Abstract
ASC-2, a multifunctional coactivator, forms a steady-state complex, named ASCOM (for ASC-2 COMplex), that contains the histone H3-lysine-4 (H3K4)-methyltransferase MLL3 or its paralogue MLL4. Somewhat surprisingly, given prior indications of redundancy between MLL3 and MLL4, targeted inactivation of the MLL3 H3K4-methylation activity in mice is found to result in ureter epithelial tumors. Interestingly, this phenotype is exacerbated in a p53+/− background and the tumorigenic cells are heavily immunostained for γH2AX, indicating a contribution of MLL3 to the DNA damage response pathway through p53. Consistent with the in vivo observations, and the demonstration of a direct interaction between p53 and ASCOM, cell-based assays have revealed that ASCOM, through ASC-2 and MLL3/4, acts as a p53 coactivator and is required for H3K4-trimethyation and expression of endogenous p53-target genes in response to the DNA damaging agent doxorubicin. In support of redundant functions for MLL3 and MLL4 for some events, siRNA-mediated down-regulation of both MLL3 and MLL4 is required to suppress doxorubicin-inducible expression of several p53-target genes. Importantly, this study identifies a specific H3K4 methytransferase complex, ASCOM, as a physiologically relevant coactivator for p53 and implicates ASCOM in the p53 tumor suppression pathway in vivo.
Activating signal cointegrator-2 (ASC-2; also named AIB3, TRBP, TRAP250, NRC, NCOA6, and PRIP) is a coactivator of numerous nuclear receptors, and other transcription factors (1). Its physiological importance as a key coactivator, most notably for several nuclear receptors, has been demonstrated in studies with various ASC-2 mouse models (1, 2). Mechanistically, ASC-2 has been shown to interact directly with DNA-binding transcription factors and to be associated with cofactors involved in histone H3K4 methylation. H3K4 methylation is an evolutionarily conserved mark for transcriptionally active chromatin that counters the generally repressive chromatin environment imposed by H3K9 or H3K27 methylation in higher eukaryotes (3). Notably, H3K4 trimethylation is associated with promoters and early transcribed regions of active genes (4, 5).
Enzymes that carry out H3K4 methylation include yeast Set1 (ySet1) and mammalian Set1, MLL1, MLL2, MLL3/HALR, MLL4/ALR, Ash1, and Set7/9 (6). These proteins contain a conserved SET domain that is associated with an intrinsic histone lysine-specific methyltransferase activity (3, 6), and the Set1 and MLL enzymes form a family of evolutionarily conserved complexes that are collectively named Set1-like complexes (6). ASC-2 is an integral component of the first-described mammalian Set1-like complex that we named ASCOM (for ASC-2 complex) and that contains MLL3 or MLL4 (7, 8). We and others have shown that ASCOM is indeed an H3K4 methyltransferase complex (7–10). More recent studies identified additional ASCOM-specific components that include PTIP, PTIP-associated 1 (PA1), and UTX (9, 10). UTX has been found to be an H3K27 demethyase (11–14). Thus, ASCOM contains 2 distinct histone modifiers that are linked to transcriptional activation.
To elucidate the physiological role of ASCOM, we recently established a homozygous mouse line designated MLL3Δ/Δ (8). In these mice, wild-type MLL3 is replaced by a mutant MLL3 that contains an in-frame deletion of a 61-aa catalytic core region in the MLL3 SET domain and that is expressed and incorporated into ASCOM (8). Unlike ASC-2-null mice, which die at approximately E9.5-E13.5 (1), MLL3Δ/Δ mice show only a partial embryonic lethality (8). Interestingly, genetic data are also indicative of interactions between MLL3 and ASC-2, because MLL3Δ/Δ and isogenic ASC-2+/− (15) mice share at least 3 phenotypes that include stunted growth, decreased cellular doubling rate, and lower fertility (8).
Importantly, ASC-2 was originally identified on the basis of its gene-amplification and overexpression in human breast and other cancers (16–18). However, it has been unclear whether ASC-2 is involved in any aspect of pro- or antitumorigenesis, particularly with the multitude of target transcription factors for ASC-2 (1). In this report, we find redundant but crucial roles for ASCOM-MLL3 and ASCOM-MLL4 in transactivation by the tumor suppressor p53. Moreover, we show that targeted inactivation of MLL3 H3K4-methylation activity in the mouse results in ureter epithelial tumors accompanied by an increased level of DNA damage and that ASCOM appears to be important for DNA damage-induced expression of p53-target genes. Thus, our results show that ASCOM serves as a tumor suppressive coactivator complex in vivo, at least in part through its ability to function as a coactivator of the tumor suppressor p53.
Results
Targeted Inactivation of MLL3 H3K4 Methyltransferase Activity in the Mouse Causes Ureter Epithelial Tumors.
