Abstract
DnaK is the canonical Hsp70 molecular chaperone protein from Escherichia coli. Like other Hsp70s, DnaK comprises two main domains: a 44-kDa N-terminal nucleotide-binding domain (NBD) that contains ATPase activity, and a 25-kDa substrate-binding domain (SBD) that harbors the substrate-binding site. Here, we report an experimental structure for wild-type, full-length DnaK, complexed with the peptide NRLLLTG and with ADP. It was obtained in aqueous solution by using NMR residual dipolar coupling and spin labeling methods and is based on available crystal structures for the isolated NBD and SBD. By using dynamics methods, we determine that the NBD and SBD are loosely linked and can move in cones of ±35° with respect to each other. The linker region between the domains is a dynamic random coil. Nevertheless, an average structure can be defined. This structure places the SBD in close proximity of subdomain IA of the NBD and suggests that the SBD collides with the NBD at this area to establish allosteric communication.
Keywords: allostery, dipolar couplings, dynamics, NMR, structure
Hsp70 (heat shock 70 kDa) chaperone proteins are central to protein folding, refolding, and trafficking in organisms ranging from Archae to Homo sapiens, both at normal and at stressed conditions (for a review, see ref. 1). Recently, Hsp70s have been linked to breast and colon cancer (2) and to diseases such as Alzheimer's (3), Parkinson's (4), and Huntington's (5) diseases. In this report, DnaK, the canonical Hsp70 molecular chaperone protein from Escherichia coli, is studied. In the ADP state, DnaK, like other Hsp70s, binds to exposed hydrophobic residues of unfolded or partially misfolded proteins. Upon ATP binding, which induces an allosteric conformational change, DnaK releases the client protein (6). This process is tightly regulated by cochaperone proteins (7). DnaK consists of three subdomains. The structure of the nucleotide-binding domain (NBD, residues 1–370), was solved by crystallography (8). It competitively binds ATP and ADP and can slowly hydrolyze ATP (9). Structures for the 15-kDa substrate-binding domain (SBD, residues 390–600) were solved in different forms by crystallography (10) and NMR (11–13). It harbors the hydrophobic substrate-binding cleft. Here, this subdomain is referred to as BETA. A subsequent 10-kDa subdomain of α-helical structure (residues 510–638) was characterized by NMR (14) and crystallography (15). This subdomain, referred to as the LID, plays a key role in regulating the kinetics of substrate binding (16, 17).
Recently, structures have become available comprising both the NBD and SBD. Our group has used NMR methods to determine the global 3D solution structure of an NBD–SBD construct (residues 1–501) of Thermus thermophilus DnaK (18). A crystal structure of Bos taurus Hsc70 (residues 1–554 and E213A/D214A) was reported (19). A crystal structure for Geobacillus kaustophilus DnaK (residues 1–509) was determined (20). Furthermore, a crystal structure of Saccharomyces cerevisiae Hsp110 (2–659), which is a Hsp70 homolog permanently locked in the ATP state, has been reported (21). However, none of these structures is compatible with any of the others. The location where the SBD docks to the NBD differs by tens of angstroms. In one case, NBD and SBD are not docked at all. Moreover, some of the structures are in nonnative dimer form (G. kaustophilus DnaK and S. cerevisiae Hsp110) or interact with themselves in a nonnative way (B. taurus Hsc70). The closest to wild-type (WT) DnaK is a NBD–SBD construct of the E. coli DnaK (1–552) L542Y/L543E, which does not bind to itself and does not dimerize. With this construct, Swain et al. (22) show with NMR in solution that the NBD and linker are docked in the ATP state, but not in the ADP state, where they found the linker to be flexible. In addition, from the comparison of unassigned NMR spectra, they suggest that NBD and SBD are relatively independent in the ADP state, but not in the ATP state. However, no structures were determined in that otherwise seminal work.
Here, we report the solution conformation for the full-length, WT E. coli DnaK (1–638) and for a truncation (1–605), while complexed to substrate peptide (NRLLLTG) and ADP, determined by using NMR techniques. In this report, we show conclusively that the NBD, SBD, and linker move relatively independently of each other in this state of the protein. However, the motion of SBD with respect to NBD is restricted to a cone of ≈70° opening angle. By using NMR residual dipolar coupling (RDC) analysis (23) and spin labeling, we show that within this cone there is a preferred orientation of SBD with respect to NBD that can be defined with a ±3° orientational and ±5 Å translational precision. The relative locations of NBD and SBD in this state imply that the SBD preferentially collides with subdomain IA of the NBD, in the vicinity of the IA–IIA interface. Because E. coli DnaK is highly homologous to the human Hsp70s (24), we expect that our findings here are relevant for the human proteins as well.
Results
Dynamic Differences Between the Domains.
The effective rotational correlation times for the NBD and BETA/LID units in E. coli DnaK ADP/NRLLLTG are 30 ns and 22 ns, respectively. The intensities of the cross-peaks in the 3D HNCO-TROSY data are much smaller than those of the cross-peaks of the BETA/LID domain [see supporting information (SI) Appendix, Fig. S1]. Together, these data show that the NBD and BETA/LID domains of E. coli DnaK in the ADP/NRLLLTG state have different mobilities and move relatively independently. The HNCO data also show that SBD subdomains BETA (400–500) and LID (500–600) move together as a single rigid unit. Large HNCO intensities are observed for the resonances of the NBD–SBD linker residues (379–397). This demonstrates a large amount of flexibility for these residues. The lack of dispersion in NMR chemical shifts shows that the flexible linker has a random coil conformation.
Time-Averaged Structure.
