Abstract
Orthogonal, parallel and independent, systems are one key foundation for synthetic biology. The synthesis of orthogonal systems that are uncoupled from evolutionary constraints, and selectively abstracted from cellular regulation, is an emerging approach to making biology more amenable to engineering. Here, we combine orthogonal transcription by T7 RNA polymerase and translation by orthogonal ribosomes (O-ribosomes), creating an orthogonal gene expression pathway in Escherichia coli. We design and implement compact, orthogonal gene expression networks. In particular we focus on creating transcription–translation feed-forward loops (FFLs). The transcription–translation FFLs reported cannot be created by using the cells' gene expression machinery and introduce information-processing delays on the order of hours into gene expression. We refactor the rRNA operon, uncoupling the synthesis of the orthogonal 16S rRNA for the O-ribosome from the synthesis and processing of the rest of the rRNA operon, thereby defining a minimal module that can be added to the cell for O-ribosome production. The minimal O-ribosome permits the rational alteration of the delay in an orthogonal gene expression FFL. Overall this work demonstrates that system-level dynamic properties are amenable to rational manipulation and design in orthogonal systems. In the future this system may be further evolved and tuned to provide a spectrum of tailored dynamics in gene expression and investigate the effects of delays in cellular decision-making processes.
Keywords: directed evolution, orthogonal ribosome, protein engineering, synthetic biology
Synthetic biology aims to embed engineered systems into cells to perform new and potentially useful functions (1–10). A central challenge in synthetic biology is to engineer and embed synthetic systems in cells that take full advantage of the host cell's abilities, but are not limited by the host cells' regulatory networks or evolutionary history. Nowhere is this challenge more acute than in the fundamental process of gene expression, in which genetic information is copied and decoded to produce the networks of molecules that mediate biological function.
Selective abstraction of gene expression in synthetic networks from cellular gene expression and its associated regulatory processes would release biology for more effective engineering. The construction of orthogonal gene expression systems, in which the operation of converting the information in DNA into proteins is executed entirely by components that are functionally insulated from the endogenous gene expression machinery, would provide a compact solution to the selective abstraction of gene expression. Gene expression in bacteria relies on the coupled transcription of a DNA template by a DNA-dependent RNA polymerase (RNAP) and translation of the transcript by ribosomes. We realized that orthogonal gene expression might be achieved by the discovery, invention, and integration of components for orthogonal transcription and translation.
We previously described the evolution and characterization of orthogonal ribosome (O-ribosome)–orthogonal mRNA (O-mRNA) pairs in Escherichia coli (11). In these pairs the O-ribosome efficiently and specifically translates its cognate O-mRNA, which is not a substrate for the endogenous ribosome. The specificity of O-ribosomes for the translation of an O-mRNA arises from the altered 16S rRNA in the O-ribosome that recognizes an altered Shine-Dalgarno sequence in the leader sequence of the O-mRNA in the initiation phase of translation (12). In previous work O-ribosomes have been evolved that decode genetic information in orthogonal mRNAs in new ways. In combination with orthogonal aminoacyl-tRNA synthetases and tRNAs that recognize unnatural amino acids the evolved O-ribosomes have allowed us to begin to undo the “frozen accident” of the natural genetic code and direct the efficient incorporation of unnatural amino acids into proteins encoded on O-mRNAs (13, 14). O-ribosomes have also been used to create new translational Boolean logic functions that would not be possible to create by using the essential cellular ribosome (15) and to define functionally important nucleotides in the structurally-defined interface between the 2 subunits of the ribosome (16).
T7 RNAP is a small (99 kDa) DNA-dependent RNAP derived from bacteriophage T7 (17–19). The polymerase efficiently and specifically transcribes genes bearing a T7 promoter (PT7). In the absence of T7 RNAP the promoter does not direct transcription by endogenous polymerases in E. coli (20). T7 RNAP and its cognate promoters are therefore a natural orthogonal polymerase–promoter pair for transcription in E. coli.
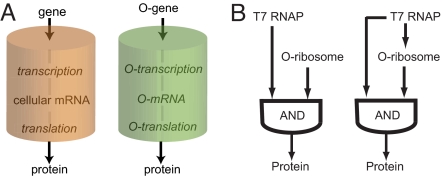
We realized that it might be possible to direct the transcription and translation of a gene by using a T7 promoter and an O-ribosome binding site (O-rbs) to create an orthogonal transcription translation system. The resulting gene would be transcribed only to its corresponding mRNA in the presence of T7 RNAP and would be translated only to its encoded protein product in the presence of the O-ribosome, creating an orthogonal gene expression pathway in the cell (Fig. 1A) that relies on AND logic (Fig. 1B). Because T7 RNAP and the O-ribosome, unlike the cell's endogenous RNAPs and ribosome, are not responsible for the synthesis of the cell's proteome from its genome, the orthogonal gene expression pathway opens the possibility of inventing and exploring new modes of gene regulation.
Fig. 1.