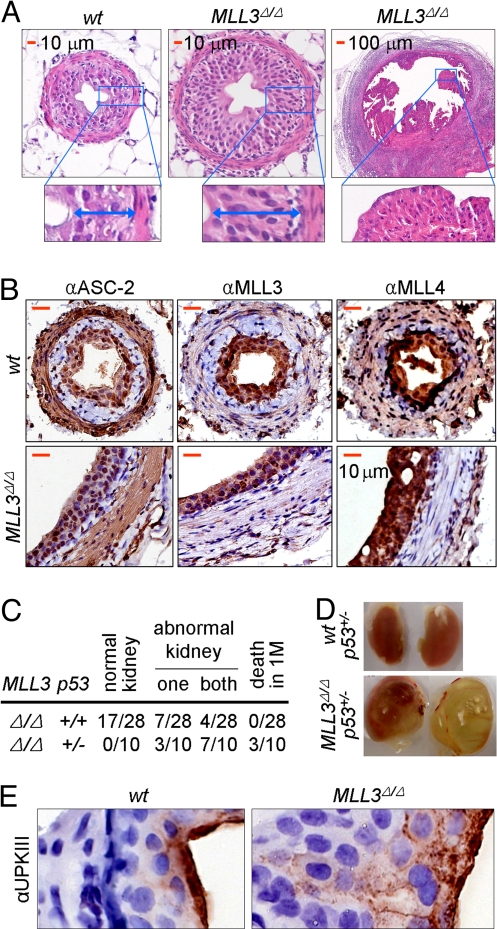
Our recently established MLL3Δ/Δ mice showed not only the aforementioned 3 phenotypes (stunted growth, decreased cellular doubling rate, and lower fertility) shared by isogenic ASC-2+/− mice (15), but also 2 additional phenotypes. First, all MLL3Δ/Δ mice had very little white fat, which led us to identify crucial roles for ASCOM in adipogenesis (19). Second, within 4 months after birth, ≈50% of MLL3Δ/Δ mice displayed unusual hyperproliferation and tumors in the innermost layer of ureter cells (i.e., urothelial cells) located close to the renal pelvis (Fig. 1A and Fig. S1A). In this layer of cells, MLL3 is highly enriched together with ASC-2 and MLL4 (Fig. 1B). Remarkably, in a p53+/− background, 100% of the MLL3Δ/Δ mice developed this tumorigenic phenotype within 1–2 months after birth (Fig. 1 C and D). The affected kidneys of MLL3Δ/Δ mice also displayed features associated with primary vesicoureteral reflux, a hereditary disease affecting ≈1% of pregnancies and representing a leading cause of renal failure in infants (20). These include hydronephrosis and kidney abnormalities such as expanded renal pelvic space, and effacement of medullary structure and thinning of cortex with expanded interstitial compartment and dilated and atrophied tubules (Fig. 1D and Fig. S1B). In addition, an Indian Ink solution instilled into the bladders of anesthetized mice back-flowed into the ureters (Fig. S2A). Similar phenotypes have also been observed in mice lacking uroplakin protein UPII or UPIII (21, 22). Uroplakins form 2-dimensional crystals (urothelial plaques) that cover >90% of the apical urothelial surface and contribute to its stabilization and to the permeability barrier function of the urothelium (20–22). Although intense staining of the apical urothelial surface by antibodies against uroplakin III and other uroplakins was evident in wild-type mice, only diffuse cytoplasmic and basal-lateral cell periphery staining without apical enrichment was observed in MLL3Δ/Δ mice (Fig. 1E, Fig. S2B). This suggests that the apical urothelial surface is severely disturbed in MLL3Δ/Δ mice. More studies are needed to determine whether hyperproliferative urothelial cells lead to the primary vesicoureteral reflux-like phenotypes in MLL3Δ/Δ mice as a result of perturbed apical urothelial surfaces. Nonetheless, because the p53 haploinsufficiency exacerbates the mutant phenotype, these results raise the interesting possibility that the ASCOM-MLL3 complex may serve as a coactivator for the tumor suppressor p53.
Fig. 1.
Inactivation of MLL3 H3K4 methyltransferase activity results in urothelial tumors. (A) Analysis of 3- to 3.5-month-old wild-type and MLL3Δ/Δ mouse ureters, using hematoxylin and eosin-stained paraffin sections revealed hyperproliferation and tumors of urothelial cells. (B and E) Paraffin-sections of 3- to 3.5-month-old wild-type and MLL3Δ/Δ mouse urothelia were deparaffinized and stained immunohistochemically, using antibodies to ASC-2, MLL3, and MLL4 (B) and uroplakin III (E). (Scale bars: 10 μm.) (C) The kidney abnormality appears to involve p53, as evident from the comparative analysis of 1- to 4-month-old MLL3Δ/Δ mice with wild-type and p53+/− background. (D) Kidney abnormalities such as hydronephrosis are evident in MLL3Δ/Δ mice. MLL3Δ/Δ:p53+/− compound mice with defects in both kidneys are shown.
ASCOM as a Key Player in DNA Damage Response.
