Abstract
Alanyl-tRNA synthetase (AlaRS) specifically recognizes the major identity determinant, the G3:U70 base pair, in the acceptor stem of tRNAAla by both the tRNA-recognition and editing domains. In this study, we solved the crystal structures of 2 halves of Archaeoglobus fulgidus AlaRS: AlaRS-ΔC, comprising the aminoacylation, tRNA-recognition, and editing domains, and AlaRS-C, comprising the dimerization domain. The aminoacylation/tRNA-recognition domains contain an insertion incompatible with the class-specific tRNA-binding mode. The editing domain is fixed tightly via hydrophobic interactions to the aminoacylation/tRNA-recognition domains, on the side opposite from that in threonyl-tRNA synthetase. A groove formed between the aminoacylation/tRNA-recognition domains and the editing domain appears to be an alternative tRNA-binding site, which might be used for the aminoacylation and/or editing reactions. Actually, the amino acid residues required for the G3:U70 recognition are mapped in this groove. The dimerization domain consists of helical and globular subdomains. The helical subdomain mediates dimerization by forming a helix–loop–helix zipper. The globular subdomain, which is important for the aminoacylation and editing activities, has a positively-charged face suitable for tRNA binding.
Keywords: crystal structure, dimerization domain, aminoacyl-tRNA synthetase, proofreading, wobble base pair
Aminoacyl-tRNA synthetases (aaRSs) catalyze the ligation of cognate amino acids and tRNAs, and thus establish the genetic code in protein biosynthesis. They are modular proteins composed of an aminoacylation domain and a few additional domains for discrete functions, such as tRNA binding, oligomerization, and amino acid proofreading (1, 2). The 20 aaRSs are divided into 2 classes, I and II, based on the 2 unrelated types of aminoacylation domains (3, 4). The aminoacylation reaction occurs at the catalytic site on the aminoacylation domain, and the reaction generally consists of 2 steps: the initial activation of the amino acid with ATP to generate the aminoacyl-adenylate, followed by the transfer of the aminoacyl moiety to the 3′ end of the tRNA. Although the aminoacylation is generally accurate, several aaRSs cannot completely avoid the misactivation of a noncognate amino acid, when it is similar to the cognate one. To solve this problem, these aaRSs use a proofreading mechanism, in which the incorrect products are hydrolyzed at the active site in the editing domain.
Alanyl-tRNA synthetase (AlaRS) is one of the class II aaRSs and consists of 4 domains: the N-terminal class II aminoacylation domain, the tRNA-recognition domain, the editing domain, and the C-terminal oligomerization (dimerization or tetramerization) domain (Fig. 1A) (1, 2). AlaRS occupies a special position in the history of aaRS research. Escherichia coli AlaRS was among the first aaRSs that were cloned, sequenced, and characterized genetically and biochemically (1, 5, 6). tRNAAla conserves a unique G3:U70 wobble base pair in the acceptor stem, and this base pair dictates the tRNA identity toward AlaRS (7, 8). This remarkable finding, that a small number of nucleotide residues serve as the predominant determinant for the tRNA identity, accelerated the search for the identity determinants of other aaRS–tRNA pairs. It was also striking that the predominant identity determinant of a tRNA exists in the acceptor–stem duplex, rather than the anticodon and the discriminator base (9, 10). Actually, AlaRS can aminoacylate small, isolated portions of tRNA, such as a “minihelix” and a “microhelix,” as long as they have the G3:U70 base pair (11). The G3:U70 base pair is considered to be recognized from the minor groove side (12, 13).
Fig. 1.
The structures of AlaRS-ΔC and AlaRS-C. (A) Domain organizations of A. fulgidus AlaRS, AlaRS-ΔC, and AlaRS-C. Shown are the aminoacylation domain (green), Mid1 (blue) and Mid2 (cyan) in the tRNA-recognition domain, the β-barrel (yellow) and editing-core (orange) subdomains in the editing domain, and the helical (midnight blue) and globular (light blue) subdomains in the dimerization domain. (B) A ribbon representation of AlaRS-ΔC. The model is colored as in A, and the N-terminal addition (AddA1) and insertions (InsA1 and InsA2) are highlighted in purple and brown, respectively. Motif 1 in the aminoacylation domain (violet), the helix–loop in the editing domain (salmon), and the linker connecting the tRNA-recognition and editing domains (red) are shown. Ala-SA in the aminoacylation site and the editing-site zinc ion are depicted as cpk models. (C) The AlaRS-C dimer is shown as a ribbon model. One molecule of the dimer is colored as in A, and the other molecule is colored gray.