An orthogonal gene expression network. (A) Orthogonal gene expression is insulated from cellular gene expression. (B) The orthogonal AND function (Left) and an orthogonal transcription–translation FFL (Right).
Studies on transcriptional gene regulatory networks and other information-processing networks, including the worldwide web, electronic circuits, and the neuronal network of Caenorhabditis elegans, indicate that the type 1 coherent feed-forward loop (FFL) is an important module in these networks (21). The FFL consists of 3 components (X, Y, and Z) in which X directly activates Y, and both X and Y are required to activate Z (21, 22). Given the importance of transcriptional FFLs in controlling the timing of gene expression, an important property in almost all aspects of natural and synthetic biology from cell cycle control, developmental control, and circadian control to synthetic dynamic circuits, switches and oscillators (7), we became interested in synthesizing and characterizing orthogonal transcription–translation FFLs. We define an orthogonal transcription–translation FFL (Fig. 1B), as a network in which an orthogonal RNAP (T7 RNAP) transcribes the orthogonal rRNA necessary for the production of O-ribosomes and transcribes a mRNA bearing an O-rbs. The O-ribosome then translates the orthogonal message to produce the output protein. Unlike natural transcriptional FFLs, in which 2 transcription factors act to produce a single transcript (22), the orthogonal transcription–translation FFL is activated at sequential, but coupled steps in gene expression, leading to a short cascade (Figs. 2A and 3A).
Fig. 2.
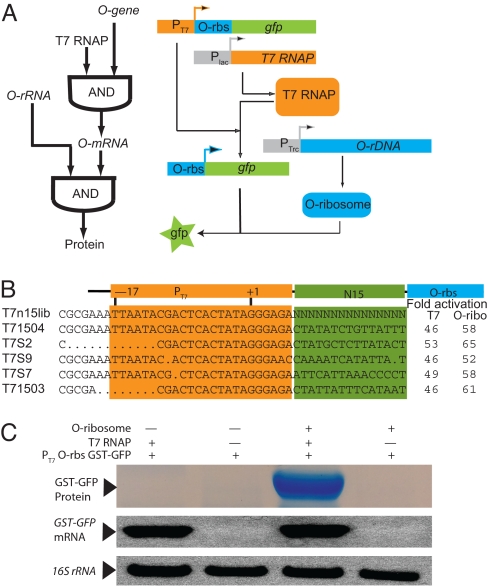
The construction of an orthogonal AND function. (A) The logic of orthogonal gene expression (Left). A molecular strategy (Right) for the realization of orthogonal gene expression. Plac, lac promoter; T7 RNAP, the gene encoding T7 RNAP; PT7, T7 promoter; gfp, gene encoding the GFP; Ptrc, a hybrid of the tryptophan and lactose promoters; O-rDNA, encodes an orthogonal ribosomal RNA. (B) Sequences selected after 3 rounds of FACS sorting for O-ribosome-dependent translation of a T7 transcript. T7S2 was represented 3 times and T71503 was represented twice among the best clones for GFP expression. (C) Orthogonal transcription–translation shows AND logic. The expression construct pT7 O-rbs GST-GFP contains the regulatory sequences from T71504 (in B) upstream of a genetic fusion between GST and sfGFP. (Top) Expressed and purified GST-GFP. (Middle) A Northern blot to detect sfGFP mRNA. (Bottom) A loading control for total RNA.
Fig. 3.
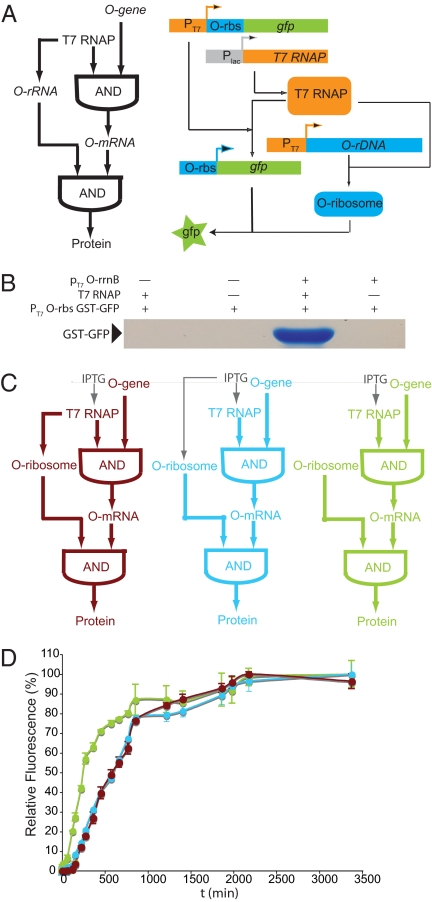
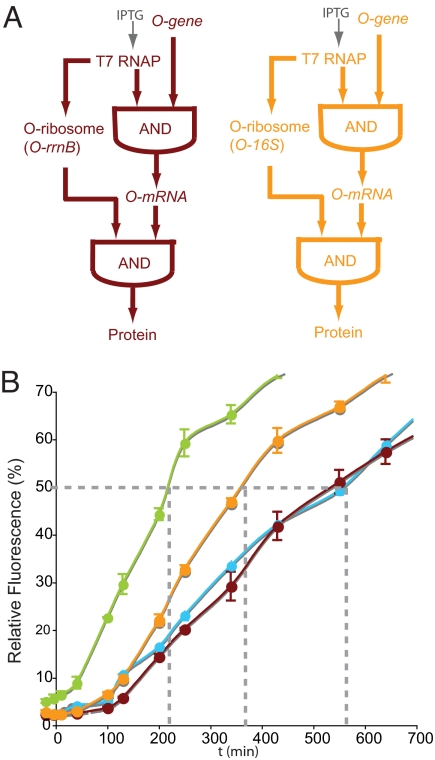
Synthesis and characterization of an orthogonal transcription translation FFL. (A) (Left) Logic of the orthogonal transcription–translation FFL. (Right) A molecular strategy for the realization of an orthogonal transcription–translation FFL. Labels are as in Fig. 2. (B) The orthogonal transcription–translation FFL displays AND logic. pT7 O-rrnB is the rrnB operon that produces O-16S rRNA, 23S rRNA, and 5S rRNA. Cells were harvested 5 h after the addition of 1 mM IPTG. (C) The orthogonal genetic circuits constructed and examined for altered dynamics. Maroon, FFL; blue, simple AND; green, AND