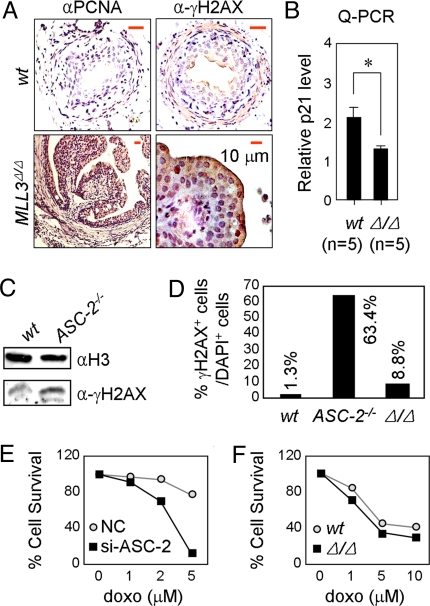
Based on the importance of p53 in DNA damage repair (23), an earlier demonstration of DNA damage-induced H3K4 methylation on a p53 target gene (24), and the potential role of ASCOM-MLL3 as a coactivator complex for p53, we examined the level of proliferation and DNA damage in tumorigenic urothelial cells of MLL3Δ/Δ mice. The terminally differentiated wild-type urothelial cells are expected to cease proliferation. However, tumorigenic urothelial cells of MLL3Δ/Δ mice showed strong immunostaining for PCNA, a mark for cellular proliferation (Fig. 2A), consistent with an impaired function of p53 in blocking cell cycle progression. Relative to urothelial cells in wild-type mice, the tumorigenic urothelial cells of MLL3Δ/Δ mice also showed more intense immunostaining for γH2AX (Fig. 2A), a mark for damaged DNA. This is suggestive of a role for MLL3 in repairing damaged DNA through the ASCOM complex, presumably through coactivation of p53 target genes whose products are involved in DNA repair (23). Although these genes have not yet been examined, a Q-PCR analysis of ureter mRNAs revealed that expression of p21, a prominent p53 target gene, was significantly lower in MLL3Δ/Δ mice relative to wild-type mice (Fig. 2B). Two additional observations related to the ASC-2 subunit of ASCOM are further suggestive of a p53 coactivator function for this complex. First, E.9.5 ASC-2−/− MEFs displayed an increased level of γH2AX relative to wild-type cells (Fig. 2C). In addition, when cells were challenged with 0.1 μM doxorubicin for 24 h and allowed to recover for 3 h, ASC-2−/− cells showed a significantly increased number of γH2AX-positive cells compared with wild-type cells (Fig. 2D), clearly demonstrating a reduced ability of ASC-2−/− cells to repair damaged DNA. These results raise the interesting possibility that ASC-2 may directly regulate expression of p53 target genes needed for DNA repair, although it also is possible that ASC-2 may act through other transcription factors involved in DNA repair. Second, HeLa cells expressing siRNA against ASC-2 showed a markedly increased sensitivity to increasing amounts of doxorubicin (Fig. 2E). Under these conditions, MLL3Δ/Δ cells showed more γH2AX-positive cells than did wild-type cells but significantly less than did ASC-2-null cells (Fig. 2D). Correspondingly, E9.5 MLL3Δ/Δ MEF cells were slightly, albeit reproducibly, more sensitive to doxorubicin than were wild-type cells (Fig. 2F). Overall, these results suggest that both ASCOM-MLL3 and ASCOM-MLL4 may play crucial roles in the DNA repair pathway by p53.
Fig. 2.
ASCOM is involved in DNA repair. (A) Paraffin-sections of 3- to 3.5-month-old wild-type and MLL3Δ/Δ mouse urothelia were deparaffinized and immunostained using antibodies to PCNA and γH2AX. All scale bars represent 10 μm. (B) 3- to 3.5-month-old wild-type and MLL3Δ/Δ ureters were tested for p21 transcript levels by Q-PCR. (C) E9.5 wild-type and ASC-2−/− MEFs were immunoblotted with H3 and γH2AX antibodies. (D) Wild-type, ASC-2−/− or MLL3Δ/Δ MEF cell lines were immunostained with DAPI and γH2AX antibodies and the number of γH2AX-positive cells were counted compared with DAPI-positive cells. (E and F) HeLa cells with control si or si-ASC-2 (E) or E9.5 wild-type or MLL3Δ/Δ MEFs (F) were seeded in 48-well plates, treated with an increasing amount of doxorubicin, harvested, and counted. Shown is a representative experiment.
ASCOM as a Key Coactivator of p53.