An E. coli AlaRS fragment comprising the aminoacylation and tRNA-recognition domains (the N-terminal 461 residues) can specifically aminoacylate tRNAAla (1). The crystal structure of the corresponding fragment (AlaRS-N) from the bacterium Aquifex aeolicus was reported (14, 15). It revealed that AlaRS does not dimerize through the aminoacylation domain, in contrast to the other class II aaRSs. The structures of amino acid- and ATP-bound AlaRS-N revealed how the cognate alanine and the noncognate glycine and serine interact with the aminoacylation site. AlaRS is one of the aaRSs that use the proofreading mechanism, in that mischarged products, such as Gly-tRNAAla and Ser-tRNAAla, are transferred to the editing domain, where the ester bond is hydrolyzed (2). A defect in the AlaRS editing activity causes cell death in the mouse nervous system (16). It was recently reported that the E. coli AlaRS editing domain possesses a region, distinct from the N-terminal domains, that recognizes the G3:U70 base pair (17). Therefore, AlaRS may transfer the acceptor stem of tRNAAla from the first binding site in the aminoacylation domain to the second site in the editing domain, in contrast to the other editing aaRSs (classes I and II), which have been proposed to shuttle the flexible single-stranded CCA terminus of the tRNA between the aminoacylation and editing catalytic sites (18–22). The C-terminal domain of AlaRS is not only essential for the oligomerization, but also important for the aminoacylation and editing reactions (17, 23). Small proteins homologous to the AlaRS editing domain, designated AlaX, are found in many organisms (24, 25). They are active in the trans hydrolysis of misacylated tRNAAla in vitro (24). The crystal structures of AlaX-S (specific to Ser-tRNAAla) and AlaX-M (specific to Ser-tRNAAla and Gly-tRNAAla) from the archaeon Pyrococcus horikoshii have been reported (26, 27).
The structures of the editing and oligomerization domains, the basis of oligomerization, and the domain arrangement in the full-length AlaRS have remained elusive. We previously succeeded in the crystallization of 2 fragments of AlaRS from the archaeon Archaeoglobus fulgidus, AlaRS-ΔC, comprising the aminoacylation, tRNA-recognition, and editing domains, and AlaRS-C, comprising the dimerization domain (23). In the present study, we determined their crystal structures at 2.2- and 3.2-Å resolutions, respectively. The AlaRS-ΔC structure revealed a unique arrangement of the editing domain, relative to the aminoacylation/tRNA-recognition domains, and the archaea-specific insertions/deletions. The AlaRS-C structure provided the basis of dimerization, via the formation of a helix–loop–helix zipper (HLHZ). The structures suggested the domain organization in the full-length AlaRS dimer, and thus could serve as a platform for future analyses of how the aminoacylation/tRNA-recognition domains and the editing domain of AlaRS independently recognize the G3:U70 base pair of tRNAAla.
Results
Structure Determination.
A. fulgidus AlaRS is a homodimer of 906 amino acid residue polypeptides (23). It was genetically divided into 2 parts, AlaRS-ΔC (residues 1–739) and AlaRS-C (residues 736–906) (23), and both structures were solved (Table S1). AlaRS-ΔC is composed of the class-II specific aminoacylation domain, the tRNA-recognition domain, and the editing domain. The structure of AlaRS-ΔC complexed with an alanyl-adenylate analog, 5′-O-[N-(l-alanyl)sulfamoyl] adenosine (Ala-SA), was determined at 2.2 Å (Fig. 1B). The crystallographic asymmetric unit contained 1 AlaRS-ΔC molecule. The refined model has R and Rfree factors of 21.5% and 26.4%, respectively. AlaRS-C comprises the dimerization domain, and its structure was determined at 3.2 Å (Fig. 1C). Two AlaRS-C molecules form a homodimer, and there are 4 dimers in the asymmetric unit. The refinement converged to R and Rfree factors of 20.5% and 27.6%, respectively (Table S1).
The Aminoacylation Domain.
The A. fulgidus AlaRS-ΔC structure revealed the aminoacylation domain (residues 1–257), composed of a central antiparallel β-sheet (β1–β8 and β10) and 5 α-helices (α1–α5), which is typical of the class-II aaRSs. It is superposable on that of A. aeolicus AlaRS-N, with an rmsd of 1.9 Å for 179 Cα atoms. A. fulgidus AlaRS possesses an archaea-specific N-terminal extension (AddA1, residues 1–58), including α1, α2, β1, and β2 (Fig. 2 and Fig. S1A). β1 and β2 are integrated in the central β-sheet, and thus AddA1 is part of the aminoacylation domain. Although the bacterial and eukaryal AlaRSs lack AddA1, they instead possess 2 insertions (depicted as InsB/E1 and InsB/E2 in A. aeolicus AlaRS), which occupy the corresponding space. A. fulgidus AlaRS also contains an insertion (InsA1, residues 226–232) including β9 (Fig. 2 and Fig. S1A). Lys-229, at the tip of InsA1, seems to occupy the position of Lys-73 in the E. coli enzyme, which cross-links to tRNAAla (28).