Despite its dynamic properties, a remarkably precise average structure can be defined for E. coli DnaK ADP/NRLLLTG by using RDC analysis (23) and spin labeling. We based our analysis on the X-ray structures of the isolated E. coli DnaK NBD (25) and E. coli DnaK SBD (10). The hybrid solution X-ray conformation was determined with the best precision for E. coli DnaK, residues 1–605, in aqueous buffer with ADP, inorganic phosphate, and substrate peptide NRLLLTG (pH 7.2), 27 °C. WT E. coli DnaK contains another 33 residues at the C terminus. The NMR TROSY spectrum reveals that these residues are not structured in solution (spectra not shown). A preliminary RDC analysis of the WT data showed that the relative orientations of the SBD and NBD were identical (within experimental error, see Fig. 1) to those in the DnaK (1–605) construct. Hence, the 1–605 construct is, in structural terms, a bona fide full-length Hsp70 chaperone. In addition, the construct has ATP hydrolysis activity and DnaJ stimulation of that activity identical to that of WT DnaK (see SI Appendix, Figs. S4 and S5). To date, no function has been ascribed to the 33-aa C terminus of E. coli DnaK.
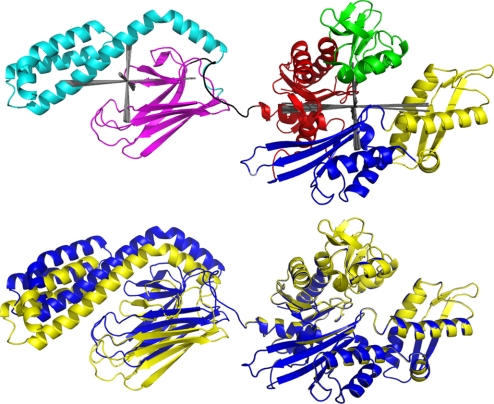
Fig. 1.
Hybrid NMR RDC structure of E. coli DnaK. (Upper) Hybrid NMR RDC structure of E. coli (1–605) with ADP, orthophosphate, and the substrate peptide NRLLLTG bound. Red, NBD IA; green, NBD IB; blue, NBD IIA; yellow, NBD IIB; magenta, SBD BETA; cyan, SBD LID; black, NBD–SBD linker. The orientations and experimental uncertainties of the alignment tensors are shown in gray. (Lower) Hybrid NMR RDC structure of E. coli DnaK (1–605, blue) and WT E. coli DnaK (1–638, yellow) superposed on NBD IA, IB, and IIA. Both molecules have ADP, orthophosphate, and the substrate peptide NRLLLTG bound.
The following steps in the hybrid structure determination procedure can be distinguished: NMR backbone resonance assignments, RDC measurements, paramagnetic relaxation enhancement measurements, calculations and refinements. With 638 residues, E. coli DnaK is one of the largest proteins to have its NMR backbone resonances assigned; it was carried out by assembling assignments of isolated domains. For RDC measurements, DnaK was diffused into a nondenaturing, uncharged 3% cross-linked polyacrylamide gel (26). The gel was stretched to provide an anisotropic environment allowing the measurements of RDCs. Several hundred 15N, 1H RDCs in the range of −15 to +20 Hz were measured. Of the measured RDCs, 152 could be confidently assigned for the NBD, 98 for the SBD, and 22 for the LID. This number of RDCs is more than sufficient to obtain the relative orientations of the DnaK domains for which structures are known independently (23). A 2.8-Å resolution crystal structure of E. coli DnaK NBD (residues 3–383) complexed with GrpE (25) was used as a starting point for the structure of the NBD in DnaK (1–605). A 2.0-Å crystal structure (10) of E. coli DnaK SBD (residues 389–607, containing BETA and LID) complexed with the peptide NRLLLTG was taken as a starting point for the structure of the SBD in the DnaK (1–605). First, the relative orientations of the NBD subdomains IA, IB, IIA, and IIB and the SBD subdomains BETA and LID were determined from the RDC data. As Table S2 in the SI Appendix shows, domains IA, IB, and IIA are similarly oriented within experimental error. The orientation of domain IIB differs from these by 20°. Individually, the BETA and LID subdomains are similarly oriented, within experimental error. On the basis of this, it was decided to treat the SBD BETA and LID subdomains as a single unit, and the NBD domains IA, IB, and IIA as a single unit. The object became to orient SBD, NBD(IA,IB,IIA), and NBD(IIB) relative to each other. The statistics of the RDC data analysis are reported in Table 1.
Table 1.
RDC calculation statistics
| Subdomains | PDB | NRDCa | DA, Hzb | DR/DAc | α, ° | β, ° | γ, ° | rmsd, Hzd | Qe | Szz, ×104 | Syy, ×104 | Sxx, ×104 | GDO, ×104f | ηg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NBD IA-IB-IIA best fit | E. coli DnaK 1DKG.pdb | 108 | −11.62 | 0.29 | 80.27 | 79.16 | 93.75 | 5.28 | 0.43 | −9.54 | 6.81 | 2.73 | 9.83 | 0.43 |
| Error selfh | 0.63 | 0.07 | 44.74 | 2.15 | 50.55 | 0.30 | 0.03 | 0.52 | 0.49 | 0.53 | 0.50 | 0.10 | ||
| Error mci | 0.80 | 0.07 | 40.02 | 1.98 | 38.79 | 0.41 | 0.04 | 0.66 | 0.49 | 0.58 | 0.64 | 0.10 | ||
| NBD IIB best fit | E. coli DnaK 1DKG.pdb | 29 | −12.09 | 0.22 | 97.98 | 64.16 | 78.93 | 5.32 | 0.43 | −9.93 | 6.61 | 3.32 | 10.11 | 0.33 |
| Error selfh | 1.45 | 0.16 | 47.68 | 3.66 | 13.25 | 0.77 | 0.07 | 1.19 | 1.08 | 1.46 | 1.11 | 0.24 | ||
| Error mci | 1.14 | 0.11 | 45.59 | 5.52 | 80.27 | 79.16 | 93.75 | 0.94 | 1.05 | 0.98 | 0.96 | 0.17 | ||
| SBD BETA-LID best fit | E. coli DnaK 1DKX.pdb | 79 | −14.38 | 0.31 | 30.54 | 81.86 | 120.07 | 2.67 | 0.19 | −11.81 | 8.62 | 3.19 | 12.22 | 0.46 |
| Error selfh | 0.44 | 0.03 | 34.46 | 1.39 | 33.40 | 0.37 | 0.03 | 0.36 | 0.25 | 0.34 | 0.33 | 0.05 | ||
| Error mcj | 0.29 | 0.02 | 41.00 | 0.87 | 41.28 | 0.25 | 0.02 | 0.24 | 0.18 | 0.27 | 0.22 | 0.04 |
Best fits were computed using a grid-search program optimizing DA, DR/DA, and the three tensor orientation angles α, β, and γ.