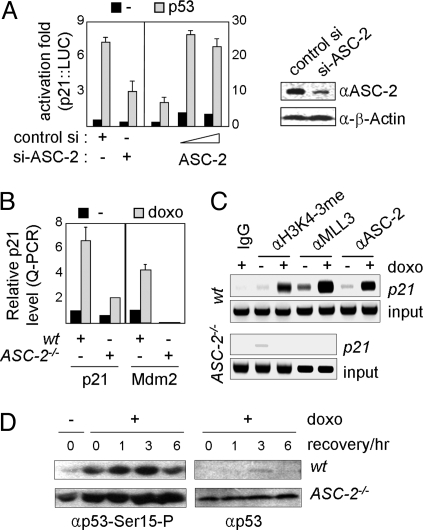
In support of the role for ASC-2, through ASCOM, as a p53 coactivator, p53-mediated transactivation of p21:LUC, a reporter directed by p53-response elements (p53REs) in the p21 promoter, was impaired by siRNA against ASC-2 (8) in HeLa cells (Fig. 3A Left), whereas it was enhanced by ectopically expressed ASC-2 (Fig. 3A Right). Correspondingly, doxorubicin-induced expression of endogenous p21 and Mdm2, 2 p53 target genes, was severely impaired in primary E9.5 ASC-2−/− MEFs compared with E9.5 wild-type MEFs (Fig. 3B). Moreover, chromatin immunoprecipitation (ChIP) experiments revealed doxorubicin-dependent recruitment of ASC-2 and MLL3 to endogenous p21-p53REs in a wild-type MEF cell line but not in an ASC-2−/− MEF cell line (Fig. 3C). Recruitment of ASC-2 and MLL3 to p21-p53REs was correlated with enhanced H3K4 trimethylation, but this methylation was also abolished in an ASC-2−/− MEF cell line (Fig. 3C). Interestingly, in an ASC-2−/− MEF cell line, phosphorylation of p53 at Ser-15, a mark for p53 activation by ataxia telangiectasia mutated (ATM) (23), was observed even before doxorubicin treatment and further enhanced by doxorubicin treatment (Fig. 3D). These results suggest that ASC-2−/− cells have an intact ATM signaling pathway for p53 activation and that p53 is constitutively activated, likely reflecting the DNA-damaged state of the cells. In addition, p53 appeared to be considerably more stabilized in the ASC-2−/− MEF cell line than in the wild-type MEF cell line (Fig. 3D), consistent with the down-regulated expression of Mdm2 in ASC-2−/− cells.
Fig. 3.
ASCOM as a coactivator of p53. (A) p21:LUC reporter, either without or with a p53 expression vector, was transfected into HEK293 cells along with control siRNA or ASC-2-siRNA and into HeLa cells along with an increasing amount of ASC-2. Cells were then incubated for 12 h and tested for luciferase activity. Down-regulation of ASC-2 is as shown in immunoblotting compared with β-actin as control. (B) E9.5 MEFs from wild-type and ASC-2−/− mice were treated with vehicle or 0.1 μM doxorubicin for 24 h and allowed to recover for 1 h, after which they were then tested for p21 and Mdm2 transcript levels by Q-PCR. (C) Wild-type and ASC-2−/− MEF cell lines were subjected to ChIP assays for H3K4 trimethylation and MLL3/ASC-2 recruitment during treatment with vehicle or 0.4 μM doxorubicin for 8 h. (D) Wild-type and ASC-2−/− MEF cell lines treated with 5 μM doxorubicin for 2 h and allowed to recover for 0–6 h were immunoblotted with antibodies against p53 phosphorylated at Ser-15 and p53.
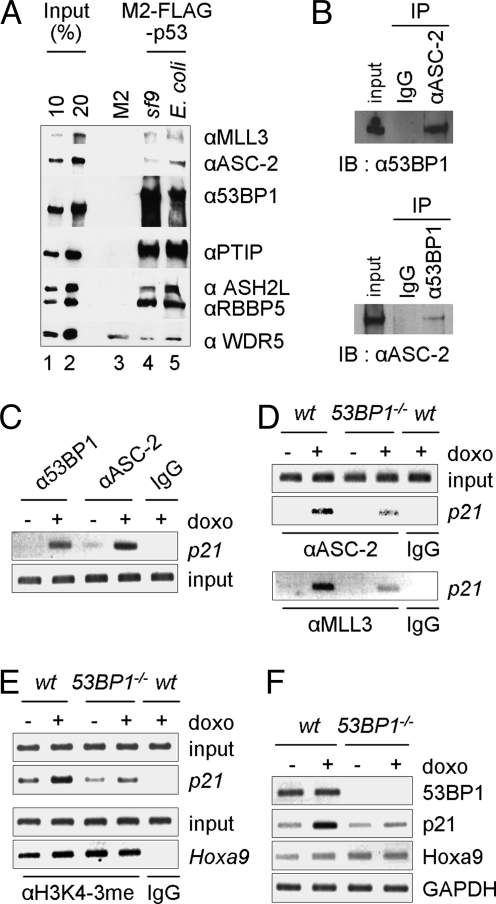
In further support of the coactivator role of ASCOM for p53, we found that an affinity purified ASCOM (19, Fig. S3A) directly interacts with immobilized p53. As shown in Fig. 4A, this complex bound equally well to M2-agarose-immobilized p53 purified from Sf9 cells or from Escherichia coli but not to M2-agarose. Apart from binding of subunits (ASH2L, RBBP5 and WDR5) common to the various Set1-like complexes, MLL3 and other subunits (ASC-2 and PTIP) unique to the MLL3/MLL4 complexes also showed specific binding. The molecular basis underlying this association does not involve ASC-2, because it failed to interact with p53. Interestingly, our purified complex contains 53BP1 (Fig. S3A), a protein originally identified as a p53-interacting coactivator (25–27) and subsequently found to be a major DNA repair effector (for a review, see ref 28). Consistent with the recent finding by Cho et al. (9), our chromatographic purification revealed that at least a fraction of nuclear 53BP1 copurifies with ASCOM (Fig. S3B). We also found that 53BP1 is readily coimmunoprecipitated with ASC-2 and vice versa (Fig. 4B) and that a C-terminal region of 53BP1 directly interacts with the N-terminal region of ASC-2 (Fig. S3 C–E). Moreover, 53BP1 was recruited, along with ASC-2, to p21-p53REs in response to doxorubicin (Fig. 4C). These results support the previous finding that 53BP1 also functions as a coactivator of p53 (26, 27), and suggest an interesting adaptor role for 53BP1 in linking p53 and ASCOM. Indeed, doxorubicin-induced recruitment of ASC-2 and MLL3 to p21-p53REs (Fig. 4D), H3K4-trimethylation of p21-p53REs (Fig. 4E) and expression of p21 (Fig. 4F) were significantly reduced in a 53BP1−/− MEF cell line relative to a wild-type MEF cell line. In contrast, H3K4-trimethylation and expression of Hoxa9, a target gene of the MLL1 complex (29), were indistinguishable between the 2 cell types (Fig. 4 E and F). Importantly, recruitment of ASC-2/MLL3 to p21-p53REs was reduced but not abolished in the 53BP1−/− MEF cell line (Fig. 4D), suggesting the existence of yet other redundant pathways for ASCOM recruitment by p53. These results suggest that 53BP1 is at least in part responsible for tethering ASCOM to p53.
