Abstract
Eukaryotic protein synthesis begins with mRNA positioning in the ribosomal decoding channel in a process typically controlled by translation initiation factors. Some viruses utilize an internal ribosome entry site (IRES) in their mRNA to harness ribosomes independently of initiation factors. We show here that a ribosome conformational change induced upon Hepatitis C viral IRES binding is necessary but not sufficient for correct mRNA positioning. Using directed hydroxyl radical probing to monitor the assembly of IRES-containing translation initiation complexes, a critical step has been defined in which mRNA is stabilized upon initiator tRNA binding. Surprisingly, however, this stabilization occurs independent of the AUG codon, underscoring the importance of initiation factor-mediated interactions that influence decoding channel configuration. These results reveal how an IRES RNA supplants some, but not all, of the functions normally carried out by protein factors during protein synthesis initiation.
Protein synthesis in all cells begins with binding and positioning of a messenger RNA (mRNA) on the small ribosomal subunit. In eukaryotes, the 5’ 7-methyl guanosine cap structure on most mRNAs triggers an assembly of translation factors that recruit the 40S ribosomal subunit. This highly regulated process, leading to 60S subunit joining and formation of an active 80S ribosome, requires at least twelve initiation factors composed of roughly 28 polypeptides1,2. Some viral mRNAs lack a 5’ cap and instead include a structured RNA sequence called an internal ribosome entry site (IRES) in the 5' untranslated region (UTR), which functions in place of some or all of the canonical initiation factors3,4. Some of the most detailed information to date for this type of mechanism has come from the study of the IRES found in the Hepatitis C virus (HCV) mRNA5,6. An efficient 40S subunit initiation complex on the HCV IRES requires only two initiation factors, eIF2 and eIF3 (ref.7). The eIF2 complex recruits the initiator tRNA (Met-tRNAi), while the much larger eIF3 complex enhances 40S subunit initiation complex formation on the HCV IRES, in part by stabilizing the eIF2•GTP•Met-tRNAi complex (ternary complex) on the 40S subunit8–10.
In both initiation pathways, mRNA recruitment and decoding occur in the mRNA binding channel, which is situated between the head, body and platform of the 40S subunit. This binding region comprises the channel through which the mRNA enters the 40S subunit, the ribosome decoding sites (aminoacyl (A) site, peptidyl (P) site and exit (E) site), and the exit channel through which the mRNA leaves the 40S subunit11 (Fig. 1a). The entry channel is occluded in empty 40S subunits, leading to the proposal that a conformational change is required for mRNA loading12–14. Indeed, cryo-EM derived structures of 40S-HCV IRES complexes revealed a structural change in 40S subunits bound to the wild-type IRES whereby the mRNA entry channel appeared more open relative to unbound 40S subunits13. Domain II of the HCV IRES (Fig. 1b) was shown to be responsible for the 40S structural change, correlating with toeprinting data that indicated a requirement of domain II for mRNA entry into the binding channel in the absence of initiation factors15,16. Subsequent cryo-EM reconstructions of the 40S subunit bound to the cricket paralysis viral IRES or to initiation factors eIF1 and eIF1A showed similar conformational changes in the 40S subunit head, hinting at a common mechanism of mRNA loading by viral IRESs and cap-dependent initiation factors14,17.