Fig. 2.
The aminoacylation and tRNA-recognition domains. (A) The aminoacylation and tRNA-recognition domains of A. fulgidus AlaRS, colored as in Fig. 1B, are shown. (B) The A. aeolicus AlaRS-N structure, shown in the same orientation. The 2 regions missing in A. fulgidus (InsB/E1 and InsB/E2) are colored brown, and Mid2 is shown in gold.
A clear electron density corresponding to Ala-SA is visible in the active-site cleft (Fig. 1B and Fig. S2). The manner of interaction with Ala-SA in the aminoacylation active site is described in SI Text.
The tRNA-Recognition Domain.
The α-helix-rich middle domain of AlaRS-ΔC (residues 258–484), which is supposed to interact with the tRNA acceptor arm, is composed of 11 α-helices and a 2-stranded short parallel β sheet (Fig. 1B and Fig. S1A). This domain can be divided into 2 subdomains, designated here as Mid1 (residues 258–419) and Mid2 (residues 420–484). These subdomain structures in A. fulgidus AlaRS are similar to their counterparts in A. aeolicus AlaRS (14), as revealed by the rmsds of 2.4 and 2.3 Å, respectively. Mid1 tightly contacts the aminoacylation domain by hydrophobic interactions, whereas Mid2 protrudes from the rest of the protein body. The α-helix (α13) connecting Mid1 and Mid2 is continuous, whereas the corresponding helix is distorted in the middle in A. aeolicus AlaRS-N. Thus, Mid2 in A. fulgidus AlaRS-ΔC is oriented outward by ≈20°, compared with that of A. aeolicus AlaRS-N (Fig. S3). It is further tilted by ≈50°, because the last α-helix of Mid2 is connected to the editing domain by the linker. In the beginning of the subdomain, the Mid1, archaeal AlaRSs possess an insertion of ≈50 amino acid residues, which is missing in the bacterial AlaRSs. In the A. fulgidus AlaRS-ΔC structure, this insertion (InsA2, residues 277–330) assumes a helix–loop–helix structure (α8–α9) and is bent back to form part of the active-site cleft. The presence of this insertion seems to be incompatible with the tRNA interaction manner proposed previously for the bacterial AlaRS (14), as discussed below.
The Editing Domain.
The editing domain of A. fulgidus AlaRS (residues 501–737) consists of 2 subdomains, the N-terminal β-barrel subdomain (residues 501–588) and the C-terminal α/β subdomain (the editing core, residues 589–737), composed of 2 central α-helices (α17 and α18, residues 590–614 and 642–657, respectively) sandwiched by 3- and 4-stranded antiparallel β-sheets (Fig. 1B and Fig. S1B). The editing domain is superposable on P. horikoshii AlaX-M [Protein Data Bank (PDB) ID code 2E1B], with an rmsd of 1.5 Å for 202 Cα atoms. The α/β subdomain also superposes well on P. horikoshii AlaX-S (PDB ID code 1WXO) lacking the β-barrel subdomain, with an rmsd of 1.4 Å for 134 Cα atoms.
A cavity is formed at the subdomain interface. At the bottom, His-600, His-604, His-707, and Cys-703 coordinate a metal ion, which is supposed to be the editing active center (Fig. S4A). The tetrahedral coordination and the intense anomalous peak observed at the metal site suggested that the metal is a zinc, as in the case of the AlaX proteins. Thr-603, Gln-620, Gln-682, and Gln-701 constitute the cavity wall. His-600, Thr-603, His-604, Gln-620, His-707, and Cys-703 are conserved among the AlaRSs. Glu/Gln occupies the position corresponding to Gln-701. Gln-620, Gln-682, and Gln-701 correspond to Thr-30, Asp-92, and Asp-114, respectively, in P. horikoshii AlaX-S, which are involved in interactions with serine (Fig. S4B) (27). In AlaX-S, Thr-33 is also involved in the serine interaction, but AlaRS lacks the corresponding residue. The conserved Thr-603 in AlaRS could structurally compensate for the absence of this residue. The glycine-rich loop of the β-barrel subdomain resides at the entrance of the cavity and might interact with 3′ end of the tRNA.
Position of the Editing Domain.