aNumber of dipolar restraints used.
bDA = [Szz − (Syy + Sxx)/2]* DMAX, where DMAX is the full dipolar coupling (22 KHz).
cDR = (Sxx − Syy) * DMAX.
drmsd of RDC fit.
eQ = RMSD/.
fGDO = .
gη = (Sxx − Syy)/Szz.
hThirty fits were computed for NBD and SBD each, using on average 60% of the RDC data, randomly picked (self-validation).
iForty sets of synthetic RDC data, corresponding to the actual available RDC data for NBD IA, IB, and IIA, with a Monte Carlo random error of 15 degrees in NH orientation and a random RDC measurement error of 2 Hz were analyzed.
jForty sets of synthetic RDC data, corresponding to the actual available RDC data for the SBD, with a random error of 5° degrees in NH orientation and a random RDC measurement error of 2 Hz were analyzed.
Relative Domain Position.
The model of the average structure in solution as shown in Fig. 1 was obtained as follows. Stage 1 entailed orienting the two domains such that the principal axis system of the alignment tensors coincide, followed by a translation of one of the domains to a position that is in agreement with the covalent structure; that is, the SBD has to be to the “left” of the NBD in the representation of Fig. 1. Still, there are two possibilities: either domain can be flipped by 180° along the long (zz) axis of the tensor. This ambiguity was solved with a spin labeling experiment. The mutation V210C was introduced in DnaK (1–605). The function of DnaK, as measured by an ATP hydrolysis assay, was not affected by this mutation (see SI Appendix, Figs. S4 and S5). (1-Oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)Methanethiosulfonate spin label (27) (MTSL) was covalently attached to this residue. MTSL causes a broadening of 1H NMR resonances beyond detection for protons that are closer than 15 Å (28). Fig. 2 shows that the label attached at V210C broadened the ends of SBD β-strands 2 and 3 and the loop in between. In addition, a hydrophobic patch on the “face” of the SBD was broadened. The broadening data could not be analyzed quantitatively because a control experiment using free MTSL showed broadening to the same hydrophobic patch on the face of the SBD. Apparently, the spin label at V210C can dynamically access multiple sites; this is not surprising in light of the dynamic nature of the DnaK molecule itself.
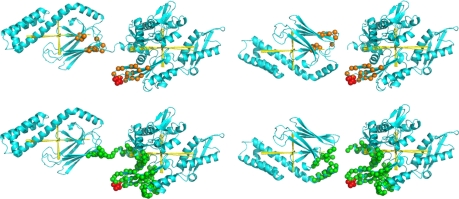
Fig. 2.
Results of MTSL spin labeling. (upper left) NH resonances that disappeared from the TROSY spectrum (orange spheres) when V210C (red spheres) is spin labeled with MTSL, indicated on E. coli DnaK with the correct SBD orientation. (upper right) NH resonances that disappeared from the TROSY spectrum (orange spheres) when V210C (red spheres) is spin labeled with MTSL, indicated on E. coli DnaK with the wrong SBD orientation. (lower left) NH within 25 Å (green spheres) of V210C (red spheres), indicated on E. coli DnaK with the correct SBD orientation. (lower right) NH within 25 Å (green spheres) of V210C (red spheres), indicated on E. coli DnaK with the wrong SBD orientation.
In fully extended side chain conformation, the nitroxide atoms of the spin label are 9.5 Å away from the Ca atom of the cysteine to which it is attached. Hence resonances as far as 25 Å from the Ca atom of the labeled cysteine can be affected. Fig. 2 shows excellent correspondence between the experimental broadening and residues in the range of 25 Å of V210C. Certainly, the experimental broadening pattern is not compatible at all with a NBD–SBD structure with the SBD “up-side down.” Hence, the spin labeling studies show that the orientation as presented in Fig. 1 must be the correct one and that the NBD and SBD are on average not much further apart than indicated; otherwise, no resonances of the SBD could have been affected by spin labeling on the NBD.
Further Refinement.
The stretched polyacrylamide gel used for the RDC experiments contains elongated pores that host the protein. An elongated protein like DnaK aligns (for ≈0.2% of the time) with its long axis parallel to the long axis of the pores. In such a case of pure sterical alignment (as opposed to charge-induced or susceptibility-induced alignment), one may use the program PALES to predict the alignment tensor of several trial models for the full-length protein (29). The PALES program only uses the shape of the protein to predict the orientation of the overall alignment tensor very precisely and is sensitive to ±5-Å translation in the x and y direction perpendicular to the long (Szz) axis (see SI Appendix, Table S4). The experimentally determined rhombicity of the alignment tensor was used to estimate the ratio of the long and short axis of the protein model. The best agreement between measured and observed tensor orientation and rhombicity is found when the coordinate center of the SBD is 0, −10, −65 Å displaced from the coordinate center of the NBD in x–z, respectively (see Fig. 1).