Fig. 4.
Interactions of ASCOM with p53 and identification of 53BP1 as an adaptor between ASCOM and p53. (A) Direct binding of ASCOM to p53. The purified complex (ref. 19 and Fig. S3A) was incubated with M2-agarose or M2-agarose-immobilized Flag-p53 fusion proteins expressed in and purified either from Sf9 cells or from E. coli. Bound proteins were scored by immunoblot with the indicated antibodies. (B) 53BP1 is coimmunoprecipitated by ASC-2 antibody from HEK293 cells and vice versa. (C–E) HepG2 cells (C) or wild-type and 53BP1−/− MEF cell lines (D and E) were subjected to ChIP assays for MLL3/ASC-2 recruitment and H3K4 trimethylation after treatment with vehicle or 0.4 μM doxorubicin for 8 h. (F) RT-PCR analyses of 53BP1, p21, Hoxa9, and GAPDH expression were carried out for wild-type and 53BP1−/− MEF cell lines.
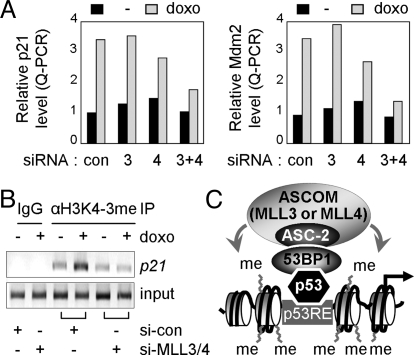
Finally, in further support of at least partially redundant functions for MLL3 and MLL4 in p53 transactivation, the doxorubicin-induced expression level of p21 and Mdm2 was minimally affected by each siRNA alone but more significantly impaired by joint expression of both siRNAs (Fig. 5A). Doxorubicin-induced H3K4-trimethylation of p21-p53REs was also impaired by inclusion of both siRNAs but not by either individual siRNA alone (Fig. 5B).
Fig. 5.
Redundant functions for MLL3 and MLL4 in p53-mediated activation. (A) HepG2 cells were transfected with control siRNA, siRNA against MLL3 or MLL4s, or siRNAs for both MLL3 and MLL4. Two days after transfection, cells were treated with vehicle or 0.5 μM doxorubicin for 24 h, and they were then tested for p21 and Mdm2 transcript levels by Q-PCR. (B) HepG2 cells expressing control siRNA or siRNAs for both MLL3 and MLL4 were subjected to ChIP assays for H3K4 trimethylation during treatment with vehicle or 0.5 μM doxorubicin for 24 h. (C) Working model. ASCOM-MLL3 and ASCOM-MLL4 are recruited to p53RE-bound p53, at least in part, through 53BP1 and thereby carry out H3K4 trimethylation of p53 target genes.
Collectively, these results demonstrate that ASCOM-MLL3 and ASCOM-MLL4 function as redundant but crucial H3K4-trimethylating coactivator complexes for p53.
Discussion
ASC-2, a coactivator with potential links to cancer (16–18), resides in a steady-state complex, ASCOM, that contains the H3K4 methyltransferase MLL3 or its paralogue MLL4 (7, 8). To establish potentially interrelated functions of ASC2 and MLL3/4, we have used a variety of in vitro and in vivo assays (8, 19, 30). Interestingly, our recent analysis of a homozygous mouse line that expresses an enzymatically inactivated mutant form of MLL3 demonstrated genetic interactions between ASC-2 and MLL3 (8, 15). Therefore, and because these animals show only a partial embryonic lethality (8) as opposed to the early embryonic lethality of ASC-2-null mice (1), they serve as an excellent model system to study the physiological function of ASCOM. Surprisingly, our current study has revealed that the MLL3 mutant mice spontaneously develop ureter epithelial tumors and, further, that ASCOM-MLL3 and ASCOM-MLL4 act as redundant but crucial p53 coactivators and are required for H3K4 trimethyation and expression of endogenous p53 target genes in response to the DNA damaging agent doxorubicin. This study provides documentation of a physiologically relevant H3K4 methyltransferase coactivator complex for p53. Importantly, our results also implicate ASCOM in the p53 tumor suppression pathway in vivo.