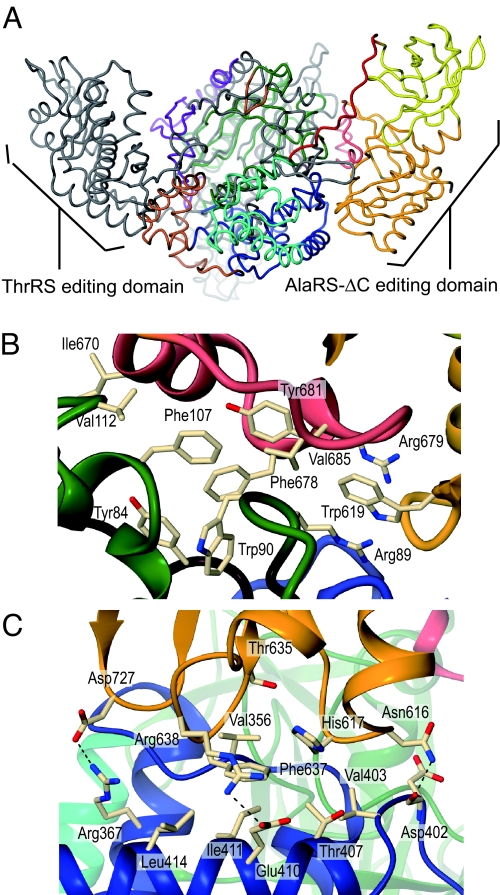
The editing domain of A. fulgidus AlaRS is connected to the last α-helix of the tRNA-recognition domain by a 38-Å-long loop, consisting of 16 amino acid residues (residues 485–500). The editing domain contacts the aminoacylation domain to form a hydrophobic core (Fig. 3B). Ile-670, Tyr-681, Phe-678, and Val-685, from a helix–loop structure (residues 669–689) in the editing domain, interact with Tyr-84 in motif 1 (residues 61–88), Trp-90, Phe-107, and Val-112 of the aminoacylation domain. Arg-89, Trp-619, and Arg-679 are stacked. The editing domain also contacts Mid1 of the tRNA-recognition domain (Fig. 3C). His-617, Thr-635, and Phe-637 in the editing domain and Val-356, Val-403, Thr-407, Ile-411, and Leu-414 in Mid1 form a hydrophobic core. Asn-616, Arg-638, and Asp-727 in the editing domain interact with Asp-402, Glu-410, and Arg-367, respectively, in Mid1.
Fig. 3.
The editing domain. (A) The position of the AlaRS editing domain. The structure of AlaRS-ΔC, depicted as a tube model, is superposed on that of ThrRS by the aminoacylation domain. The AlaRS-ΔC model is colored as in Fig. 1B, and ThrRS is colored gray. (B) The interface of the editing and aminoacylation domains. The helix–loop in the editing domain and motif 1 in the aminoacylation domain are colored salmon and brown, respectively. The residues involved in the interactions are shown as white stick models. (C) The interface of the editing and tRNA-recognition domains. The editing core subdomain and Mid1 are colored orange and blue, respectively.
It is remarkable that the position of the editing domain relative to the aminoacylation domain differs from those in other class II aaRSs with reported structures. For example, compared with E. coli threonyl-tRNA synthetase (ThrRS) (20), the editing domain resides on the opposite side of the aminoacylation domain in AlaRS-ΔC (Fig. 3A). The aminoacylation active site is ≈37 Å away from the editing active site in A. fulgidus AlaRS, which is comparable with the ≈39 Å distance in ThrRS (20, 29). We previously obtained a 3.7-Å dataset from an AlaRS-ΔC crystal belonging to a different space group (23). The structure was solved by molecular replacement, using the present AlaRS-ΔC structure. The position of the editing domain relative to the aminoacylation domain and their interface are the same.
In most cases, the dimerization of class II aaRSs is mediated through motif 1 (30, 31). Nevertheless, A. aeolicus AlaRS-N reportedly does not form a dimer, because the tRNA-recognition domain hinders dimerization through motif 1 (14). Consistent with this finding, A. fulgidus AlaRS-ΔC motif 1 does not mediate dimerization, but interacts with the helix–loop structure (residues 668–688) in the editing domain to form an interdomain interface. It is interesting to note that AlaX-M, which lacks the helix–loop structure, exists as a monomer in solution (26). In contrast, the helix–loop mediates the homodimerization of AlaX-S (27).
The C-Terminal Dimerization Domain.