A key element determining the validity of the PALES computation is whether the two domains are each independently aligned or whether the alignment represents the average shape of the molecule as a whole. We distinguish these extremes by computing theoretical alignments for the different isolated domains and comparing those with the experimental data for the domains in the context of the full-length protein. As SI Appendix, Table S4 shows, the predicted alignment angles of the NBD by itself deviate ≈30° from the experimental alignment seen for NBD in full-length DnaK. The NBD in full-length DnaK thus senses the presence of the SBD. Hence, it is justified to conceive of the dynamic ensemble DnaK in the ADP/peptide bound state as shown in Fig. 1 as a “structure” aligned on the basis of its time-averaged structure. The coordinates for this model are deposited in the PDB (accession code 2kho).
Reconciliation of Dynamic and Structural Data.
Having deduced the time-averaged conformation of DnaK in solution, we may propose a model for the interdomain dynamics. By using available equations describing the rotational diffusion of ellipsoids (30), we calculate an average rotational correlation time of 50 ns for a rigid molecule of the shape shown in Fig. 1. Using the same equations, we calculate correlation times of 18 and 9.5 ns for the isolated NBD and SBD, respectively. However, as mentioned before, the experimental rotational correlation times of for the NBD and BETA/LID in the context of the full-length protein are 30 and 22 ns, respectively. In the SI Appendix, we reconcile all measured and calculated correlation times with a dynamic model in which NBD and SBD diffuse in a cone of opening angle of ±35° with respect to each other.
With model calculations, we show that the averaging of the RDCs over this large amplitude of motion still results in a RDC dataset, that is, within experimental error, compatible with a single structure at the average position shown in Fig. 1 (see Figs. S2 and S3 and text in the SI Appendix).
Discussion
Overall Findings.
It was reported that the NMR TROSY spectrum of E. coli DnaK (1–552) L542Y/L543E, in the ADP–peptide state superposes well on TROSY spectra of the corresponding isolated domains (22). These data suggested that the NBD and SBD had little or no contact in this state. Narrow resonances for the interdomain linker argued that this conserved polypeptide was flexible. These authors (22) did not carry out dynamic analysis, leaving open the question of how motionally restricted the domains are in the ADP state and whether there are functionally important domain–domain interactions. The present rigorous analysis of full-length DnaK shows conclusively that the domains are independent and mobile in the ADP–peptide state.
This was taken to suggest that NBD and SBD are independent in this state, even though those data are also compatible with a static structure with a small interface. The data obtained in our work show conclusively that the domains are independent and mobile, also for WT DnaK. Moreover, we have determined that the relative motion of NBD and SBD is restricted in a ±35° cone, and more importantly, that the time-averaged structure places the SBD in a position in which it is poised to contact subdomain IA of the NBD in a well-defined orientation. In addition, we show that the LID and BETA subdomains are docked in the ADP–peptide state and that residues 606–638 are disordered in solution.
Comparison with Mutagenesis Data.
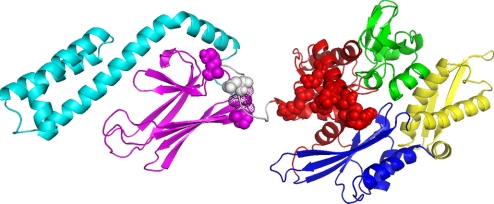
The residues in E. coli DnaK and other Hsp70s that, when mutated, leave the ATP hydrolysis and substrate-binding properties of DnaK intact but that abrogate or attenuate allosteric communication between the domains, are shown in Fig. 3. The figure shows that all of these mutations map in the collision interface as suggested in the current work. The average structure suggests that the NBD samples the SBD surface at the loop L2,3 between β-strands 2 and 3, residues 410–420, and L6,7 between β-strands 6 and 7, residues 479–482. Several mutations in the SBD that affect the allosteric function of E. coli DnaK have been mapped to the loop L2,3 region: DnaK K414I shows an absence of allostery (31); mutation of Pro-419 (DnaK numbering) in Hsp70s leads to defects in function (32, 33). Residues Thr-417 and Ile-418 in the same loop display strong millisecond dynamics in the isolated E. coli DnaK SBD/apo, but not in isolated SBD with NRLLLTG bound (11). This shows a structural/dynamic coupling between the substrate-binding cleft and L2,3. The mutagenesis and dynamics data strongly suggest that the structural/dynamic state of the surface of loop L2,3 is critical for signaling the presence or absence of substrate in the SBD to the NBD. The conserved hydrophobic linker likely plays a role in the sampling process. In the current ensemble of E. coli DnaK describing the ADP/NRLLLTG state, the linker is disordered. This result is not novel; it has been demonstrated before from limited tryptic digestion (34), amide proton exchange (35), mutagenesis (36), and NMR (22).
Fig. 3.
Mutagenesis and structure. The residues in E. coli DnaK, which when mutated affect the NBD–SBD interdomain communication, are represented as spheres in the colors of the subdomains to which they belong. These residues are, on the NBD: Y145A, N147A, and D148A (48); P143G and R151A (49); K155D and R167D (36). On the linker one has D393A (36). On the SBD they are K414I (31) and P419 (32, 33). Residues 417 and 418 SBD that show significant line broadening in the peptide-free form but not in the peptide bound form of the isolated SBD (11) are shown as dot surfaces.
Comparison with Other Structural Information.