Multiple lines of evidence further support the tumor suppressor function for ASCOM. First, in an interesting study carried out in mouse models (31), haploid inactivation of ASC-2 was found to accelerate polyoma middle-T antigen-induced mammary tumorigenesis. Second, PTIP, which has been proposed to play important roles in cellular responses to DNA damage (32, 33), recently was identified as an additional component of ASCOM (9, 10). In this regard, it is important to note that the tumor suppressor p53 plays a key role in countering the adverse effects of DNA damage (23), which otherwise can be lethal or lead to oncogenic transformation, and that DNA damage induces the transcriptional activity of p53 via damage sensors such as ATM (23). It thus is interesting that PTIP appears to be required for ATM-mediated phosphorylation of p53 at Ser-15 and for DNA damage-induced up-regulation of the cyclin-dependent kinase inhibitor p21 (34). Correspondingly, loss-of-function studies in mice indicate that PTIP is essential for the maintenance of genomic stability (32). One intriguing future challenge is to test whether this function of PTIP occurs in the context of ASCOM, particularly because ASC-2 null cells show increased phosphorylation of p53 at Ser-15. Regardless, and as our results suggest that ASCOM acts as a tumor suppressive coactivator complex for p53, ASCOM, like PTIP, may also play crucial roles in the maintenance of genomic stability. Indeed, ASC-2 and MLL3/4 appear to be important for DNA damage-induced expression of p53-target genes, and ureter epithelial tumors of the MLL3 mutant mice are accompanied by an increased level of DNA damage. Third, a number of recent reports also suggest a tumor suppressive role for MLL3. Thus, MLL3 was identified as a gene displaying somatic mutations in breast and colorectal cancers (35); and subsequent evaluations also identified somatic mutations of MLL3 in glioblastoma, melanoma, and pancreatic carcinoma (36). Notably, MLL3 is located on 7q36, a region for common chromosomal aberrations in myeloid leukemic patients (37, 38).
Because a majority of tumors are associated with a loss of p53 function, and in view of the role of ASCOM as a p53 coactivator, the seeming propensity of MLL3Δ/Δ mice to selectively develop urothelial tumors may appear paradoxical. However, a more careful examination may also uncover other types of tumors. In fact, ASCOM may guard against a broad range of epithelial tumors, as noted above, and, for instance, acceleration of polyoma middle-T antigen-induced mammary tumorigenesis by haploid inactivation of ASC-2 (31) may similarly involve genomic instability caused by the impaired ability of ASCOM to support p53 transactivation. Nonetheless, it also is possible that, along with a general redundancy between ASCOM-MLL3 and ASCOM-MLL4, there may be a more specific (and as yet unknown) function of MLL3 in the urothelium that selectively leads to urothelial tumors in MLL3Δ/Δ mice. It should also be noted that the mutant form of MLL3 in MLL3Δ/Δ mice lacks H3K4 methyltransferase activity but is incorporated into the ASCOM complex (8) and thus has the potential to act in a dominant negative fashion against functional MLL4-containing ASCOM complexes by competing for binding to p53 or other transcription factors. In such a case, if ASCOM-MLL3 were the only critical Set1-like coactivator complex for p53 in urothelial cells, genetic removal of p53 would not be expected to enhance the ureteral tumor phenotype of MLL3Δ/Δ mice. Therefore, on the assumption that MLL3Δ/Δ/ASCOM has a significant dominant negative effect, our results raise the possibility that p53 also uses other Set1-like complexes, most likely ASCOM-MLL4, at least in urothelial tumor suppressor function. With the newly established MLL4 mutant mice in the laboratory, along with mice with the floxed allele of ASC-2 (39), we are poised to clarify these issues in the future.
The notion of an ASCOM coactivator function for p53 is strongly supported by our demonstration of a direct interaction of ASCOM (containing ASC-2, MLL3/4, 53BP1, PTIP, and other ASCOM subunits) with p53. Interestingly, ASC-2 appears to play a crucial role in effecting MLL3/4 function on p53 target genes, because our results reveal that doxorubicin-induced ASCOM-MLL3 and ASCOM-MLL4 recruitment, corresponding H3K4 trimethylation, and activation of p53 target genes depends on ASC-2 (Fig. 5C). However, the precise role of ASC-2 in p53 transactivation is not clear. Our search for the mechanistic basis underlying the association of ASCOM with p53 led to the surprising finding that 53BP1 is recruited to p21-p53REs. Moreover, the timing of this recruitment precisely overlaps that of ASC-2 recruitment, suggesting corecruitment of these 2 proteins. These results support the previously proposed role for 53BP1 as a coactivator of p53 (26, 27). Our proposal for an adaptor role for 53BP1 between p53 and ASCOM (Fig. 5C) is further supported by our finding that 53BP1 and ASC-2 are readily coimmunopurified from cellular extracts and that 53BP1 appears to directly interact not only with p53 (25) but also with ASC-2. These results suggest that ASC-2 may play a critical role in linking the core MLL3/4 H3K4-methyltransferase unit to 53BP1, which in turn may facilitate ASCOM binding to p53 (Fig. 5C). However, because a significant level of ASC-2/MLL3 is still recruited to p21-p53REs in 53BP1 null cells, there are likely additional mechanisms for ASCOM recruitment. Thus, the detailed molecular basis underlying the recruitment of ASCOM to p53 remains to be further delineated.
