Abstract
The bone morphogenetic protein (BMP)-1/tolloid metalloproteinases are evolutionarily conserved enzymes that are fundamental to dorsal–ventral patterning and tissue morphogenesis. The lack of knowledge regarding how these proteinases recognize and cleave their substrates represents a major hurdle to understanding tissue assembly and embryonic patterning. Although BMP-1 and mammalian tolloid (mTLD) are splice variants, it is puzzling why BMP-1, which lacks 3 of the 7 noncatalytic domains present in all other family members, is the most effective proteinase. Using a combination of single-particle electron microscopy, small-angle X-ray scattering, and other biophysical measurements in solution, we show that mTLD, but not BMP-1, forms a calcium-ion-dependent dimer under physiological conditions. Using a domain deletion approach, we provide evidence that EGF2, which is absent in BMP-1, is critical to the formation of the dimer. Based on a combination of structural and functional data, we propose that mTLD activity is regulated by a substrate exclusion mechanism. These results provide a mechanistic insight into how alternative splicing of the Bmp1 gene produces 2 proteinases with differing biological activities and have broad implications for regulation of BMP-1/mTLD and related proteinases during BMP signaling and tissue assembly.
Keywords: procollagen C-proteinase, chordin, small angle X-ray scattering
Bone morphogenetic protein (BMP)-1 (procollagen C-proteinase-1; PCP-1) and mammalian tolloid (mTLD/PCP-2) are alternatively spliced products of the Bmp1 gene (1). Together with mammalian tolloid like-1 (mTLL-1) and mTLL-2, they comprise a small group of zinc- and calcium-dependent proteinases, fundamental to tissue patterning and extracellular matrix (ECM) assembly. The BMP-1/TLD family is conserved in species ranging from Drosophila to humans, and their importance is highlighted by the embryonic lethal phenotype of Bmp1/Tll1 homozygous null mice, which display heart malformations and abnormal procollagen processing (2).
In vertebrates, BMP-1/TLD proteinases are involved in the biosynthetic processing of a range of ECM precursors, including major and minor fibrillar collagens (3–5), the collagen and elastin cross-linking enzyme prolysyl oxidase (6), cellular anchoring proteins prolaminin-5 and procollagen VII (7, 8), and the small leucine-rich proteoglycans osteoglycin and probiglycan (9, 10). BMP-1/TLD proteinases also release a number of TGF-β superfamily members, including BMP-2 and BMP-4, growth and differentiation factors (GDF) 8/11, and TGFβ1 from their corresponding latent complexes. This activity modulates dorsal ventral patterning, growth of skeletal muscle and neural tissue, and cellular behavior, respectively (11–14). These dual roles have fuelled speculation that BMP-1/TLD proteinases orchestrate ECM assembly by means of signaling by TGF-β-like proteins (15).
BMP-1/TLD proteinases contain an N-terminal protease domain followed by CUB (complement, Uegf, and BMP-1) and calcium-ion-binding EGF-like domains. The noncatalytic domains appear to restrict proteolytic activity in terms of substrate specificity and efficiency, because when secreted alone, the BMP-1/mTLD protease domain cleaves additional sites in previously characterized substrates, and cleaves other matrix proteins such as fibronectin, which are left intact by full-length BMP-1 (16). The protease domains of BMP-1 and mTLL-1 also process probiglycan with better kinetics than the full-length enzymes (17).
BMP-1 is a more efficient proteinase than mTLD for almost all tested substrates in vitro. Recently, Stöcker and coworkers (18) proposed an explanation for the functional difference between BMP-1 and mTLD, suggesting that binding affinity to procollagen increases toward the C terminus of the molecule. Intriguingly, they also reveal that fragments containing EGF domains bind procollagen more strongly than those containing only CUB domains. However, this result creates a paradox, because in an earlier study it was observed that removal of either or both of the EGF domains from mTLD converts it into a more efficient C-proteinase and results in the acquisition of chordinase activity (19).
To address this conflict and provide a greater understanding of the mechanism of action of tolloids, we investigated the structures of BMP-1 and mTLD. We show here that mTLD, but not BMP-1, forms a dimer at physiological calcium ion concentrations. We also find that mTLD molecules lacking CUB4 and CUB5 domains are still able to form dimers and have improved chordinase activity. Based on these data, we propose a model whereby mTLD activity is restricted by a substrate exclusion mechanism that requires the presence of the C-terminal CUB domains.
Results
The mTLD, but Not BMP-1, Forms a Ca2+-Dependent Dimer.
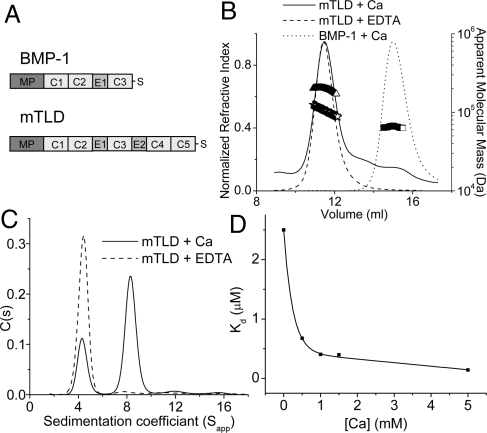
The size and oligomeric status of BMP-1 and mTLD were first analyzed by multiangle laser light scattering (MALLS) (SDS/PAGE of purified proteins is shown in Fig. S1A). In the presence of 1 mM CaCl2, mTLD had a molecular mass of 196,400 Da, approximately twice that predicted from amino acid sequence (Fig. 1B and Table 1). However, in the presence of EDTA, the molecular mass was reduced to 103,600 Da. These initial data gave the first indication that mTLD forms a dimer in solution. BMP-1 had a molecular mass of 64,920 Da in the presence of 1 mM CaCl2, slightly smaller than that expected for a monomer (Table 1), but MS analysis showed protein sequence present from all domains (protease domain to CUB3) (Fig. S1B).