with constitutive O-rRNA production. (D) O-ribosome production causes an identical delay in the kinetics of orthogonal gene expression in the simple AND function and in the FFL. Percentage of maximal sfGFP expression produced from pT7 O-rbs GST-GFP as a function of time >60 h after the addition of 1 mM IPTG. Green solid circles, BL21 (DE3) (produce T7 RNAP from an IPTG inducible promoter), pSC101* O-ribosome, pT7 O-rbs GST-GFP. Blue solid circles, BL21 (DE3) pTrc RSF O-ribosome (produces O-rRNA from an IPTG inducible Trc promoter), pT7 O-rbs GST-GFP. Maroon solid circles, BL21 (DE3), pT7 RSF O-ribosome, pT7 O-rbs GST-GFP. In this and subsequent figures the error bars represent the SD of at least 3 independent trials and the growth of cells containing all combinations of constructs was comparable as judged by following the OD600 (Figs. S4 and S5). Cells that lacked either T7 RNAP or the O-ribosome exhibited only backgorund fluorescence (Fig. S6).
Here, we describe the implementation of an orthogonal gene expression pathway in cells. We use orthogonal transcription and translation to create orthogonal gene expression networks, including transcription–translation FFLs and examine their dynamic properties. We demonstrate that the transcription–translation networks, which could not be created by using host polymerases or endogenous ribosomes, allow the introduction of distinct delays into gene expression that have not been demonstrated in natural systems. In the process of creating these networks we refactor (23) the rRNA operon (rrnB) to uncouple O-16S rRNA synthesis and processing from the synthesis and processing of the rest of the rrnB and define a minimal module for O-ribosome production in cells. The minimal O-ribosome allows us to rationally alter the delay in gene expression.
Results
Selection of a T7 Promoter O-rbs System.
To implement orthogonal gene expression we required a module that would respond specifically and efficiently to T7 RNAP and the O-ribosome (Fig. 2A). To create an orthogonal transcription–translation gene regulation module we first inserted the O-rbs and its immediate flanking sequence into a standard T7 expression vector (pET17b) in place of the wild-type ribosome binding site, 5′ to a GFP ORF (Fig. S1). When transformed into cells containing the O-ribosome, no GFP expression was observed above background levels, suggesting that this initial construct does not support gene expression. The insertion of additional 5′ sequence derived from the O-rbs vector (spacer 1) did not lead to O-ribosome-dependent expression of GFP, nor did deletion of the spacer 2 region in the T7 transcript derived from the pET vector. These experiments suggested that a linear combination of the pET leader sequence and the O-rbs sequence are not compatible with high-level GFP expression. Because both the T7 promoter and the O-rbs are active in several other contexts, and transcription and translation are physically-coupled processes in bacteria, it seemed reasonable that the sequence between the promoters and ribosome binding sites might be important for efficient gene expression. We therefore decided to combinatorially optimize the sequence between the T7 promoter and the O-rbs for T7 RNAP-dependent and O-ribosome-dependent gene expression.