A. fulgidus AlaRS forms a dimer through an interaction between the C-terminal dimerization domains of the 2 molecules (23). AlaRS-ΔC, lacking the dimerization domain, exists as a monomer in solution (23). We first determined the structure of the isolated dimerization domain of A. fulgidus AlaRS, AlaRS-C (Fig. 1C). The structure revealed that the dimerization domain consists of a long helical subdomain and a globular subdomain. The helical subdomain contains 2 α-helices of 32 and 53 Å and a linker in between. This subdomain exclusively mediates the dimer interaction to form a characteristic HLHZ (Fig. 4A and Fig. S5). Val-744, Met-747, Leu-750, and Leu-751 in α20, and Leu-765, Val-769, Phe-772, Phe-773, Trp-776, Gln-779, Ile-783, Leu-786, Val-789, Ile-790, Leu-793, and Ile-797 in α21, in 1 protomer of the dimer, respectively, form leucine-zipper-like interactions with their counterparts in the other monomer. The N-terminal portion of α21 (Pro-762, Leu-765, Pro-766, and Val-769) in 1 protomer also interacts with the C-terminal portion of α20 (Ala-754, Ile-757, and Leu-758). The linker mediates the formation of the hydrophobic core at the α20–α21 junction, where the coiled-coil is twisted (Fig. 4A). Similar HLHZ structures are also present in several transcriptional regulators, such as the Myc protooncogene product and its relatives (32).
Fig. 4.
The C-terminal dimerization domain. (A) The dimer interactions via the helical subdomains. An HLHZ formed in the dimer is shown in a stereoview. One molecule is colored midnight blue, and the side chains are shown as red stick models. The other molecule is colored gray. (B) The globular subdomain is shown in a ribbon representation. Basic amino acid residues forming the basic patch are shown as stick models. The conserved Gly-rich segment (870KGSGGGR876) is highlighted in red.
The C-terminal globular subdomain is composed of a 6-stranded antiparallel β-sheet and 3 α-helices (Fig. 4B and Fig. S1B). This subdomain tightly packs against α21 of the HLHZ to form a hydrophobic core. Trp-794, Leu-798, and Met-799, in α21, and Val-811, Val-815, Leu-829, Leu-838, and Phe-851, in the globular subdomain, are involved in the hydrophobic interactions. One surface of the globular subdomain is positively charged, by the contributions of Lys-855, Arg-859, Arg-863, Arg-867, Lys-870, Arg-876, and Lys-877. It is interesting that the conserved glycine-rich segment (870KGSGGGR876) forms a β-strand that is part of the β-sheet (Fig. 4B). A structural similarity search using the DALI server (33) revealed that the globular subdomain is similar to that of the ssDNA 5′-3′ exonuclease RecJ (34) and exopolyphosphatase (35), with high Z scores of 11.5 and 9.1, respectively.
Discussion
tRNA Interactions.
In the crystal structures of tRNA-bound class-II aaRSs, including ThrRS, seryl-tRNA synthetase, and aspartyl-tRNA synthetase, the amino acid acceptor arm of the tRNA binds to a common site on the class II aminoacylation domain (20, 36, 37). The binding site corresponds to the groove formed between Mid1 and Mid2 in the tRNA-recognition domain of A. fulgidus AlaRS. However, if the tRNA acceptor arm binds to the groove of A. fulgidus AlaRS via the common mode (mode 1), then the nucleotides at positions 1–5 and 68–73 have a serious steric clash with the archaea-specific insertion of a helix–loop–helix (InsA2) within the groove (Fig. 5). To avoid the putative clash, a drastic conformational change should occur. Because InsA2 interacts with α7 and α10 to form a hydrophobic core, the reorientation of InsA2 seems to be unlikely. When the tRNA relocates to the other tRNA-binding site for editing, it should dissociate to move over the protuberant Mid2 subdomain and then rebind. However, an alternative mode (mode 2) is that the tRNA acceptor stem binds to a groove formed between the Mid2 subdomain and the editing domain (“alternative groove”) of A. fulgidus AlaRS (Fig. 5). In the present structure, the linker connecting Mid2 and the editing domain is located in the alternative groove, but the linker appears to be quite flexible and to change its conformation upon tRNA binding. The alternative groove has entrances to both the aminoacylation and editing active sites, which would facilitate tRNA relocation between them. Consequently, the alternative mode 2 is more likely than the common mode 1 for A. fulgidus AlaRS.
Fig. 5.
Models of tRNA binding. A tRNA model was created by superposition of the aminoacylation domains of AlaRS-ΔC and the E. coli ThrRS·tRNAThr complex, and the acceptor-arm portion of the tRNA (residues 1–7 and 66–76) is shown as a dark-yellow transparent surface model (mode 1). The second tRNA model, bound to an alternative tRNA-binding site, is also depicted as a blue surface model (mode 2). The A76 residues in the first and second models are highlighted in yellow and blue, respectively. Amino acid residues important for the aminoacylation or editing activity (17, 28, 38, 39, 45, 46) are shown as stick models. Ala-SA in the aminoacylation site and the editing-site zinc ion are depicted as cpk models. A stereo version of this figure is presented as Fig. S6.
