555. doi: 10.1128/JB.186.2.543-555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang AS, Honig B. On the pH dependence of protein stability. J Mol Biol. 1993;231:459–474. doi: 10.1006/jmbi.1993.1294. [DOI] [PubMed] [Google Scholar]
- 28.Pelton JG, Torchia DA, Meadow ND, Roseman S. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 1993;2:543–558. doi: 10.1002/pro.5560020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geierstanger B, Jamin M, Volkman BF, Baldwin RL. Protonation behavior of histidine 24 and histidine 119 in forming the pH 4 folding intermediate of apomyoglobin. Biochemistry. 1998;37:4254–4265. doi: 10.1021/bi972516+. [DOI] [PubMed] [Google Scholar]
- 30.Grey MJ, et al. Characterizing a partially folded intermediate of the villin headpiece domain under non-denaturing conditions: Contribution of His41 to the pH-dependent stability of the N-terminal subdomain. J Mol Biol. 2006;355:1078–1094. doi: 10.1016/j.jmb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Horovitz A, Serrano L, Avron B, Bycroft M, Fersht AR. Strength and co-operativity of contributions of surface salt bridges to protein stability. J Mol Biol. 1990;216:1031–1044. doi: 10.1016/S0022-2836(99)80018-7. [DOI] [PubMed] [Google Scholar]
- 32.Xiao F, et al. The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant J. 2007;52:595–614. doi: 10.1111/j.1365-313X.2007.03259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finkelstein A. Proton-coupled protein transport through the anthrax toxin channel. Philos Trans R Soc London Ser B. 2009;364:209–215. doi: 10.1098/rstb.2008.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falnes PO, Sandvig K. Penetration of protein toxins into cells. Curr Opin Cell Biol. 2000;12:407–413. doi: 10.1016/s0955-0674(00)00109-5. [DOI] [PubMed] [Google Scholar]
- 35.Krantz BA, Trivedi AD, Cunningham K, Christensen KA, Collier RJ. Acid-induced unfolding of the amino-terminal domains of the lethal and edema factors of anthrax toxin. J Mol Biol. 2004;344:739–756. doi: 10.1016/j.jmb.2004.09.067. [DOI] [PubMed] [Google Scholar]
- 36.McIlvaine TC. A buffer solution for colorimetric comparison. J Biol Chem. 1921;49:183–186. [Google Scholar]
- 37.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Goddard TD, Kneller DG. San Francisco: University of California; 2008. Sparky 3. ver. 3.115. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.