To optimize the T7 O-rbs construct we first created a 109 member library in which a 15-nt stretch 3′ to the T7 promoter (20) and 5′ to the O-rbs is randomized to all possible combinations (Fig. 2B). The resulting library (T7n15GFP) was screened for O-ribosome-dependent expression from the T7 promoter by multiple rounds of FACS (for FACS data see Fig. S2). In a first round of positive FACS sorting we screened for expression of GFP in the presence of O-ribosomes and T7 RNAP. We transformed the T7n15GFP library into cells that constitutively produce O-ribosomes (by virtue of expressing O-rRNA from pSC101*O-ribosome) that express T7 RNAP. We collected fluorescent cells and isolated the pool of T7n15GFP variants. To remove T7n15GFP variants from the pool that direct expression of GFP by the wild-type ribosome, we performed a second round of negative FACS sorting. We transformed the pool of T7n15GFP library members from the positive FACS sort into cells that do not produce O-ribosomes, but do express T7 RNAP and wild-type ribosomes and collected cells that do not express GFP, and are therefore not fluorescent. We isolated the resulting T7n15GFP clones and performed a third round of positive FACS sorting. The fluorescence of 96 T7n15GFP clones surviving all 3 rounds of sorting was examined in cells containing T7 RNAP and the O-ribosome. The 8 clones exhibiting the greatest fluorescence were examined further. The expression of GFP from all 8 clones was strongly O-ribosome-dependent and T7 RNAP-dependent (Fig. 2B and Fig. S2B). Sequencing the T7n15GFP variants revealed 5 distinct sequences. In all of the selected sequences the randomized region was very rich in A and T. The selected sequences may minimize spurious RNA–RNA interactions with the O-rbs-containing sequence. However, because transcription and translation are coupled in bacteria, we cannot rule out more sophisticated explanations in which these sequences modulate coupling. Although all of the selected sequences retain the −9 to +1 sequence most important for transcriptional efficiency in vitro (24), 4 of these sequences contain deletions in the −11 to −17 region of the promoter (Fig. 2B) where most point mutations have a modest effect on the efficiency of the T7 promoter in vitro (24). Indeed Northern blots of the T7 RNAP-dependent GFP transcript demonstrate that comparable transcript accumulates with each of the selected T7N15lib sequences (Fig. S2C). A single selected sequence (T71504) had a wild-type T7 promoter and displayed excellent O-ribosome dependence, so we decided to characterize this sequence in more detail.
To begin to demonstrate the portability of the selected T7 promoter O-rbs combination and confirm that the system shows Boolean AND logic we replaced the GFP gene with a GST-GFP fusion (creating pT7 O-rbs GST-GFP). The GST-GFP fusion protein was produced only in the presence of both O-ribosomes and T7 RNAP, as demonstrated by both the level of GFP fluorescence and the purification of GST-GFP from cells that contain the O-ribosome and T7 RNAP, but not from cells containing any other combination of O-ribosomes and T7 RNAP. In addition, the GST-GFP mRNA was produced only in the presence of T7 RNAP (Fig. 2C). These experiments demonstrate that we have created a genetic element that is heritable in, but unreadable by, the host cell. This genetic element is efficiently transcribed and translated by the orthogonal polymerase and ribosome.
Synthesis of a Transcription–Translation FFL.
To construct a transcription–translation FFL (Fig. 3A) we required an O-16S rRNA that is transcribed from a T7 promoter and assembled into O-ribosomes. Because the mutations in the 3′ end of the O-16S sequence differentiate the translational initiation sites of natural and O-ribosomes, production of the O-ribosome requires the synthesis, processing, and incorporation of O-16S rRNA into 70S ribosomes (25, 26). With a single characterized exception (27–29), ribosomal RNA is produced in a single transcript (25). This transcript generally contains a 5′ leader sequence, the 16S rDNA, a spacer that may contain a tDNA, the 23S rDNA, and the 5S rDNA with or without additional tDNAs. The primary transcript is cleaved by a number of ribonucleases, some of which (most notably RNase III) can act cotranscriptionally, and is processed and assembled into ribosomes (26). Processing enzymes are known to be required to different extents for processing different parts of the transcript. For example, RNase III is required for correct end processing of 23S rRNA, but it is dispensable for correct end processing of 16S rRNA (26). To test whether a version of rrnB-producing O-16S rRNA can produce O-ribosomes for the synthesis of an orthogonal transcription–translation FFL we cloned rrnB containing the O-16S sequence onto a T7 promoter in an RSF vector (creating pT7 RSF O-ribosome). We followed the production of GST-GFP in cells containing T7 RNAP, pT7 O-rbs GST-GFP, and pT7 RSF O-ribosome over 60 h and the production of GST-GFP in control cells that did not contain T7 RNAP or pT7 RSF O-ribosome.
We found that the orthogonal transcription–translation FFL leads to the expression of GST-GFP that strictly depends on both T7 RNAP and pT7 RSF O-ribosome. In the presence of T7 RNAP, pT7 RSF O-ribosome, and pT7 O-rbs GST-GFP, GST-GFP was purified in good yield. However, in the absence of T7 RNAP or pT7 RSF O-ribosome no GST-GFP was purified (Fig. 3B). The GFP fluorescence of cells confirms the accumulation of GST-GFP is pT7 RSF O-ribosome and T7 RNAP-dependent (Figs. S3–S6). These data demonstrate that we have synthesized an orthogonal transcription–translation FFL that displays AND logic.