One computes 39.6 Å for the radius of gyration (Rg) of the structure shown in Fig. 1. This may be compared with SAXS data obtained more than a decade ago: the Rg of E. coli DnaK in the ADP state was found to be 37–38 Å (37), and the Rg of B. taurus Hsc70 in the ADP state is between 35 and 42 Å (38), depending on analysis methods. The correspondence of our structure with these data are good but likely coincidental. WT E. coli DnaK contains an additional 33 dynamic and disordered residues at its C terminus compared with DnaK (1–605). Bovine Hsc70 is 54 residues longer than DnaK (1–605). These extensions should tend to make the Rg of WT DnaK and WT Hsc70 larger than that of DnaK (1–605). However, the samples used for the SAXS experiments did not contain peptide substrate. Recently, it has become apparent that the NBD and SBD of DnaK are mostly docked in the absence of peptide, even in the presence of ADP (22). This would tend to decrease the Rg of the SAXS samples compared with DnaK (1–605) ADP/NRLLLTG.
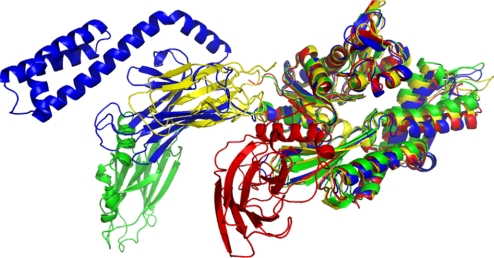
Several structures of two-domain Hsp70 NBD–SBD constructs have been published in the last few years. However, none of these structures is compatible with any of the others, as is shown in Fig. 4. In contrast, the presently reported E. coli DnaK conformation corresponds remarkably well with the NMR-RDC solution structure of T. thermophilus DnaK (1–507), even though the latter was lacking the complete LID domain and contained the mutations ΔT428, A429E (E. coli DnaK count) (18). There is, however, an important dynamic difference: the NBD and SBD of T. thermophilus DnaK move as a single unit, and the linker is buried and structured (18). The T. thermophilus data were recorded at 50 °C, a full 25 °C lower than the physiological temperature for the species (39). Likely, the rigidity observed in the earlier structure may be an artifact of the low relative temperature, absence of peptide, and/or the construct.
Fig. 4.
Comparison of structures. Comparison of the Hsp coordinates as obtained for E. coli DnaK (1–605) with other published structures for multidomain constructs in the Hsp70 family. The Ca positions of the corresponding residues in the NBDs were superimposed. Dark blue, hybrid solution X-ray conformation of E. coli DnaK in the ADP/peptide state; yellow, T. thermophilus DnaK in the ADP/apo state (18); green, self-binding G. kaustophilus DnaK dimer in the ADP state (20); red, self-binding B. taurus Hsc70 (19); cyan, S. cerevisiae Hsp110 dimer in the ATP state (21).
In character, a crystal structure for a dimer of Geobacillus kaustophilus DnaK (1–509) (20) is closest to our result (see Fig. 4). In that structure, the NBD and SBD are not docked either. However, the reported positions of NBD and SBD are likely artifactual: the substrate-binding cleft of the SBD of one monomer interacts with the NBD–SBD linker of the other monomer.
A crystal structure of a mutated (E208A/D209A), truncated (D552–610), and self-binding NBD–SBD construct (E. coli count) of human Hsc70 (19) is not compatible with our solution data at all: the SBD in the crystal structure is rotated by 90° and interacts not only with subdomain IA, but also IIA (see Fig. 4). It is difficult to rationalize these differences as being caused by intrinsic differences between E. coli and T. thermophilus DnaK on the one hand, and Hsc70 Homo sapiens on the other hand, because the sequence homology is so high.
Allosteric Model.
The present work shows, conclusively, that the ADP- and peptide-bound state of E. coli DnaK E. coli ADP/NRLLLTG is characterized by “noncommunication” between the domains. For the domains to communicate in the allosteric cycle, they must make contact in the ATP state. Recently, a crystal structure for a yeast Hsp110 dimer, locked in the ATP state, was solved (21). Hsp110 has strong Hsp70 homology, and NBD and SBD are indeed docked in an area generally similar to the area of close apposition for the SBD and NBD we find for E. coli and T. thermophilus DnaK (see Fig. 4).
In Hsp110 in the ATP state, the hydrophobic NBD–SBD linker is buried in a hydrophobic groove between subdomains IA and IIA, close to the NBD–SBD interface. This groove is closed in other crystal structures of Hsp70 proteins and homologs. It was suggested that this difference is key to the allosteric mechanism. We have recently been able to prove this hypothesis for the isolated NBD of T. thermophilus DnaK (40). By using RDC NMR we detect major rotations of NBD subdomains IA and IIA with respect to each other when comparing the ADP and AMPPNP states. These rotations result in an open groove in the ATP (AMPPNP) state and an occluded groove in the ADP state. In the current work, we find that the orientations of subdomains IA, IB, and IIB are indistinguishable from those in the DnaK crystal structure, in which the groove is closed. We thus conclude the groove is also closed in the solution conformation of WT DnaK ADP/NRLLLTG. This accounts for the undocked state of the NBD and SBD and for the random, mobile state of the NBD–SBD linker.
Taking together the results of several groups accumulated over 2 decades allows the following model for the allosteric communication to be proposed: the present work shows that in the ADP/peptide state, SBD + linker collides with and samples the surface areas of subdomains IA and IIA of the NBD and that the IA/IIA cleft is closed. The X-ray work on Hsp110 ATP (21), the NMR work (22) on E. coli DnaK (1–552), and the NMR work on T. thermophilus DnaK (40) strongly suggest that in the ATP state, the NBD surface groove opens up. When this happens, we propose that the NBD–SBD collisions become productive, which place the linker in the groove and force loops L2,3 and L5,6 of the SBD to dock on the IA area. Several years ago, we showed that the structure and dynamics of loop L2,3 of E. coli DnaK SBD are allosterically linked to the presence/absence of the substrate in the substrate-binding cleft (11). Hence, in principle, the complete allosteric path from nucleotide-binding site to substrate-binding site can now be delineated.