For the bacterial AlaRS from A. aeolicus, the tRNA was docked in mode 1 (14). Because A.aeolicus AlaRS lacks the archaea-specific insertion (InsA2), tRNA binding is not hindered. In addition, the acceptor stem could be proximal to Asp-398, corresponding to Ala-409 in E. coli AlaRS, which is thought to be indirectly involved in the G3·U70 interaction (38). The aminoacylation and tRNA-recognition domains of A.aeolicus AlaRS were successfully separated from the editing domain for crystallography (14), whereas the corresponding aminoacylation/tRNA-recognition fragment and the editing domain fragment of E. coli AlaRS were both prepared for functional studies (17, 38). In contrast, in the case of the archaeal AlaRS from A. fulgidus, it was difficult to prepare the corresponding fragments, probably because of the hydrophobic interaction between the aminoacylation/tRNA-recognition domains and the editing domain (Fig. 3). Therefore, the bacterial AlaRSs might have the editing domain in a different location from that in the archaeal AlaRS, relative to the aminoacylation/tRNA-recognition domains, thus allowing the tRNA to shift easily between the 2 active sites. In E. coli AlaRS, the aminoacylation/tRNA-recognition domains and the editing domain are both able to recognize the G3·U70 base pair in the tRNA acceptor stem (17), involving Arg-314 on the tRNA-recognition domain and Arg-693 on the editing domain of E. coli AlaRS (17, 39). Intriguingly, in the present A. fulgidus AlaRS structure, Arg-371 (Mid1) and Arg-731, which correspond to the G3·U70 recognition residues Arg-314 and Arg-693, respectively, are close to the putative tRNA binding site in the alternative groove (Fig. 5, mode 2). The tRNA-recognition domain including Arg-371 resides on the minor groove side of the acceptor stem, and this binding mode (mode 2) is more preferable for the minor groove recognition of the G3·U70 base pair than mode 1 (12, 13). Therefore, we cannot exclude the possibility that the bacterial AlaRSs have a similar domain arrangement to that of A. fulgidus AlaRS, and bind tRNA via mode 2. However, the editing domain could interact with the G3·U70 base pair in the major groove (Fig. 5). Arg-371 and Arg-731 are too far from each other to simultaneously interact with the G3·U70 base pair. Our model in mode 2 is compatible with the fact that E. coli AlaRS aminoacylates the 3′-OH of A76 (40).
The C-terminal dimerization domain of A. fulgidus AlaRS is also crucial for the tRNA interaction, because the deletion of the domain dramatically reduces the aminoacylation activity (23). The A. fulgidus AlaRS-C structure reveals that the globular subdomain of the dimerization domain possesses a positively-charged face, including the conserved Gly-rich segment (Fig. 4B). The globular subdomain, therefore, is a candidate for the tRNA-binding site, to support aminoacylation reactions. For E. coli AlaRS, the region of residues 808–875 is a nonspecific tRNA-binding site (17), which includes the Gly-rich segment.
The Full-Length AlaRS Structure.
The present structures of AlaRS-ΔC and AlaRS-C provide clues to infer the full-length AlaRS structure. The distance between the N termini in the AlaRS-C dimer is only 14 Å, which could restrict the positions of the other domains. Because the editing domain C terminus is connected to the dimerization domain N terminus, the 2 editing domains in the dimer should be close to each other, whereas the aminoacylation and tRNA-recognition domains would be distant from the 2-fold axis. In the AlaRS-ΔC crystal structure, the editing domain interacts back-to-back with that of the symmetry-related molecule correlated by the crystallographic 2-fold axis. Met-650, Ile-656, Leu-657, and Met-716 mediate the interaction, and the buried surface area is ≈400 Å2. In the crystal lattice, the distance between the editing-domain C-termini is ≈19 Å, which allows their connection to the dimerization-domain N termini without a large conformation change. Overall, the full-length AlaRS dimer is likely to assume a butterfly-like structure (Fig. 6). Although this model still requires validation, it could serve as a platform for future analyses.
Fig. 6.
A model of the full-length AlaRS dimer. Two copies of AlaRS-ΔC, which are correlated by the crystallographic 2-fold axis, and an AlaRS-C dimer, are shown. The N termini of AlaRS-C were placed near the C termini of AlaRS-ΔC. The model was colored as in Fig. 1.
Materials and Methods
Protein Preparation.
See SI Text.
Crystallization and Data Collection.
See SI Text.
Structure Determination and Refinement.