One information-processing feature of certain transcriptional FFLs is their capacity to mediate delays in response to input signals (22). To investigate whether orthogonal transcription–translation FFLs mediate delays in orthogonal gene expression we compared the kinetics of gene expression for cells containing T7 RNAP, pT7 O-rbs GST-GFP, and pT7 RSF O-ribosome with cells in which only the production of T7 RNAP is inducible [pT7 RSF O-ribosome is replaced with a plasmid that constitutively produces O-rrnB (pSC101* O-ribosome)] or cells in which both O-rrnB and T7 RNAP are produced from inducible, T7 RNAP-independent promoters (pT7 RSF O-ribosome is replaced with pTrc RSF O-ribosome, which produces O-rrnB from a Trc promoter using the host RNAP) (Fig. 3C).
We found that orthogonal gene expression is fastest for cells that constitutively produce O-ribosomes (time taken to reach half-maximal expression, t1/2 = 220 min) (Fig. 3D). These cells are poised to translate the O-rbs GST-GFP transcript, and the accumulation of GST-GFP protein is therefore only limited by production of T7 RNAP and its transcription of pT7 O-rbsGST-GFP. Cells containing pTrc RSF O-ribosome show a long delay in gene expression (t1/2 = 580 min, delay = 360 min) relative to cells that constitutively produce O-ribosomes. Upon induction of these cells, O-rrnB must be transcribed and processed, and the resulting O-rRNA must be assembled into O-ribosomes. These steps account for the delay observed. Because the Trc promoter is not as strong as the P1P2 promoter on constitutively-produced O-rRNA the maximal expression of the O-GST-GFP is ≈50% of that realized when O-rRNA is constitutively produced on the P1P2 promoter (Fig. S3).
Cells containing pT7 RSF O-ribosome also show a delay in gene expression of 360 min relative to cells that constitutively produce O-ribosomes (t1/2 = 580 min, delay = 360 min). The orthogonal transcription translation FFL and the simple inducible AND system both show the same t1/2, and the same long delay relative to the system in which only transcription is inducible. This finding indicates that, in contrast to previously-characterized transcription factor FFLs that introduce distinct delays with respect to the corresponding simple AND function (22), the rate of gene expression in the orthogonal transcription translation system is independent of whether the O-ribosome is in series (FFL) or parallel (AND) with T7 RNAP, although we cannot rule out that there are smaller differences in the timing of gene expression at early time points after induction. We envisioned 2 limiting scenarios that might lead to the identical kinetics of the simple inducible AND system and the FFL: (i) the production of O-ribosomes from O-rrnB may be much slower than any other step in GST-GFP production, leading to GST-GFP expression kinetics that are insensitive to whether O-ribosome production is in series (FFL) or in parallel (simple AND) with T7 RNAP; and (ii) the transcription of O-rRNA is rate-determining for O-ribosome formation and using the faster T7 RNAP for transcription of O-rrnB cancels out the delay introduced by requiring T7 RNAP synthesis and accumulation before O-rrnB synthesis.
The final expression level with the orthogonal transcription translation FFL is twice that of the system in which O-rRNA is produced from a P1P2 promoter on a low copy plasmid (Fig. S3).
Engineering the FFL Delay via the Discovery of a Minimal O-rRNA.
We realized that because rRNA is produced on a long primary transcript (25) (Fig. 4A), but O-ribosomes only require the production of O-16S rRNA, it might be possible to minimize the transcript. A correctly-processed O-16S rRNA transcript would assemble into functional ribosomes with genomically-encoded 23S rRNA, 5S rRNA, and ribosomal proteins. A minimal O-16S transcript would be shorter and therefore be transcribed more quickly than the full-length O-rrnB transcript. Moreover, a minimal O-16S transcript would decrease the number of processing steps required to produce O-16S rRNA for O-ribosomes and would remove the requirement for processing steps in other parts of the operon. In particular 23S processing steps might be limiting for the release of O-16S from O-rrnB transcribed by T7 RNAP, because it has been reported that 23S rRNA produced by T7 RNAP [which is 5 times faster than host polymerases (17, 18)] is not efficiently cotranscriptionally processed and is incorporated into nonfunctional 50S subunits (30). If either transcription or processing of O-16S RNA from pT7 RSF O-ribosome is rate limiting, then a minimal O-16S transcript might decrease the observed delay in the orthogonal transcription–translation FFL.
Fig. 4.