Fig. 1.
mTLD, but not BMP-1, forms a calcium-dependent dimer. (A) Domain structure of BMP-1 and mTLD. MP, metalloprotease domain; C, CUB domain; E, EGF-like domain; S, specific region. (B) Summary of MALLS data. In the presence of calcium, mTLD has a molecular mass (triangles) of 196,400 ± 6,284 Da (experimental errors from polydispersity), almost twice that expected for a monomer (100,276 Da). In the presence of EDTA, the apparent molecular mass is reduced to 103,600 ± 2,797 Da (stars). In the presence of calcium, BMP-1 has a molecular mass of 64,920 ± 1,556 Da (squares), similar to that expected for a monomer (70,523 Da). (C) C(s) analysis of mTLD derived from sedimentation velocity analytical ultracentrifugation. (D) The strength of the mTLD self-association was determined at a range of calcium ion concentrations by using sedimentation equilibrium analytical ultracentrifugation.
Table 1.
Summary of experimental data
| Protein and ion | Rh, nm (MALLS) |
Rh, nm (AUC) |
Rg, nm (SAXS) | M, Da (MALLS) |
s20,W, S |
f/fo | ||
|---|---|---|---|---|---|---|---|---|
| Peak 1 | Peak 2 | Peak 1 | Peak 2 | |||||
| mTLD Ca | 5.5 ± 0.1 | 5.57 ± 0.27 | 5.36 ± 0.20 | ND | 196,400 ± 6,284 | 8.89 ± 0.51 | 4.62 ± 0.18 | 1.45 |
| mTLD EDTA | 4.8 ± 0.1 | 5.04 ± 0.21 | 5.4 | 103,600 ± 2,797 | 4.91 ± 0.29 | 1.65 | ||
| BMP-1 Ca | ND | 3.51 ± 0.23 | ND | 64,920 ± 1,556 | 4.58 ± 0.32 | 1.32 | ||
| BMP-1 EDTA | ND | ND | 3.5 | 61,130 ± 2,017 | ND | ND | ||
| mTLDTE2 Ca | 3.5 ± 0.2 | 5.28 ± 0.21 | 3.78 ± 0.30 | ND | 106,300 ± 1,063 | 6.78 ± 0.29 | 4.74 ± 0.35 | 1.53 |
| mTLDTE2 EDTA | ND | 3.97 ± 0.29 | ND | 78,450 ± 3,922 | 4.51 ± 0.36 | 1.45 | ||
The experimental data were determined by multiangle laser light scattering (MALLS), analytical ultracentrifugation (AUC), and small-angle X-ray scattering (SAXS). Rh, hydrodynamic radius; Rg, radius of gyration; M, molecular mass; s20,w, sedimentation coefficient in water at 20 ° C; f/fo, frictional coefficient; ND, not determined. The theoretical molecular masses of the mature forms including purification tags calculated from amino acid sequence are 100,276 Da (mTLD), 72,585 Da (mTLDTE2), and 70,523 Da (BMP-1).
To investigate mTLD self-association, we used sedimentation velocity analytical ultracentrifugation (AUC). In the presence of 1 mM CaCl2 (Fig. 1C), 2 distinct species are visible. The predominant species accounts for 66% of total protein, whereas the minor species accounts for 27% of total protein. These 2 distinct species have molecular mass estimates of 187 and 90 kDa, respectively, in agreement with dimer and monomer. To assess the calcium ion dependence of this interaction, samples were analyzed in the presence of EDTA. Under these conditions, the dimer species is absent, but the monomer remains.
The mTLD Dimer Is the Most Abundant Physiological Form.
To assess the strength of mTLD self-association, we determined its dissociation constant (Kd) at a range of calcium ion concentrations by using sedimentation equilibrium AUC (Fig. 1D). In the absence of externally applied calcium ions, the Kd was 2.5 μM, suggesting a dynamic reversibly associating system. After the addition of calcium ions, a rapid increase in association was observed, even at levels as low as 0.5 mM. This finding indicates that the most abundant form at physiological calcium concentrations is likely to be the dimer.
BMP-1 Adopts a Compact Conformation.
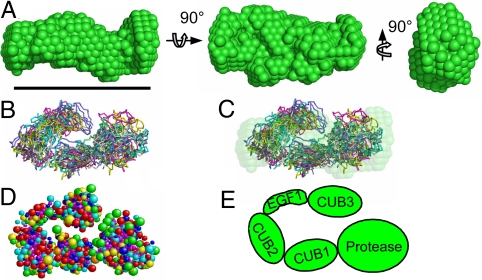
The structure of BMP-1 was first examined in the presence of EDTA by using small-angle X-ray scattering (SAXS). Under these conditions the radius of gyration (Rg) was 3.5 nm, and the pair distribution function suggested a maximal particle dimension (Dmax) of 11.3 nm and indicated an elongated particle (Fig. S2) (20). Ab initio modeling using DAMMIN (21) was used to generate the most probable shape, which has approximate dimensions 11.3 × 6.2 × 4.6 nm (Fig. 2A). To provide an estimation of the domain organization, rigid body modeling was performed by SASREF (22), using domains that share sequence similarity to those found in BMP-1 (Materials and Methods) (Fig. 2B). In these models, BMP-1 adopts a compact conformation that is similar in overall shape to the ab initio model (Fig. 2C).