The structure of AlaRS-ΔC was solved by the single-wavelength anomalous dispersion method. The Se-site and initial phase determinations and solvent flattening were performed with the AutoSHARP program (41). All 15 of the Se sites were identified. Density modification and initial model building using the RESOLVE program placed 51% of the amino acid residues, and the remaining residues were built manually with the COOT program (42, 43). Structure refinement was carried out with the CNS program (44). A randomly-chosen 5% of the data were set aside for cross-validation. The refinement included several rounds of simulated-annealing, positional, and individual B factor refinements. The refinement converged to final R and Rfree factors of 21.5% and 26.4%, respectively (Table S1). In the Ramachandran plot, 87.4%, 11.9%, and 0.6% of the residues fell in the most favored, additional allowed, and generously allowed regions, respectively. No residues were in the disallowed region.
The structure of AlaRS-C was solved by the SAD method with the AutoSHARP program (41). Of the 56 Se sites, 48 were identified. Model building was performed manually by using the COOT program (42). Refinement was done with the CNS program, and the R and Rfree factors for the final model are 20.5% and 27.6%, respectively (Table S1). In the Ramachandran plot, 88.5%, 11.2% and 0.2% of the residues fell in the most favored, additional allowed, and generously allowed regions, respectively. No residues were in the disallowed region.
Supplementary Material
Acknowledgments.
We thank the staffs of the Photon Factory (Tsukuba, Japan) and SPring-8 BL41XU (Hyogo, Japan) beam lines for assistance with our data collection. This work was supported in part by a Ministry of Education, Culture, Sports, Science, and Technology Global Centers of Excellence Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms), a Ministry of Education, Culture, Sports, Science, and Technology Grant-in-Aid for Scientific Research, and the Ministry of Education, Culture, Sports, Science and Technology Targeted Proteins Research Program. R.F. was supported by Research Fellowships from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2ZTG and 2ZVF).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901572106/DCSupplemental.
References
- 1.Jasin M, Regan L, Schimmel P. Modular arrangement of functional domains along the sequence of an aminoacyl tRNA synthetase. Nature. 1983;306:441–447. doi: 10.1038/306441a0. [DOI] [PubMed] [Google Scholar]
- 2.Beebe K, Ribas De Pouplana L, Schimmel P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 2003;22:668–675. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cusack S. Eleven down and nine to go. Nat Struct Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 4.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 5.Jasin M, Regan L, Schimmel P. Dispensable pieces of an aminoacyl tRNA synthetase which activate the catalytic site. Cell. 1984;36:1089–1095. doi: 10.1016/0092-8674(84)90059-x. [DOI] [PubMed] [Google Scholar]
- 6.Putney SD, et al. Primary structure of a large aminoacyl-tRNA synthetase. Science. 1981;213:1497–1501. doi: 10.1126/science.7025207. [DOI] [PubMed] [Google Scholar]
- 7.Hou YM, Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 8.McClain WH, Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science. 1988;240:793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 9.Vasil'eva IA, Moor NA. Interaction of aminoacyl-tRNA synthetases with tRNA: General principles and distinguishing characteristics of the high-molecular-weight substrate recognition. Biochemistry (Moscow) 2007;72:247–263. doi: 10.1134/s0006297907030029. [DOI] [PubMed] [Google Scholar]
- 10.Beuning PJ, Musier-Forsyth K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 1999;52:1–28. doi: 10.1002/(SICI)1097-0282(1999)52:1<1::AID-BIP1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 11.Francklyn C, Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989;337:478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 12.Musier-Forsyth K, Schimmel P. Functional contacts of a transfer RNA synthetase with 2′-hydroxyl groups in the RNA minor groove. Nature. 1992;357:513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- 13.Musier-Forsyth K, et al. Specificity for aminoacylation of an RNA helix: An unpaired, exocyclic amino group in the minor groove. Science. 1991;253:784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- 14.Swairjo MA, et al. Alanyl-tRNA synthetase crystal structure and design for acceptor-stem recognition. Mol Cell. 2004;13:829–841. doi: 10.1016/s1097-2765(04)00126-1. [DOI] [PubMed] [Google Scholar]
- 15.Swairjo MA, Schimmel PR. Breaking sieve for steric exclusion of a noncognate amino acid from active site of a tRNA synthetase. Proc Natl Acad Sci USA. 2005;102:988–993. doi: 10.1073/pnas.0409024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 17.Beebe K, Mock M, Merriman E, Schimmel P. Distinct domains of tRNA synthetase recognize the same base pair. Nature. 2008;451:90–93. doi: 10.1038/nature06454. [DOI] [PubMed] [Google Scholar]