Finally, mammals have additional, related H3K4 trimethyltransferase complexes. Interestingly, an MLL1 complex with common core subunits that are also present in the ASCOM MLL3 and MLL4 complexes was shown to exhibit modest p53 coactivator function in vitro (29). This may appear to be inconsistent not only with our in vivo ChIP results, which suggest that ASCOM is a critical player in H3K4 methylation at p21-p53REs, but also with the doxorubicin sensitivity of ASC-2 and MLL3 mutant cells, which suggests that ASCOM is a major H3K4 methyltransferase complex for, at least, the p53-mediated DNA repair program. However, because the spectrum of p53 target genes is much more complex, we cannot exclude possible roles for other H3K4 methyltransferase complexes, perhaps even in conjunction with an MLL3/4 complex, on a select subset of p53-target genes. Similarly, ASCOM-MLL3 and ASCOM-MLL4, in addition to their shared p53-target genes, might also control a select sets of p53 target genes, respectively.
In summary, this study implicates ASC-2 and MLL3 in the p53 tumor suppression pathway, and identifies ASCOM as a major H3K4 methyltransferase coactivator complex for the tumor suppressor p53. Our results present this complex as a possible new therapeutic target for development of anticancer drugs. For instance, by delivering antagonists of MLL3/4 H3K4 methyltransferase activity to cancer cells with wild-type MLL3/4, it may be possible to make them more sensitive to irradiation therapy. In addition, by delivering agonists to normal or early tumorigenic cells with intact MLL3/4, it may be possible to help them resist tumorigenic insults such as DNA damaging agents.
Materials and Methods
Plasmids and Transfections.
The reporter p21:LUC, the transfection indicator pRSV-ß-gal, the mammalian ASC-2 expression vector and plasmids encoding siRNAs against ASC-2, MLL3 and MLL4 were as described (7, 8, 40). Transfection and luciferase assays were performed in triplicate as described in ref. 41, and the results were normalized to the LacZ expression. Similar results were obtained in 2 independent experiments.
Q-PCR.
Total RNA was isolated from cells after lysis in TRIzol reagent according to the manufacturer's protocol (Invitrogen), and SYBR Green Q-PCRs were performed as described in refs. 41 and 42. The primer sequences are available on request.
ChIPs.
Soluble chromatin was prepared and immunoprecipitated with the indicated antibodies as described in ref. 43. The final DNA extractions were amplified by using pairs of primers that encompass the p53-responsive element in the p21 promoter and the early transcribed region of p21. The primer sequences are available on request.
RNA Interference.
Three days after transfection of cells with the previously described siRNA constructs (8), ASC-2, MLL4, and MLL3 expression levels were determined by Q-PCR and by immunostaining with anti-ASC-2 antibody (Fig. 3A). The siRNA sequences are available on request.
ASCOM Binding to p53.
FLAG-tagged full length p53 fusion proteins that were expressed either in Sf9 cells or in E. coli were coupled to M2-agarose beads (20 μL) and incubated with the purified ASCOM (1.0 μg) for 5 h at 4 °C in BC200 [40 mM Tris (pH 7.9), 200 mM KCl, 20% glycerol, 0.05% Tween 20, 0.5 mM PMSF, 1.0 μM leupeptin, 1.0 μM pepstatin, and 2.0 μM MG132]. After extensive washing of the beads in BC200, bound proteins were eluted by boiling the beads in 2% SDS and analyzed by SDS/PAGE and immunoblotting.
Histology.
Mouse ureters and kidneys were excised, embedded in paraffin, sectioned in 5-μm-thick slices, and immunostained with indicated antibodies or stained with hematoxylin and eosin, as described in refs. 22 and 41.
Supplementary Material
Acknowledgments.
We thank Drs. Tung-Tien Sun (New York University, New York), Junjie Chen (Yale University, New Haven, CT), Kai Ge (National Institutes of Health, Bethesda, MD), Sung Yun Jung, and Jun Qin for reagents and technical help. This work was supported by National Institutes of Health Grants DK064678 (to J.W.L.) and CA129325 and DK071900 (to R.G.R.) and by the Starr Foundation (Q.-H.Y. and R.G.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902873106/DCSupplemental.