Experimental Procedures
Plasmids for T7-based expression of E. coli DnaK1–638, DnaK1–605, and DnaK1–388 were constructed by PCR subcloning into the vector pET22b using the plasmid pJK as a template. Site-directed mutagenesis was accomplished by using QuikChange (Stratagene) methodology. Expression and purification of isotopically labeled DnaK and its variants were accomplished as described in ref. 18, in which the protein was at all times stabilized with proteolysis inhibitors. The proteins were denatured in 6 M guanidine hydrochloride to allow for complete protonation of the backbone amides and were then refolded by rapid dilution.
For assignments and dynamics, we used 150–250 μM samples of isotope-labeled DnaK in 25 mM Tris·HCl, 10 mM KCl, 5 mM MgCl2 10 mM DTT, 5 mM ADP, 10 mM potassium phosphate, 0.2% sodium azide, and 2 mM peptide NRLLLTG (pH 7.2). Experiments were performed at 27 °C on a Varian Inova 800 equipped with a triple-resonance cold probe. Backbone assignment was obtained by assembling assignments of domains of DnaK. The isolated NDB (387 aa) was assigned de novo by using 3D HNCA, HNCOCA, HNCO, HNCACO, HNCACB, and HNCOCACB TROSY with 2H decoupling. The combined data collection time was ≈14 days. These assignments were fully traced in HNCA and HNCOCA 3D spectra of DnaK (1–605). Available assignments for the DnaK BETA domain in the presence of NRLLLTG (13) and LID domain (14) were also traced in 3D spectra of the full-length protein. The assignment process was aided at all steps by the inclusion of information from a series of five (Asp, Ile, Leu, Val, Phe) residue-specifically labeled DnaK proteins (41).
15N autorelaxation data for DnaK (1–605) were derived from standard [15N,1H]R1-TROSY and [15N,1H]R2-TROSY experiments at 800 MHz (42).
For RDC measurements, a 4-mm-diameter, cylindrical section of polyacrylamide gel (3% T, 2.5% C) was equilibrated in H2O, dehydrated to ≈20% of its original weight, and then added to a sample of refolded 1H,13C, 2H-DnaK containing ADP, potassium phosphate, and peptide as described above. The samples were allowed to equilibrate at room temperature for ≈72 h. This yielded a homogeneous sample as was judged from stained cross-sections. The rehydrated sample was compressed into an open-ended NMR tube using a funnel (New Era). The top and bottom of the sample were sealed with inserts, and the sample was allowed to equilibrate at room temperature for an additional 48 h before data collection. The sample displayed a small solvent 2H2O quadrupolar splitting of ≈0.3 Hz. Duplicate series of J,D-modulated 15N,1H-TROSY experiments (43, 44) were collected, with κ delays of 0, 0.5, 1.0, and 1.5. The data were added, and RDCs were extracted from the slopes of linear fits to plots of κ vs. dipolar shift, assuming a constant J-coupling constant of 91 Hz. The analysis yielded 231 assigned RDCs, listed in SI Appendix, Table S5. A separate RDC sample was made with full-length DnaK (1–638) ADP/NRLLLTG. Because this sample was less concentrated and because the spectra are dominated by TROSY resonances of the unstructured tail, the analysis yielded only 183 assigned RDCs, listed in SI Appendix, Table S6. All spectra were processed with NMRPipe (45) and analyzed by using SPARKY (46).
The NH RDC were fitted individually for the SBD and NBD IA,IB,IIA and NBD IIB. The coordinates of the SBD were taken directly from the file 1DKX.pdb. The coordinates of the NBD were taken from the file 1DKG.pdb. By using SwissModel, the missing loops were modeled, and the structure was regularized with the GROMOS minimizer in SwissModel. The REDCAT RDC software was initially used (47). However, to have more control over the data fitting, error determination, and self-validation, we used in-house written programs. The program carries out a complete grid search of five parameters: the orientation of the alignment tensor with respect to the protein coordinates (rotating the three Euler angles in 10° increments), the axial alignment magnitude DA (within a range after initial tries), and the rhombicity, Dr/Da, from 0 to 1 in 0.2 increments. The program subsequently optimizes the best grid point of these five parameters the target function (sum of the squared differences between calculated and observed RDCs) by using a Monte Carlo minimization. After culling outlying RDCs (identified in SI Appendix, Tables S7 and S8) we obtain a rmsd of fit of 5 Hz for the NBD and 2 Hz for the SBD.
Simulations (SI Appendix, Table S3) with artificial data showed that the obtained rmsd of fit could be simulated with a 2-Hz uncertainty on the RDCs and a 15° uncertainty on the directions of the NH vectors for the NBD (2 Hz and 5°, respectively, for the SBD). On this basis, we carried out 40 calculations per subdomain in which we added a random errors in the range −2 to 2 Hz to the experimental RDC data and ±15 (NBD) and ±5 (SBD) to the NH vector directions. These error ranges are listed in Table 1 as “error mc.”
Test calculations showed that a self-validation at the 60% level of one input dataset with a certain known error reproduces the uncertainties in the output parameters as obtained from many input datasets with the same known error. The internal consistency of the selected RDCs was evaluated by using a self-valuation procedure. In this procedure, 60% of the experimentally available RDCs for the (sub)domain under investigation were chosen at random, and an alignment was calculated on the basis of that information. This was repeated 30 times for each (sub)domain. The variations in the alignment parameters (Da, Da/Dr, and the three Euler alignment angles) for these computations are listed in Tables 1 and SI Appendix, Table S2 as “error self.”