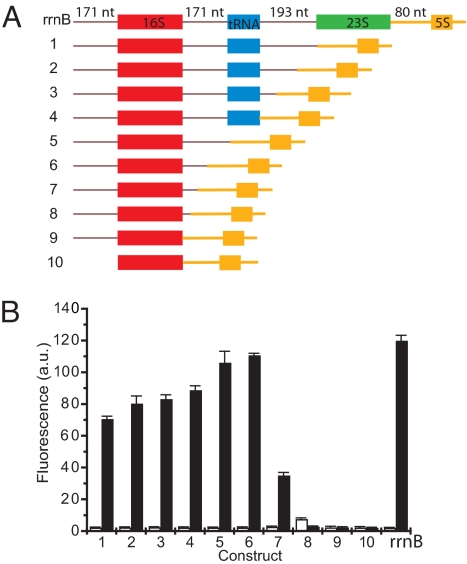
Creation of a minimal O-ribosome. (A) Schematic of the truncations examined for the production of active O-ribosomes. (B) The activity of O-ribosomes produced from each truncation construct (constructs 1–10, filled bars) compared with the full-length operon (O-rrnB). Fluorescence was measured in cells containing pXR1 (a tetracycline-resistant p15A plasmid that directs GFP expression from a constitutive promoter and O-rbs). The empty bars show the expression of GFP produced when pXR1 is combined with wild-type ribosomes in the absence of O-ribosomes.
To create a minimal O-16S expression construct we prepared a series of deletion mutants (Fig. 4A and Fig. S7) in pTrc O-ribosome [a version of rrnB that is transcribed from the IPTG-inducible pTrc promoter and contains the O-16S sequence in the rrnB operon (11)]. We assayed the function of these deletion mutants by their ability to form O-ribosomes and produce GFP from a gene with a constitutive promoter and an O-rbs (pR22).
Deletion of the 23S rRNA from pTrc O-ribosome led to a decrease in GFP fluorescence to half that of the full-length operon. However, further deletion of the spacer and tRNA led to rescue of the GFP fluorescence to levels close to that observed for the full-length operon. The maximally active truncated operons (Fig. 4B, constructs 5 and 6) contain the 5′ leader sequence of 16S rRNA and the region of the spacer immediately 3′ to 16S rRNA that is believed to form a base-paired helix with the 5′ leader sequence in the primary transcript (Fig. S7) (25). This helix contains RNase III sites that are cleaved to release a 16S rRNA-containing fragment from the primary transcript in rrnB. Deletion of sequences close to these RNase III sites leads to a loss of functional O-ribosome production, as judged by the drastic decrease in GFP signal (Fig. 4B, constructs 7–10). These experiments refactor the O-rrnB operon and define a minimal O-16S expression construct (Fig. 4, construct 6). The minimal O-16S expression construct reduces the transcript required to produce O-ribosomes to 50% of its original length, from 5,486 to 2,247 nt. The minimal O-ribosome construct will take less time to transcribe than rrnB, moreover it uncouples O-16S rRNA synthesis from 23S rRNA synthesis and tRNA synthesis, and therefore uncouples O-16S rRNA processing from processing steps that may limit the production of O-16S rRNA from O-rrnB. The decreased transcription time and the potentially decreased processing time could act together to decrease the time required to produce a functional O-ribosome.
To investigate whether the minimal O-16S leads to altered gene expression kinetics we assembled an orthogonal transcription translation FFL in which the minimal O-16S sequence was transcribed from a T7 promoter in the presence of T7 RNAP and assembled into functional O-ribosomes (Fig. 5A). T7 RNAP transcribes pT7 O-rbs GST-GFP, and the resulting mRNA is translated by the O-ribosome. Expression of GST-GFP strictly depends on T7 polymerase and the plasmid encoding O-16S rRNA from the T7 promoter (pT7 RSF O-16S), as judged by GFP fluorescence (Fig. S6). These data confirm the construction of the FFL depends on both inputs.
Fig. 5.
The FFLs with the minimal O-ribosome and the progenitor O-ribosome have identical topologies but mediate distinct delays. (A) The FFL using the progenitor O-ribosome (maroon) and the minimal O-ribosome (orange). (B) The delays in gene expression created by the orthogonal transcription translation networks. Orange solid circles, BL21 (DE3), pT7 RSF O-16S, pT7 O-rbs GST-GFP. The green blue and maroon curves are reproduced from Fig. 3 for comparison. The time taken to reach 50% of maximal expression, used to quantify the delay (22), is indicated.
We measured the kinetics of gene expression in the FFL by inducing the production of T7 RNAP with IPTG and after the increase in fluorescence as a function of time (Fig. 5B). Comparison of the FFL to a system in which O-ribosomes are constitutively produced and that is simply regulated by transcription demonstrates that the orthogonal transcription–translation FFL introduces a delay of 170 min relative to the simple transcription regulation case (t1/2 = 390 min, delay = 170 min). This delay is approximately half the length of that observed with the full-length rrnB on a T7 promoter to produce O-16S rRNA, suggesting that processing outside the of 16S sequence or transcription of rRNA determines the kinetics of gene expression. These experiments demonstrate that the minimal O-ribosome construct alters the delay in gene expression.
Discussion
We have combined orthogonal transcription by T7 RNAP and orthogonal translation by O-ribosomes to create an orthogonal gene expression pathway in the cell. This pathway specifically directs the decoding of genetic information from an orthogonal gene that is heritable in the host, but is unreadable by the host. Orthogonal genes might form one basis for creating nontransmissible, safe synthetic genetic circuits.