Fig. 2.
Structure of BMP-1. (A) Low resolution ab initio model derived from SAXS data. (B) Six of 8 rigid body models (varying colors) generated by SASREF, also shown superimposed with the ab initio model (C). (D) Representative rigid body model converted into a bead model using SOMO. (E) Proposed BMP-1 structure. (Scale bar, 10 nm; A–D.)
To validate these findings, shell and bead models were constructed based on the ab initio and rigid-body SAXS models using HYDROPRO (23) and SOMO software (24), respectively. These models were compared with hydrodynamic measurements determined by sedimentation velocity of BMP-1 in the presence of calcium ions. Models derived from the ab initio (Fig. 2A) and rigid-body models (Fig. 2D) fit well with the observed hydrodynamic properties (Table S1), suggesting that the compact shape of these models accurately reflects the solution structure of BMP-1. In comparison, a model generated from a linear array of BMP-1 domains is significantly larger.
The mTLD Monomer Adopts a Horseshoe-Like Conformation.
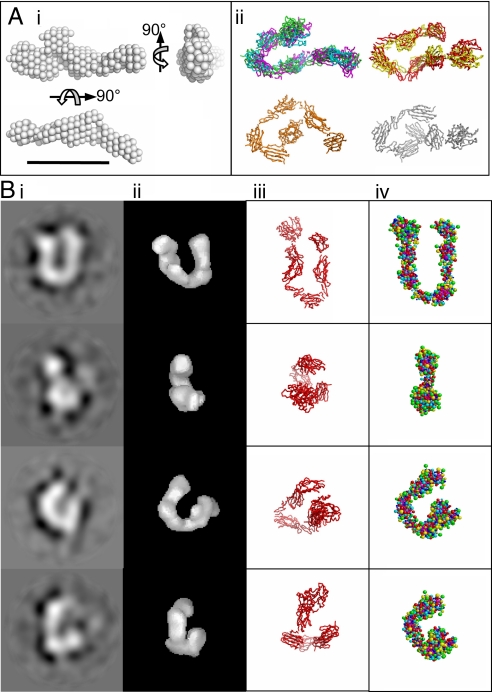
By using SAXS, the mTLD monomer was determined to have a molecular mass of 99 kDa, Rg of 5.4 nm, and Dmax of 18.5 nm. Similarly to BMP-1, the pair distribution function suggests an elongated particle (Fig. S2). Ab initio modeling revealed the most probable shape had approximate dimensions of 18 × 7 × 3 nm (Fig. 3Ai), correlating well with structures generated by rigid body modeling, which exhibit a hairpin-like structure (Fig. 3Aii).
Fig. 3.
Structure of the mTLD monomer. (A) (i) Ab initio model generated from SAXS data. (ii) Eight rigid-body models generated by SASREF (different colors) segregated into groups for clarity. (B) Comparison of mTLD structure. (i) Class average images from single-particle image processing (each image is an average of ≈127 particles). (ii) TEM 3D reconstruction calculated by angular reconstitution. (iii) Representative rigid-body SAXS model. (iv) Best-fit bead model. All images in A are to scale. (Scale bar, 10 nm; box size in B, 19.4 nm.)
The mTLD monomer was also investigated independently using single particle transmission electron microscopy (TEM). A 3D reconstruction was calculated to 26-Å resolution from 3,833 particles using angular reconstitution. The 3D model generated is similar in overall shape to models produced by rigid body modeling (Fig. 3B). To provide further support for the mTLD structure, a series of bead models were generated for comparison to experimentally determined hydrodynamic measurements. The best-fit model (Fig. 3Biv) retains characteristics similar to those of the models generated by single-particle TEM and SAXS (Table 1 and Table S1).
Within the mTLD Dimer, Monomers Appear to Be Stacked Side-by-Side.
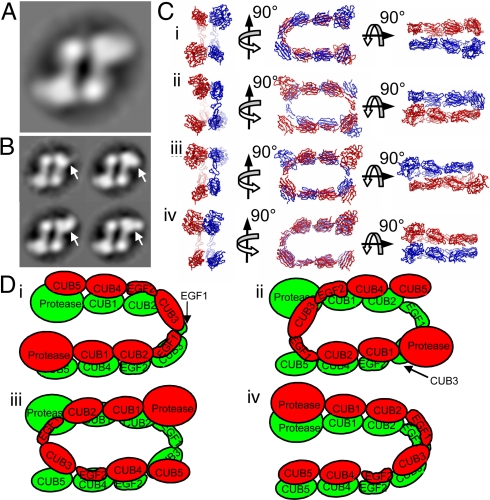
We used single-particle TEM to determine how the molecules are arranged within the mTLD dimer. After classification of 2,308 particles, it was evident that the particles were visible in only a single orientation (Fig. 4A); consequently, a 3D reconstruction was not possible. To address the possibility that smearing of the larger bulbous end of each monomer was due to flexibility, the dataset was reclassified based only on differences in this region of the right-hand side monomer. The resultant class average images (Fig. 4B) clearly show this region to adopt different conformations.