Domains IA,IB,IIA were joined with IIB (residues 228–310) by minimizing the position of IIB with respect to the original position of IIB, using translation and rotation around the Szz axis only. By using SwissProt, the chain was linked (227–228) and (310–311) and the geometry of the immediate environment minimized by using 60 steps of steepest descent minimization for residues 225–233 and 307–314, respectively.
The NBD and SBD were rotated in their experimentally determined principal axis system. This automatically aligns the alignment tensors. Once these relative orientations were established, we generated several models in which the SBD was translated in 5° increments in z,−z, y,−y, and x,−x. We used the program PALES (29) to compute the steric alignment tensor orientation of each of these models and compared it with the experimental one (see SI Appendix, Table S4).
Supplementary Material
Acknowledgments.
We thank Dr. A. V. Kurochkin for maintenance of the NMR systems used, Mr. D. S. Weaver for help with the 15N relaxation data interpretation, Dr. A. M. Al-Hashimi for stimulating discussions with respect to RDC interpretation, and Dr. G. C. Crippen for assistance with the protein modeling. This work was supported by National Institutes of Health Grants GMS GM063027 (to E.R.P.Z.) and NS059690-01A1 (to J.E.G. and E.R.P.Z.). All NMR data for this work were obtained by using a 800-MHz Varian cryogenic NMR probe funded by National Institutes of Health Grant NCCR RR-03-002.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2KHO).
This article contains supporting information online at www.pnas.org/cgi/content/full/0903503106/DCSupplemental.
References
- 1.Mayer MP, Brehmer D, Gassler CS, Bukau B. Hsp70 chaperone machines. Adv Protein Chem. 2001;59:1–44. doi: 10.1016/s0065-3233(01)59001-4. [DOI] [PubMed] [Google Scholar]
- 2.Wadhwa R, Taira K, Kaul SC. An Hsp70 family chaperone, mortalin/mthsp70/PBP74/Grp75: What, when, and where? Cell Stress Chaperones. 2002;7:309–316. doi: 10.1379/1466-1268(2002)007<0309:ahfcmm>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gestwicki JE, Crabtree GR, Graef IA. Harnessing chaperones to generate small-molecule inhibitors of amyloid-β aggregation. Science. 2004;306:865–869. doi: 10.1126/science.1101262. [DOI] [PubMed] [Google Scholar]
- 4.Chung KK, Dawson TM. Parkin and Hsp70 sacked by BAG5. Neuron. 2004;44:899–901. doi: 10.1016/j.neuron.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Novoselova TV, et al. Treatment with extracellular Hsp70/Hsc70 protein can reduce polyglutamine toxicity and aggregation. J Neurochem. 2005;94:597–606. doi: 10.1111/j.1471-4159.2005.03119.x. [DOI] [PubMed] [Google Scholar]
- 6.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 7.Diamant S, Ben-Zvi AP, Bukau B, Goloubinoff P. Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J Biol Chem. 2000;275:21107–21113. doi: 10.1074/jbc.M001293200. [DOI] [PubMed] [Google Scholar]
- 8.Flaherty KM, Wilbanks SM, DeLuca-Flaherty C, McKay DB. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild-type and mutant ATPase fragment. J Biol Chem. 1994;269:12899–12907. [PubMed] [Google Scholar]
- 9.McCarty JS, Buchberger A, Reinstein J, Bukau B. The role of ATP in the functional cycle of the DnaK chaperone system. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 10.Zhu XT, et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellecchia M, et al. Structural insights into substrate binding by the molecular chaperone DnaK. Nat Struct Biol. 2000;7:298–303. doi: 10.1038/74062. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, et al. The solution structure of the 21-kDa chaperone protein DnaK substrate binding domain: A preview of chaperone–protein interaction. Biochemistry. 1998;37:7929–7940. doi: 10.1021/bi9800855. [DOI] [PubMed] [Google Scholar]
- 13.Stevens SY, Cai S, Pellecchia M, Zuiderweg ER. The solution structure of the bacterial Hsp70 chaperone protein domain DnaK(393–507) in complex with the peptide NRLLLTG. Protein Sci. 2003;12:2588–2596. doi: 10.1110/ps.03269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertelsen EB, Zhou H, Lowry DF, Flynn GC, Dahlquist FW. Topology and dynamics of the 10-kDa C-terminal domain of DnaK in solution. Protein Sci. 1999;8:343–354. doi: 10.1110/ps.8.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou CC, et al. Crystal structure of the C-terminal 10-kDa subdomain of Hsc70. J Biol Chem. 2003;278:30311–30316. doi: 10.1074/jbc.M304563200. [DOI] [PubMed] [Google Scholar]
- 16.Buczynski G, Slepenkov SV, Sehorn MG, Witt SN. Characterization of a lidless form of the molecular chaperone DnaK: Deletion of the lid increases peptide on- and off-rate constants. J Biol Chem. 2001;276:27231–27236. doi: 10.1074/jbc.M100237200. [DOI] [PubMed] [Google Scholar]
- 17.Slepenkov SV, Witt SN. Kinetic analysis of interdomain coupling in a lidless variant of the molecular chaperone DnaK: DnaK's lid inhibits transition to the low affinity state. Biochemistry. 2002;41:12224–12235. doi: 10.1021/bi0263208. [DOI] [PubMed] [Google Scholar]