References
- 1.Mahajan MA, Samuels HH. Nuclear hormone receptor coregulator: Role in hormone action, metabolism, growth, and development. Endocr Rev. 2005;26:583–597. doi: 10.1210/er.2004-0012. [DOI] [PubMed] [Google Scholar]
- 2.Li Q, ChuMJ, Xu J. Tissue- and nuclear receptor-specific function of the C-terminal LXXLL motif of coactivator NCoA6/AIB3 in mice. Mol Cell Biol. 2007;27:8073–8086. doi: 10.1128/MCB.00451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 4.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 5.Schneider R, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 6.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 7.Goo YH, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, et al. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci USA. 2006;103:15392–15397. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Issaeva I, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 12.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 13.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 14.Hong S, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahajan MA, Das S, Zhu H, Tomic-Canic M, Samuels HH. The nuclear hormone receptor coactivator NRC is a pleiotropic modulator affecting growth, development, apoptosis, reproduction, and wound repair. Mol Cell Biol. 2004;24:4994–5004. doi: 10.1128/MCB.24.11.4994-5004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanner MM, et al. Independent amplification and frequent co-amplification of three nonsyntenic regions on the long arm of chromosome 20 in human breast cancer. Cancer Res. 1996;56:3441–3445. [PubMed] [Google Scholar]
- 17.Guan XY, et al. Hybrid selection of transcribed sequences from microdissected DNA: Isolation of genes within amplified region at 20q11–q13.2 in breast cancer. Cancer Res. 1996;56:3446–3450. [PubMed] [Google Scholar]
- 18.Lee SK, et al. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem. 1999;274:34283–34293. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- 19.Lee J, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci USA. 2008;105:19229–19234. doi: 10.1073/pnas.0810100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mak RH, Kuo HJ. Primary ureteral reflux: Emerging insights from molecular and genetic studies. Curr Opin Pediatr. 2003;15:181–185. doi: 10.1097/00008480-200304000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Hu P, et al. Ablation of uroplakin III gene results in small urothelial plaques, urothelial leakage, and vesicoureteral reflux. J Cell Biol. 2000;151:961–972. doi: 10.1083/jcb.151.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong XT, et al. Roles of uroplakins in plaque formation, umbrella cell enlargement, and urinary tract diseases. J Cell Biol. 2004;167:1195–1204. doi: 10.1083/jcb.200406025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 24.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci USA. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwabuchi K, et al. Stimulation of p53-mediated transcriptional activation by the p53-binding proteins, 53BP1 and 53BP2. J Biol Chem. 1998;273:26061–26068. doi: 10.1074/jbc.273.40.26061. [DOI] [PubMed] [Google Scholar]
- 27.Huang J, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 28.Zgheib O, et al. ATM signaling and 53BP1. Radiother Oncol. 2005;76:119–122. doi: 10.1016/j.radonc.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 29.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 30.Lee S, Lee J, Lee SK, Lee JW. Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol Endocrinol. 2008;22:1312–1319. doi: 10.1210/me.2008-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Kuang SQ, Liao L, Zhou S, Xu J. Haploid inactivation of the amplified-in-breast cancer 3 coactivator reduces the inhibitory effect of peroxisome proliferator-activated receptor gamma and retinoid X receptor on cell proliferation and accelerates polyoma middle-T antigen-induced mammary tumorigenesis in mice. Cancer Res. 2004;64:7169–7777. doi: 10.1158/0008-5472.CAN-04-1176. [DOI] [PubMed] [Google Scholar]
- 32.Cho EA, Prindle MJ, Dressler GR. BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol Cell Biol. 2003;23:1666–1673. doi: 10.1128/MCB.23.5.1666-1673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munoz IM, Jowsey PA, Toth R, Rouse J. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res. 2007;35:5312–5322. doi: 10.1093/nar/gkm493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munoz IM, Rouse J. Control of histone methylation and genome stability by PTIP. EMBO Rep. 2009;10:239–245. doi: 10.1038/embor.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 36.Balakrishnan A, et al. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res. 2007;67:3545–3550. doi: 10.1158/0008-5472.CAN-07-0065. [DOI] [PubMed] [Google Scholar]
- 37.Ruault M, Brun ME, Ventura M, Roizès G, De Sario A. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene. 2002;284:73–81. doi: 10.1016/s0378-1119(02)00392-x. [DOI] [PubMed] [Google Scholar]
- 38.Tan YC, Chow VT. Novel human HALR (MLL3) gene encodes a protein homologous to ALR and to ALL-1 involved in leukemia, and maps to chromosome 7q36 associated with leukemia and developmental defects. Cancer Detect Prev. 2001;25:454–469. [PubMed] [Google Scholar]
- 39.Zhu YJ, et al. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J Biol Chem. 2003;278:1986–1990. doi: 10.1074/jbc.C200634200. [DOI] [PubMed] [Google Scholar]
- 40.Lee SK, Kim HJ, Kim JW, Lee JW. Steroid receptor coactivator-1 and its family members differentially regulate transactivation by the tumor suppressor protein p53. Mol Endocrinol. 1999;13:1924–1933. doi: 10.1210/mend.13.11.0365. [DOI] [PubMed] [Google Scholar]
- 41.Kim SW, et al. Multiple developmental defects derived from impaired recruitment of ASC-2 to nuclear receptors in mice: Implication for posterior lenticonus with cataract. Mol Cell Biol. 2002;22:8409–8414. doi: 10.1128/MCB.22.24.8409-8414.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ledbetter MW, Preiner JK, Louis CF, Mickelson JR. Tissue distribution of ryanodine receptor isoforms and alleles determined by reverse transcription polymerase chain reaction. J Biol Chem. 1994;269:31544–31551. [PubMed] [Google Scholar]
- 43.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.