Fig. 4.
Possible models for the mTLD dimer. (A) During TEM, particles are visible in only a single orientation (shown is an average of 851 images). (B) The dataset was reclassified based on the large bulbous region of the right-hand monomer to determine whether this region was flexible. Different conformations are highlighted by white arrows. (C) The mTLD monomer model was used to generate possible models for the dimer. The best fit to AUC measurements was achieved by using a side-by-side nonstaggered arrangement. Within this arrangement, 4 distinct models are possible (shown in ribbon format) with molecules facing the same (C i and iv) or opposite (C ii and iii) directions, and in antiparallel (C i and ii) or parallel (C iii and iv) arrangement. (D) These 4 possible alternatives displayed schematically, with each monomer in a different color. (A and B, box size, 21.6 nm; C is shown on the same scale as A.)
To gain further insight into the organization of the mTLD dimer, bead models were constructed by using the monomer bead model (Fig. 3Biv). By alternating the arrangement of the monomers, a series of models were generated (Fig. S3). The best fit to experimental measurements was achieved by using side-by-side nonstaggered arrangements. When this approach is used, models in which monomers are arranged in an antiparallel (Fig. 4C i and ii) or parallel (Fig. 4C iii and iv) fashion, or with monomers facing the same (Fig. 4C i and iv) or opposite (Fig. 4C ii and iii) directions, are too similar for us to distinguish between hydrodynamically. The EM view available is suggestive of an antiparallel dimer with a side-by-side arrangement, but we cannot rule out the other arrangements on the basis of one view (Fig. 4C).
Deletion of CUB4 and CUB5 Domains Affects mTLD Dimerization and Chordinase Activity.
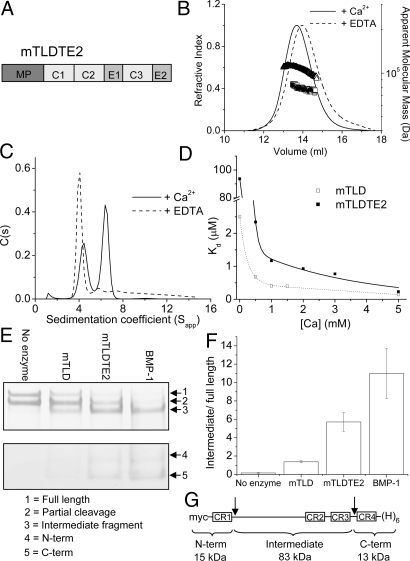
Because BMP-1 and mTLD possess identical catalytic domains, we hypothesized that the differences in substrate specificity are a consequence of dimerization in mTLD, leading to intermolecular substrate exclusion of the catalytic site. To test this possibility, we generated a mTLD molecule that terminates after EGF2 (mTLDTE2; Fig. 5A). When MALLS was used, the apparent molecular mass of mTLDTE2 in the presence of 1 mM CaCl2 was 106,300 Da, which corresponded to neither monomer (72,585 Da) nor dimer (Fig. 5B). This species was most likely a mixture of monomer and dimer, as the molecular mass was reduced to 78,450 kDa in the presence of EDTA. This result was confirmed by using sedimentation velocity AUC (Fig. 5C), which shows 2 species in the presence of 1 mM calcium with molecule mass estimates of 71 and 122 kDa.
Fig. 5.
Oligomerization and chordinase activity of mTLDTE2. (A) Domain structure of mTLDTE2. (B) Summary of MALLS data. The mTLDTE2 has a molecular mass of 106,300 Da (triangles) in the presence of 1 mM calcium ions, and 78,450 Da (squares) in the presence of EDTA. The predicted molecular mass of a monomer is 72,585 Da. (C) Sedimentation velocity AUC of mTLDTE2. (D) The strength of the mTLDTE2 self-association was determined in a range of calcium ion concentrations by using sedimentation equilibrium AUC, and is shown next to that of mTLD. (E) Cleavage of chordin. Purified chordin was incubated in the presence or absence of enzyme as indicated. For clarity, gels were cut into top and bottom layers (shown separately), which were stained to differing extents. (F) Chordinase activity expressed as the mean proportion of the intermediate fragment relative to full-length chordin obtained from 3 independent experiments (error bars represent SEM). (G) Chordin domain structure. CR, cysteine-rich repeat; myc, c-myc epitope; (H)6, hexahistidine tag. Arrows indicated BMP-1 cleavage sites.
To determine the impact that deletion of the CUB4 and CUB5 domains has on the strength of mTLD self-association, we determined the Kd of mTLDTE2 at a range of calcium ion concentrations (Fig. 5D). In the absence of externally applied calcium ions, the Kd suggests that mTLDTE2 is present almost solely as monomer. In the presence of calcium ions (0.5–3 mM), the equilibrium shifts to dimer, but the strength of the interaction is weaker than that in full-length mTLD, suggesting CUB4 and/or CUB5 contribute to some extent to self-association. However, at higher calcium ion concentrations (5 mM), the Kd is similar to that of mTLD.
To assess the impact of removal of CUB4 and CUB5 on proteolytic activity, we compared the chordinase activity of mTLDTE2 with that of BMP-1 and mTLD (Fig. 5E). In the absence of enzyme, chordin is present as the full-length, and partially processed form lacking the N terminus (based on antibody recognition). All enzymes tested processed chordin to some extent, as is evident by the loss of the full-length and the appearance of the intermediate, N- and C-terminal fragments (Fig. 5E). Removal of CUB4 and CUB5 results in a dramatic (≈4-fold) increase in mTLD chordinase activity compared with the full-length enzyme (Fig. 5F), although this activity did not meet the level observed for BMP-1.