- 18.Revington M, Zhang Y, Yip GN, Kurochkin AV, Zuiderweg ER. NMR investigations of allosteric processes in a two-domain Thermus thermophilus Hsp70 molecular chaperone. J Mol Biol. 2005;349:163–183. doi: 10.1016/j.jmb.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 19.Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20:513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang YW, Sun YJ, Wang C, Hsiao CD. Crystal structures of the 70-kDa heat shock proteins in domain disjoining conformation. J Biol Chem. 2008;283:15502–15511. doi: 10.1074/jbc.M708992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swain JF, et al. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer MW, Losonczi JA, Weaver JL, Prestegard JH. Domain orientation and dynamics in multidomain proteins from residual dipolar couplings. Biochemistry. 1999;38:9013–9022. doi: 10.1021/bi9905213. [DOI] [PubMed] [Google Scholar]
- 24.Bukau B. Regulation of the Escherichia coli heat-shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 25.Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 26.Sass HJ, Musco G, Stahl SJ, Wingfield PT, Grzesiek S. Solution NMR of proteins within polyacrylamide gels: Diffusional properties and residual alignment by mechanical stress or embedding of oriented purple membranes. J Biomol NMR. 2000;18:303–309. doi: 10.1023/a:1026703605147. [DOI] [PubMed] [Google Scholar]
- 27.Berliner LJ, Grunwald J, Hankovszky HO, Hideg K. A novel reversible thiol-specific spin label: Papain active site labeling and inhibition. Anal Biochem. 1982;119:450–455. doi: 10.1016/0003-2697(82)90612-1. [DOI] [PubMed] [Google Scholar]
- 28.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear Overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 29.Zweckstetter M, Bax A. Characterization of molecular alignment in aqueous suspensions of Pf1 bacteriophage. J Biomol NMR. 2001;20:365–377. doi: 10.1023/a:1011263920003. [DOI] [PubMed] [Google Scholar]
- 30.Woessner DE. Nuclear spin relaxation in ellipsoids undergoing rotational Brownian motion. J Chem Phys. 1962;37:647–654. [Google Scholar]
- 31.Montgomery DL, Morimoto RI, Gierasch LM. Mutations in the substrate binding domain of the Escherichia coli 70-kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J Mol Biol. 1999;286:915–932. doi: 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- 32.Burkholder WF, et al. Mutations in the C-terminal fragment of DnaK affecting peptide binding. Proc Natl Acad Sci USA. 1996;93:10632–10637. doi: 10.1073/pnas.93.20.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voisine C, et al. The protein import motor of mitochondria: Unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell. 1999;97:565–574. doi: 10.1016/s0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]
- 34.Buchberger A, et al. Nucleotide-induced conformational changes in the ATPase and substrate-binding domains of the DnaK chaperone provide evidence for interdomain communication. J Biol Chem. 1995;270:16903–16910. doi: 10.1074/jbc.270.28.16903. [DOI] [PubMed] [Google Scholar]
- 35.Rist W, Graf C, Bukau B, Mayer MP. Amide hydrogen exchange reveals conformational changes in Hsp70 chaperones important for allosteric regulation. J Biol Chem. 2006;281:16493–16501. doi: 10.1074/jbc.M600847200. [DOI] [PubMed] [Google Scholar]
- 36.Vogel M, Mayer MP, Bukau B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem. 2006;281:38705–38711. doi: 10.1074/jbc.M609020200. [DOI] [PubMed] [Google Scholar]
- 37.Shi L, Kakaoka M, Fink AL. Conformational characterization of DnaK and its complexes by small-angle X-ray scattering. Biochemistry. 1996;35:3297–3308. doi: 10.1021/bi951984l. [DOI] [PubMed] [Google Scholar]
- 38.Wilbanks SM, Chen L, Tsuruta H, Hodgson KO, McKay DB. Solution small-angle X-ray scattering study of the molecular chaperone Hsc70 and its subfragments. Biochemistry. 1995;34:12095–12106. doi: 10.1021/bi00038a002. [DOI] [PubMed] [Google Scholar]
- 39.Klostermeier D, Seidel R, Reinstein J. Functional properties of the molecular chaperone DnaK from Thermus thermophilus. J Mol Biol. 1998;279:841–853. doi: 10.1006/jmbi.1998.1816. [DOI] [PubMed] [Google Scholar]
- 40.Bhattacharya A, Kurochkin AV, Yip GNB, Zhang Y, Zuiderweg ERP. Allostery in the Hsp70 chaperones is transduced by subdomain rotations. J Mol Biol. 2009;388:475–490. doi: 10.1016/j.jmb.2009.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. Uniform and residue-specific 15N-labeling of proteins on a highly deuterated background. J Biomol NMR. 2004;29:289–297. doi: 10.1023/B:JNMR.0000032523.00554.38. [DOI] [PubMed] [Google Scholar]
- 42.Zhu G, Xia Y, Nicholson LK, Sze KH. Protein dynamics measurements by TROSY-based NMR experiments. J Magn Reson. 2000;143:423–426. doi: 10.1006/jmre.2000.2022. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Zuiderweg ER. The 70-kDa heat shock protein chaperone nucleotide-binding domain in solution unveiled as a molecular machine that can reorient its functional subdomains. Proc Natl Acad Sci USA. 2004;101:10272–10277. doi: 10.1073/pnas.0401313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang D, Tolman JR, Goto NT, Kay LE. An HNCO-based pulse scheme for the measurement of 13C[agr]-1H[agr] one-bond dipolar couplings in 15N,13C-labeled proteins. J Biomol NMR. 1998;12:325–332. doi: 10.1023/A:1008223017233. [DOI] [PubMed] [Google Scholar]
- 45.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; 2000. [Google Scholar]
- 47.Valafar H, Prestegard JH. REDCAT: A residual dipolar coupling analysis tool. J Magn Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 48.Gassler CS, et al. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc Natl Acad Sci USA. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogel M, Bukau B, Mayer MP. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol Cell. 2006;21:359–367. doi: 10.1016/j.molcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.