- 18.Fukai S, et al. Structural basis for double-sieve discrimination of l-valine from l-isoleucine and l-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 19.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat Struct Mol Biol. 2005;12:915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 20.Sankaranarayanan R, et al. The structure of threonyl-tRNA synthetase-tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 21.Silvian LF, Wang J, Steitz TA. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- 22.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat Struct Mol Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 23.Fukunaga R, Yokoyama S. Crystallization and preliminary X-ray crystallographic study of alanyl-tRNA synthetase from the archaeon Archaeoglobus fulgidus. Acta Crystallogr F. 2007;63:224–228. doi: 10.1107/S1744309107006264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahel I, Korencic D, Ibba M, Söll D. Trans-editing of mischarged tRNAs. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schimmel P, Ribas De Pouplana L. Footprints of aminoacyl-tRNA synthetases are everywhere. Trends Biochem Sci. 2000;25:207–209. doi: 10.1016/s0968-0004(00)01553-x. [DOI] [PubMed] [Google Scholar]
- 26.Fukunaga R, Yokoyama S. Structure of the AlaX-M trans-editing enzyme from Pyrococcus horikoshii. Acta Crystallogr D. 2007;63:390–400. doi: 10.1107/S090744490605640X. [DOI] [PubMed] [Google Scholar]
- 27.Sokabe M, Okada A, Yao M, Nakashima T, Tanaka I. Molecular basis of alanine discrimination in editing site. Proc Natl Acad Sci USA. 2005;102:11669–11674. doi: 10.1073/pnas.0502119102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill K, Schimmel P. Evidence that the 3′ end of a tRNA binds to a site in the adenylate synthesis domain of an aminoacyl-tRNA synthetase. Biochemistry. 1989;28:2577–2586. doi: 10.1021/bi00432a035. [DOI] [PubMed] [Google Scholar]
- 29.Dock-Bregeon AC, et al. Achieving error-free translation; the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol Cell. 2004;16:375–386. doi: 10.1016/j.molcel.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Logan DT, Mazauric MH, Kern D, Moras D. Crystal structure of glycyl-tRNA synthetase from Thermus thermophilus. EMBO J. 1995;14:4156–4167. doi: 10.1002/j.1460-2075.1995.tb00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosyak L, Reshetnikova L, Goldgur Y, Delarue M, Safro MG. Structure of phenylalanyl-tRNA synthetase from Thermus thermophilus. Nat Struct Biol. 1995;2:537–547. doi: 10.1038/nsb0795-537. [DOI] [PubMed] [Google Scholar]
- 32.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by protooncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 33.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamagata A, Kakuta Y, Masui R, Fukuyama K. The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity. Proc Natl Acad Sci USA. 2002;99:5908–5912. doi: 10.1073/pnas.092547099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ugochukwu E, Lovering AL, Mather OC, Young TW, White SA. The crystal structure of the cytosolic exopolyphosphatase from Saccharomyces cerevisiae reveals the basis for substrate specificity. J Mol Biol. 2007;371:1007–1021. doi: 10.1016/j.jmb.2007.05.066. [DOI] [PubMed] [Google Scholar]
- 36.Cavarelli J, et al. The active site of yeast aspartyl-tRNA synthetase: Structural and functional aspects of the aminoacylation reaction. EMBO J. 1994;13:327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biou V, Yaremchuk A, Tukalo M, Cusack S. The 2.9-Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 38.Ho C, Jasin M, Schimmel P. Amino acid replacements that compensate for a large polypeptide deletion in an enzyme. Science. 1985;229:389–393. doi: 10.1126/science.3892692. [DOI] [PubMed] [Google Scholar]
- 39.Ribas de Pouplana L, Buechter D, Sardesai NY, Schimmel P. Functional analysis of peptide motif for RNA microhelix binding suggests new family of RNA-binding domains. EMBO J. 1998;17:5449–5457. doi: 10.1093/emboj/17.18.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hecht SM, Chinualt AC. Position of aminoacylation of individual Escherichia coli and yeast tRNAs. Proc Natl Acad Sci USA. 1976;73:405–409. doi: 10.1073/pnas.73.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2006;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 43.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 45.Davis MW, Buechter DD, Schimmel P. Functional dissection of a predicted class-defining motif in a class II tRNA synthetase of unknown structure. Biochemistry. 1994;33:9904–9911. doi: 10.1021/bi00199a012. [DOI] [PubMed] [Google Scholar]
- 46.Shi JP, Musier-Forsyth K, Schimmel P. Region of a conserved sequence motif in a class II tRNA synthetase needed for transfer of an activated amino acid to an RNA substrate. Biochemistry. 1994;33:5312–5318. doi: 10.1021/bi00183a039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.