Discussion
Despite their key role as essential factors controlling tissue morphogenesis during development, the regulatory mechanisms governing BMP-1/TLD proteinases have remained poorly understood. In this study we have investigated the structure of BMP-1 and mTLD with the specific aim of explaining how alternative splicing can produce proteins with differing biological activity. We show that BMP-1 adopts a compact conformation that is significantly smaller than a linear array of domains. Based on the biochemical data regarding the poor specificity and high efficiency of the protease domain when secreted in isolation (16, 17), it appears likely that this compact conformation is required to restrict the access of substrates to the protease domain, to provide specificity and prevent unchecked ECM degradation.
In contrast to BMP-1, mTLD was observed as a dimer in the presence of calcium ions. Here, we report oligomerization for a member of the BMP-1/TLD family. Because mTLD activity is highly calcium ion dependent (3), we investigated the calcium ion dependence of mTLD self-association. Our results indicate mTLD has a propensity to form dimers in the presence of calcium ions, even at concentrations considerably lower than the 5–10 mM required for optimal C-proteinase activity in vitro (3), indicating the dimer to be the most likely physiologically abundant form. However, we do not discount the possibility of monomer existing in vivo at low protein concentrations. It is important to note that we do not attribute the calcium ion dependence of BMP-1/mTLD activity to differences in oligomeric status, because the isolated protease domain remains markedly calcium ion dependent (17).
Although the absence of CUB4 and CUB5 domains did affect the strength of mTLD self-association at low calcium ion concentrations, the Kd of the mTLDTE2 dimer is similar to that of the mTLD dimer at the 5 mM calcium ion concentrations used in our chordinase assay. Thus, any differences in the observed activity are not thought to be due to differences in oligomeric status. Based on our structural data alone, a number of models for the mTLD dimer are possible (Fig. 4D). Although the CUB4/5 deletion data do not rule out any of the possible mTLD dimer models, those with monomers facing opposite directions offer no easy explanation to account for the observed increased chordinase activity. We favor models in which the monomers face the same direction, because these models place the CUB4/5 domains in close proximity to the protease domain, and their deletion could make the catalytic site more accessible to substrates (Figs. 4 C and D; Fig. S3). These models provide a substrate exclusion mechanism that would be intramolecular in the parallel arrangement, but inter- and intramolecular in the antiparallel arrangement.
Of the 2 models with monomers facing the same direction, we prefer an antiparallel arrangement, because (i) this model appears most similar to the single view available by TEM, and (ii) in this model, the CUB/EGF arrangement is similar to the one that mediates homodimerization in the related mannose binding protein associated serine protease (MASP)-2 and C1s enzymes (25–28). In these related proteinases, dimerization, which is calcium ion dependent in the case of C1s, occurs through a CUB–EGF interaction and is facilitated by a head-to-tail (or antiparallel) arrangement of CUB and EGF domains (29). Based on the similarity to MASP2/C1s, and the observation that BMP-1 is unable to dimerize, whereas mTLDTE2 is, it appears that EGF2 has a major role in mTLD self-association. Although it has been suggested that CUB–EGF interactions may be involved in homo/heterodimerization in other members of the MASP2/C1s families, if our theory is correct, this mechanism may be more widespread than first believed.
Although we believe a substrate exclusion mechanism to be the primary explanation for the functional difference between BMP-1 and mTLD, it may not be the only contributing factor. For example, the chordinase activity of mTLDTE2 did not match that of BMP-1, and it may be that dimerization results in the masking of substrate binding sites on the inner face of the CUB and/or EGF domains, which would otherwise aid in the recruitment of substrates. It is noteworthy that, although previous attempts to determine the chordinase activity of mTLD have failed to detect the N- or C-terminal reaction products (19, 30), we were able to detect a low level of chordinase activity attributable to mTLD, at least in the absence of ancillary factors.
In summary, we have shown that BMP-1 adopts a compact conformation and mTLD forms a calcium-ion-dependent dimer. We propose that the protease domains of both BMP-1 and mTLD are restricted by substrate exclusion mechanisms that act to reduce protease efficiency. Our data provide functional insights into this class of essential enzymes that have critical roles in development, tissue homeostasis, and remodeling.
Materials and Methods
Production, Expression, and Purification of Recombinant Proteins.
BMP-1 and mTLD were generated as described previously (19). The mTLDTE2 and chordin were amplified by PCR as described in SI Materials and Methods and ligated into pCEP-PU vectors by using NotI and XhoI restriction enzymes; 293-EBNA cells were maintained in DMEM/F12 with Glutamax containing 10% FCS, 0.1 unit/mL penicillin, and 10 μg/mL streptomycin (growth medium) in a 37 °C incubator under a 5% CO2. For transfection, 1 μg of plasmid containing mTLDTE2 or chordin DNA was incubated with Lipofectin and added to 293-EBNA cells. After 24 h, selection was initiated by the addition of 5 μg/mL puromycin; 293-EBNA cells transfected with BMP-1 or mTLD were maintained in growth medium containing 0.25 mg/mL hygromycin B.