Because both O-ribosomes and T7 RNAP are nonessential and directed specifically to orthogonal genes, the expression of orthogonal genes can be regulated and altered in ways not possible with the cells' essential polymerases and ribosome that have been evolutionarily trapped by the requirement to synthesize the proteome and ensure cell survival. We have demonstrated that the modular combination of an orthogonal RNAP and an O-ribosome allows the construction of compact transcription–translation networks with predictable properties. These networks, which could not be created by using host polymerases or endogenous ribosomes, allow the introduction of translational delays into gene expression that are not possible in natural systems. We have created 4 orthogonal gene expression networks with different expression kinetics that control the timing of gene expression in another way. The fastest orthogonal gene expression system (t1/2 = 3.5 h) uses constitutively-produced O-ribosomes and requires induction of T7 RNAP. The slowest systems (t1/2 = 10 h) require induction of full-length rrnB, either in series (FFL) or in parallel (simple AND) with T7 RNAP. In the process of creating these networks we refactored the rrnB to uncouple O-16S rRNA synthesis and processing from the synthesis and processing of the rest of the operon, and we defined a minimal module for O-ribosome production in cells. This minimal O-ribosome allowed the creation of a FFL with an intermediate delay (t1/2 = 6.5 h).
Overall, this work creates an orthogonal gene expression system that has the properties of a parallel operating system embedded in the cell. The orthogonal gene expression system is more flexible and amenable to abstraction and engineering than natural transcription and translation (that has the evolutionarily inherited burden of decoding the genome and synthesizing the proteome), yet the orthogonal system is bootstrapped to the natural system and takes advantage of the natural systems capabilities, including the cells' nonorthogonal components for genetic encoding, and replication.
It will be interesting to investigate the properties of other compact orthogonal gene expression circuits, including orthogonal transcription–translation FFLs in which an O-ribosome, rather than an orthogonal polymerase, is the master regulator. Moreover, using mutually O-ribosomes (11, 31), other translational control strategies (32), and mutually orthogonal polymerases, novel sigma factors, and other transcriptional control strategies it will be possible to assemble combinatorially more complex systems, allowing us to simultaneously boot up multiple, mutually orthogonal parallel operating systems within the cell. It may be possible to regulate the timing of host gene expression by using the orthogonal systems to investigate the effects of altering timing on the phenotypic or cellular decision outcome of signaling. Finally, because the orthogonal gene expression system is composed of nonessential components it may be possible to use genetic selections to discover systems that display tailored delays in gene expression or other interesting and useful dynamic properties.
Materials and Methods
A full experimental section, including a more detailed version of each of the following sections, can be found in SI Text. Primers used are in Table S1.
Characterization of Orthogonal Gene Expression Kinetics.
We used BL21 (DE3), which produces T7 RNAP on addition of IPTG, or BL21-TIR, which does not produce T7 RNAP. Cells contained pT7 O-rbs GST-GFP with pRSF O-ribosome, pSC101* O-ribosome, pT7 RSF O-ribosome, or pT7 RSF O-16S, as described in SI Text. To determine the effect of the O-ribosome on gene expression the experiments were carried out by using the wild-type ribosome equivalent of the O-rRNA vectors described above. Each experiment was repeated for at least 3 independent cultures, and error bars in the figures represent the standard deviation.
Selection of an Optimized T7O-rbs System.
An optimized T7 promoter/O-rbs system was selected by 3 rounds of FACS screening (positive, negative, and positive). In the positive rounds of screening BL21 (DE3) (Novagen) containing pSC101*O-ribosome were transformed with pT7n15lib. A total of 3.7 × 107 cells were sorted and 3 × 106 cells were collected (region R1 in Fig. S2). Total plasmid DNA was isolated from cells, and pT7n15lib DNA was separated from pSC101*O-ribosome DNA by 1% agarose gel electrophoresis. The pT7n15lib DNA was extracted from the gel for use in the next round of screening or characterization of individual clones.
For negative FACS sorting pT7n15lib DNA surviving the positive sort was transformed into BL21 (DE3) containing pSC101*BD (this vector produces rrnB from the native ribosomal P1P2 promoter). In the negative FACS sort 108 cells were sorted and 88.5% of the cells were collected, because they had a level of fluorescence comparable to negative controls. The collected negative clones were amplified and their pT7n15lib DNA was resolved and extracted, as described above for positive sort clones. In the final positive sort 108 cells were sorted and 6 × 103 cells with strong fluorescence were collected.
Characterizing pT7 O-rbs GFP Expression Constructs.
Individual pT7n15lib clones were transformed into BL21 (DE3) containing either the wild-type (pTrc RSF wt ribosome or pSC101*BD) or the O-ribosome (pTrc RSF-O-rDNA or pSC101*O-ribosome). Fluorescence was quantified with a fluorescent plate reader, and the GFP values were normalized by OD600 values.