Recombinant proteins were harvested and purified initially by ion-exchange (for BMP-1) or nickel affinity chromatography (for all others) as described in SI Materials and Methods. All variants were then subjected to size-exclusion chromatography using a Superdex200 10/300GL gel filtration column in 10 mM Tris·HCl (pH 7.4) containing 0.5 M NaCl and 1 mM CaCl2. In preparation for activity assays, nickel affinity purified chordin was dialyzed into 50 mM Tris buffer (pH 7.4) containing 150 mM NaCl at 4 °C. Where needed, proteins were concentrated by using Vivaspin centrifugal concentrators (Sartorius).
MALLS.
Samples (0.5 mL at ≈0.5 mg/mL) were gel filtered using a Superdex-200 10/300 GL column in 10 mM Tris·HCl buffer (pH 7.4) containing 0.5 M NaCl in the presence of either 1 mM CaCl2 or 2 mM EDTA at 0.71 mL/min. The eluate was passed through a Wyatt EOS 18-angle laser photometer with the 13th detector replaced with a Wyatt QELS detector for the simultaneous measurement of hydrodynamic radius. This was coupled to a Wyatt Optilab rEX refractive index detector and the hydrodynamic radius, molecular mass moments, and concentration of the resulting peaks was analyzed by using Astra 5.3.2.
AUC.
All experiments were performed in 10 mM Tris (pH 7.4) containing 0.5 M NaCl and the indicated concentrations of EDTA/calcium using an XL-A ultracentrifuge (Beckman) with an An50Ti-8-hole rotor fitted either with the standard 2-sector open-filled centerpiece for sedimentation velocity or a 6-sector Epon-filled centerpiece for equilibrium studies, with quartz glass windows. Equilibrium sedimentation was performed at 4 °C, using rotor speeds producing 7,000, 12,000, and 19,000 × g (for mTLD) and 8,000, 15,000, and 22,000 × g (for mTLDTE2) with scanning at 230 and 280 nm after equilibrium was reached (14 h). Association kinetics was performed using concentrations of between 0.25 and 1.4 μM, and global analysis of the data was performed with nonlinear regression using the Sedphat program (31). Velocity sedimentation analysis was performed at either 40,000 (mTLD and BMP-1) or 48,000 (mTLDTE2) × g at 20 °C, with the sedimenting boundary monitored every 90 sec for a total of 200 scans. Protein concentrations used were 0.13 mg/mL (BMP-1), 0.38 mg/mL (mTLDTE2 in EDTA), 0.07 mg/mL (mTLDTE2 in Ca2+), and 0.28 mg/mL (mTLD). Data were interpreted with the model-based distribution of Lamm equation solutions C(s) using the software Sedfit (32). Frictional ratios (f/fo) for the monomer and dimer were calculated from the sedimentation coefficient. Bead models were generated using the atomic coordinates of homologous domains as described for SAXS rigid-body modeling. These domains were oriented in PyMOL to match the domain structure of BMP-1 or mTLD and used to build bead models with the solution modeling software SOMO.
SAXS.
SAXS data for mTLD (1 mg/mL) were collected on EMBL beamline X33 at the light source facilities DORISIII at HASYLAB/DESY (33). Data were collected on a MAR345 image plate detector using a 60-s exposure time and 2.4-m sample-to-detector distance to cover a momentum transfer interval 0.10 nm−1 < q < 5.0 nm−1. The modulus of the momentum transfer is defined as q = 4π sin θ/λ, where 2θ is the scattering angle and λ is the wavelength. The q range was calibrated by using silver behenate powder based on diffraction spacings of 58.38 Å. The scattering images obtained were spherically averaged using in-house software and buffer scattering intensities were subtracted by using PRIMUS. Molecular mass estimates were obtained by normalizing scattering to BSA. SAXS data for BMP-1 (2 mg/mL) were collected at station 2.1 of the Synchrotron Radiation Source at Daresbury Laboratory, United Kingdom. Images were collected in multiple 60-s frames at 1 and 4 m sample-to-detector distances, and the resulting profiles were merged to cover a momentum transfer interval of 0.14 nm−1 < q < 4.0 nm−1. Both BMP-1 and mTLD were maintained at 10 °C during data collection using each beamline's standard solution sample holder. The Rg, forward scattering intensity, and 1D intraparticle distance distribution function p(r) in real space were evaluated with the indirect Fourier transform program GNOM (34), and particle shapes were restored ab initio by using DAMMIN. Multiple DAMMIN runs were performed to generate 24 (mTLD) or 20 (BMP-1) similar shapes that were then combined and filtered to produce an averaged model using the DAMAVER (35) software package. Rigid-body modeling to the experimental scattering data was performed by using SASREF (best χ values are 1.37 for BMP-1 and 2.11 for mTLD). Templates included the astacin protease domain, to which the BMP-1 protease domain (residues 124–321) was modeled by using Swissmodel (36), and the EGF (residues 138–181) and CUB2 (residues 182–297) domains from rat MASP-2. These domains are 35%, 43–45%, and 24–30% identical to the mTLD protease, EGF1–2 and CUB1–5, respectively. Due to the large number of domains present in mTLD, the EGF-CUB region was treated as a single module wherever possible.
TEM and Single-Particle Analysis.
The mTLD (≈8 μg/mL) was absorbed onto glow-discharged carbon-coated grids and stained with 4% (wt/vol) uranyl acetate (pH 4.7). Grids were observed using an FEI Tecnai Twin TEM operating at 120 kV. Images were recorded on a 2,048 × 2,048 pixel CCD camera at 69,000× magnification between 0.5 and 1.58 μm defocus and processed by using Imagic5 software (37). The total number of particles in the dataset was 2,308 in the presence of CaCl2 and 3,833 in the presence of EGTA. Selected particles were band-pass-filtered with a high-frequency cut-off of 20 Å and a low-frequency cut-off of 150 Å (in the presence of calcium) or 130 Å (in the presence of EGTA). Characteristic class-sum images were used as references to align the dataset during iterative rounds of multireference alignment. Symmetry was not applied at any stage of the image analysis to either of the datasets. Euler angles were assigned to class-sum images, enabling calculation of an initial 3D reconstruction, which was then subjected to multiple rounds of iterative refinement. The resolution of the final model was determined to be 26 Å by Fourier shell correlation using a 3σ criterion.
Assay for Chordinase Activity.
To assay proteinase activity, enzyme and substrate concentrations were quantified based on comparison with known amounts of BSA by using SDS/PAGE and GeneTools software (SynGene). Purified chordin (1.25 μg) was then incubated in the presence or absence of 75 ng of mTLD, mTLDTE2, or BMP-1 in a final volume of 50 μL in 50 mM Tris buffer (pH 7.4) containing 150 mM NaCl and 5 mM CaCl2 at 37 °C for 24 h. Reactions were stopped by the addition of 4× LDS sample buffer and heating to 95 °C for 5 min. Reaction products were separated by SDS/PAGE using 4–12% Bis/Tris gels and visualized by silver staining. Chordinase assay reaction products were quantified by densitometry using SynGene software, and are represented as the mean ± SEM of 3 independent experiments.
Supplementary Material
Acknowledgments.
We thank Marge Howard and Emma Keevill in the Biomolecular Analysis Facility, Adam Huffman in the Bioinformatics Facility, and the staff in the EM facility for technical assistance (University of Manchester); the Synchrotron Radiation Source and Deutsches Elektronen Synchrotron (DESY) for beamtime, and the European Community for funding work undertaken at Deutsches Elektronen Synchrotron. R.B. is supported by Biotechnology and Biological Sciences Research Council Studentship 200513125. K.E.K. and E.G.C.-L. are supported by the Wellcome Trust. R.A.K. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812178106/DCSupplemental.
References
- 1.Takahara K, Lyons GE, Greenspan DS. Bone morphogenetic protein-1 and a mammalian tolloid homologue (mTld) are encoded by alternatively spliced transcripts which are differentially expressed in some tissues. J Biol Chem. 1994;269:32572–32578. [PubMed] [Google Scholar]
- 2.Pappano WN, Steiglitz BM, Scott IC, Keene DR, Greenspan DS. Use of Bmp1/Tll1 doubly homozygous null mice and proteomics to identify and validate in vivo substrates of bone morphogenetic protein 1/tolloid-like metalloproteinases. Mol Cell Biol. 2003;23:4428–4438. doi: 10.1128/MCB.23.13.4428-4438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hojima Y, van der Rest M, Prockop DJ. Type I procollagen carboxyl-terminal proteinase from chick embryo tendons. Purification and characterization. J Biol Chem. 1985;260:15996–16003. [PubMed] [Google Scholar]
- 4.Medeck RJ, Sosa S, Morris N, Oxford JT. BMP-1-mediated proteolytic processing of alternatively spliced isoforms of collagen type XI. Biochem J. 2003;376:361–368. doi: 10.1042/BJ20030894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unsold C, Pappano WN, Imamura Y, Steiglitz BM, Greenspan DS. Biosynthetic processing of the pro-alpha 1(V)2pro-alpha 2(V) collagen heterotrimer by bone morphogenetic protein-1 and furin-like proprotein convertases. J Biol Chem. 2002;277:5596–5602. doi: 10.1074/jbc.M110003200. [DOI] [PubMed] [Google Scholar]
- 6.Panchenko MV, Stetler-Stevenson WG, Trubetskoy OV, Gacheru SN, Kagan HM. Metalloproteinase activity secreted by fibrogenic cells in the processing of prolysyl oxidase. Potential role of procollagen C-proteinase. J Biol Chem. 1996;271:7113–7119. doi: 10.1074/jbc.271.12.7113. [DOI] [PubMed] [Google Scholar]
- 7.Veitch DP, et al. Mammalian tolloid metalloproteinase, and not matrix metalloprotease 2 or membrane type 1 metalloprotease, processes laminin-5 in keratinocytes and skin. J Biol Chem. 2003;278:15661–15668. doi: 10.1074/jbc.M210588200. [DOI] [PubMed] [Google Scholar]
- 8.Rattenholl A, et al. Proteinases of the bone morphogenetic protein-1 family convert procollagen VII to mature anchoring fibril collagen. J Biol Chem. 2002;277:26372–26378. doi: 10.1074/jbc.M203247200. [DOI] [PubMed] [Google Scholar]
- 9.Ge G, et al. Bone morphogenetic protein-1/tolloid-related metalloproteinases process osteoglycin and enhance its ability to regulate collagen fibrillogenesis. J Biol Chem. 2004;279:41626–41633. doi: 10.1074/jbc.M406630200. [DOI] [PubMed] [Google Scholar]