To demonstrate that pT7 O-rbs-GST-GFP displays Boolean AND logic we transformed BL21 (T1R) (Sigma/Aldrich) and BL21 (DE3) with pT7 Orbs-GST-GFP and either pSC101*O-ribosome or pSC101*BD. We expressed and purified the resulting GST-GFP protein and examined the protein made by SDS/PAGE. We extracted total RNA and examined the GST-GFP transcription by Northern blot analysis.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900267106/DCSupplemental.
References
- 1.Chin JW. Modular approaches to expanding the functions of living matter. Nat Chem Biol. 2006;2:304–311. doi: 10.1038/nchembio789. [DOI] [PubMed] [Google Scholar]
- 2.Sprinzak D, Elowitz MB. Reconstruction of genetic circuits. Nature. 2005;438:443–448. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- 3.de Lorenzo V, Danchin A. Synthetic biology: Discovering new worlds and new words. EMBO Rep. 2008;9:822–827. doi: 10.1038/embor.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 5.Kohanski MA, Collins JJ. Rewiring bacteria, two components at a time. Cell. 2008;133:947–948. doi: 10.1016/j.cell.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 6.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 7.Kaern M, Blake WJ, Collins JJ. The engineering of gene regulatory networks. Annu Rev Biomed Eng. 2003;5:179–206. doi: 10.1146/annurev.bioeng.5.040202.121553. [DOI] [PubMed] [Google Scholar]
- 8.Serrano L. Synthetic biology: Promises and challenges. Mol Syst Biol. 2007;3:158. doi: 10.1038/msb4100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasty J, Isaacs F, Dolnik M, McMillen D, Collins JJ. Designer gene networks: Toward fundamental cellular control. Chaos. 2001;11:207–220. doi: 10.1063/1.1345702. [DOI] [PubMed] [Google Scholar]
- 10.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 11.Rackham O, Chin JW. A network of orthogonal ribosome x mRNA pairs. Nat Chem Biol. 2005;1:159–166. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- 12.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 13.Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 14.Xie J, Schultz PG. A chemical toolkit for proteins: An expanded genetic code. Nat Rev Mol Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 15.Rackham O, Chin JW. Cellular logic with orthogonal ribosomes. J Am Chem Soc. 2005;127:17584–17585. doi: 10.1021/ja055338d. [DOI] [PubMed] [Google Scholar]
- 16.Rackham O, Wang K, Chin JW. Functional epitopes at the ribosome subunit interface. Nat Chem Biol. 2006;2:254–258. doi: 10.1038/nchembio783. [DOI] [PubMed] [Google Scholar]
- 17.Chamberlin M, Ring J. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J Biol Chem. 1973;248:2235–2244. [PubMed] [Google Scholar]
- 18.Golomb M, Chamberlin M. Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J Biol Chem. 1974;249:2858–2863. [PubMed] [Google Scholar]
- 19.Steitz TA. The structural basis of the transition from initiation to elongation phases of transcription, as well as translocation and strand separation, by T7 RNA polymerase. Curr Opin Struct Biol. 2004;14:4–9. doi: 10.1016/j.sbi.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Basu S, Maitra U. Specific binding of monomeric bacteriophage T3 and T7 RNA polymerases to their respective cognate promoters requires the initiating ribonucleoside triphosphate (GTP) J Mol Biol. 1986;190:425–437. doi: 10.1016/0022-2836(86)90013-6. [DOI] [PubMed] [Google Scholar]
- 21.Milo R, et al. Network motifs: Simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 22.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100025. 20050018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imburgio D, Rong M, Ma K, McAllister WT. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry. 2000;39:10419–10430. doi: 10.1021/bi000365w. [DOI] [PubMed] [Google Scholar]
- 25.Brosius J, Dull TJ, Sleeter DD, Noller HF. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava AK, Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- 27.Hartmann RK, Ulbrich N, Erdmann VA. An unusual rRNA operon constellation: In Thermus thermophilus HB8 the 23S/5S rRNA operon is a separate entity from the 16S rRNA operon. Biochimie. 1987;69:1097–1104. doi: 10.1016/0300-9084(87)90009-5. [DOI] [PubMed] [Google Scholar]
- 28.Hartmann RK, Ulbrich N, Erdmann VA. Sequences implicated in the processing of Thermus thermophilus HB8 23S rRNA. Nucleic Acids Res. 1987;15:7735–7747. doi: 10.1093/nar/15.19.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartmann RK, Toschka HY, Erdmann VA. Processing and termination of 23S rRNA-5S rRNA-tRNA(Gly) primary transcripts in Thermus thermophilus HB8. J Bacteriol. 1991;173:2681–2690. doi: 10.1128/jb.173.8.2681-2690.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewicki BT, Margus T, Remme J, Nierhaus KH. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J Mol Biol. 1993;231:581–593. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 31.Chubiz LM, Rao CV. Computational design of orthogonal ribosomes. Nucleic Acids Res. 2008;36:4038–4046. doi: 10.1093/nar/gkn354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate. Mol Syst Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.