- 10.Scott IC, et al. Bone morphogenetic protein-1 processes probiglycan. J Biol Chem. 2000;275:30504–30511. doi: 10.1074/jbc.M004846200. [DOI] [PubMed] [Google Scholar]
- 11.Scott IC, et al. Homologues of Twisted gastrulation are extracellular cofactors in antagonism of BMP signalling. Nature. 2001;410:475–478. doi: 10.1038/35068572. [DOI] [PubMed] [Google Scholar]
- 12.Ge G, Greenspan DS. BMP1 controls TGFbeta1 activation via cleavage of latent TGFbeta-binding protein. J Cell Biol. 2006;175:111–120. doi: 10.1083/jcb.200606058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge G, Hopkins DR, Ho WB, Greenspan DS. GDF11 forms a bone morphogenetic protein 1-activated latent complex that can modulate nerve growth factor-induced differentiation of PC12 cells. Mol Cell Biol. 2005;25:5846–5858. doi: 10.1128/MCB.25.14.5846-5858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfman NM, et al. Activation of latent myostatin by the BMP-1/tolloid family of metalloproteinases. Proc Natl Acad Sci USA. 2003;100:15842–15846. doi: 10.1073/pnas.2534946100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopkins DR, Keles S, Greenspan DS. The bone morphogenetic protein 1/Tolloid-like metalloproteinases. Matrix Biol. 2007;26:508–523. doi: 10.1016/j.matbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wermter C, et al. The protease domain of procollagen C-proteinase (BMP1) lacks substrate selectivity, which is conferred by non-proteolytic domains. Biol Chem. 2007;388:513–521. doi: 10.1515/BC.2007.054. [DOI] [PubMed] [Google Scholar]
- 17.Ge G, Zhang Y, Steiglitz BM, Greenspan DS. Mammalian tolloid-like 1 binds procollagen C-proteinase enhancer protein 1 and differs from bone morphogenetic protein 1 in the functional roles of homologous protein domains. J Biol Chem. 2006;281:10786–10798. doi: 10.1074/jbc.M511111200. [DOI] [PubMed] [Google Scholar]
- 18.Hintze V, et al. The interaction of recombinant subdomains of the procollagen C-proteinase with procollagen I provides a quantitative explanation for functional differences between the two splice variants, mammalian tolloid and bone morphogenetic protein 1. Biochemistry. 2006;45:6741–6748. doi: 10.1021/bi060228k. [DOI] [PubMed] [Google Scholar]
- 19.Garrigue-Antar L, Francois V, Kadler KE. Deletion of epidermal growth factor-like domains converts mammalian tolloid into a chordinase and effective procollagen C-proteinase. J Biol Chem. 2004;279:49835–49841. doi: 10.1074/jbc.M408134200. [DOI] [PubMed] [Google Scholar]
- 20.Svergun DI, Koch MHJ. Small-angle scattering studies of biological molecules in solution. Rep Prog Phys. 2003;66:1735–1782. [Google Scholar]
- 21.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia de la Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78:719–770. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rai N, et al. SOMO (SOlution MOdeler) differences between X-ray- and NMR-derived bead models suggest a role for side chain flexibility in protein hydrodynamics. Structure. 2005;13:723–734. doi: 10.1016/j.str.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Chen CB, Wallis R. Stoichiometry of complexes between mannose-binding protein and its associated serine proteases. Defining functional units for complement activation. J Biol Chem. 2001;276:25894–25902. doi: 10.1074/jbc.M103539200. [DOI] [PubMed] [Google Scholar]
- 26.Wallis R, Dodd RB. Interaction of mannose-binding protein with associated serine proteases: Effects of naturally occurring mutations. J Biol Chem. 2000;275:30962–30969. doi: 10.1074/jbc.M004030200. [DOI] [PubMed] [Google Scholar]
- 27.Busby TF, Ingham KC. NH2-terminal calcium-binding domain of human complement C1s- mediates the interaction of C1r- with C1q. Biochemistry. 1990;29:4613–4618. doi: 10.1021/bi00471a016. [DOI] [PubMed] [Google Scholar]
- 28.Thielens NM, et al. The N-terminal CUB-epidermal growth factor module pair of human complement protease C1r binds Ca2+ with high affinity and mediates Ca2+-dependent interaction with C1s. J Biol Chem. 1999;274:9149–9159. doi: 10.1074/jbc.274.14.9149. [DOI] [PubMed] [Google Scholar]
- 29.Gregory LA, Thielens NM, Arlaud GJ, Fontecilla-Camps JC, Gaboriaud C. X-ray structure of the Ca2+-binding interaction domain of C1s. Insights into the assembly of the C1 complex of complement. J Biol Chem. 2003;278:32157–32164. doi: 10.1074/jbc.M305175200. [DOI] [PubMed] [Google Scholar]
- 30.Scott IC, et al. Mammalian BMP-1/Tolloid-related metalloproteinases, including novel family member mammalian Tolloid-like 2, have differential enzymatic activities and distributions of expression relevant to patterning and skeletogenesis. Dev Biol. 1999;213:283–300. doi: 10.1006/dbio.1999.9383. [DOI] [PubMed] [Google Scholar]
- 31.Vistica J, et al. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 32.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roessle M, et al. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J Appl Crystallogr. 2007;40:s190–s194. [Google Scholar]
- 34.Semenyuk AV, Svergun DV. GNOM - a program package for small-angle scattering data processing. J Appl Crystallogr. 1991;24:537–540. [Google Scholar]
- 35.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.