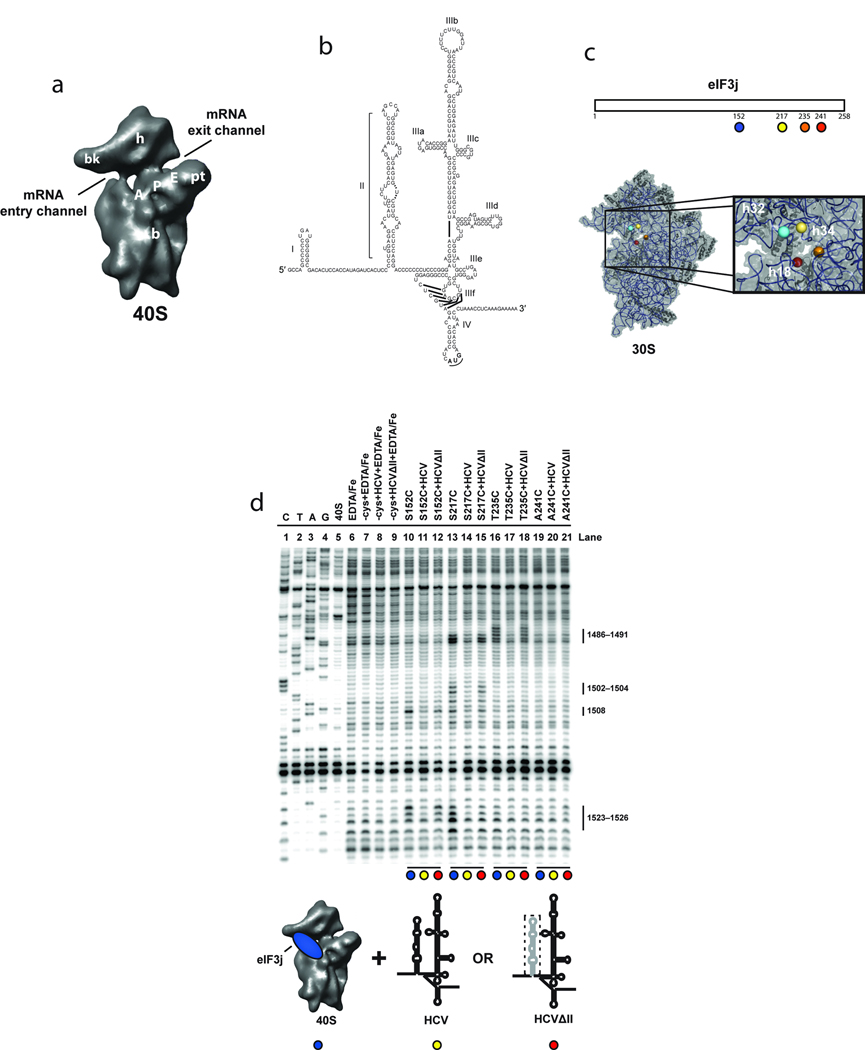
Figure 1.
Directed hydroxyl radical probing of 18S rRNA from BABE-Fe-eIF3j-40S-HCV complexes. (a) The 40S subunit structure based on a cryo-EM reconstruction13 viewed from the subunit interface with landmarks indicated: A, A-site; P, P-site; E, E-site; bk, beak; b, body; pt, platform; and h, head. (b) The 5´ UTR of the HCV mRNA consists of four domains (I–IV); the IRES domains (II–IV) with sub-domains (a–f) of domain III are indicated. (c) Representation of eIF3j indicating the positions of cysteine mutations used for BABE-Fe attachment (upper panel). Modeled positions of eIF3j amino acids in the T. thermophilus 30S crystal structure, adapted from a previous publication19 (lower panel). The boxed area provides a detailed view of the mRNA entry channel and A-site with helices 18, 32 and 34 indicated. Nucleotides cleaved in these helices for each experiment are shown in Supplementary Fig. 1a. (d) Primer extension analysis of 18S rRNA cleaved by BABE-Fe-modified eIF3j. Sequencing lanes are indicated (C, T, A, and G). Other control lanes include 40S subunits in the absence or presence of EDTA/Fe, mock-derivatized eIF3j (−cys+EDTA-Fe) in the absence (lane 7) or presence of wild type (HCV; lane 8), or domains III-IV (HCVΔII ; lane 9) of the HCV IRES RNA. Other lanes include eIF3j derivatized with BABE-Fe at the positions indicated either in the absence (lanes 10, 13, 16, and 19), or presence of HCV IRES (lanes 11, 14, 17, and 20), or HCVΔII IRES (lanes 12, 15, 18, and 21). 18S rRNA nucleotide positions of cleavage sites are indicated. Colored circles indicate components added in each reaction as depicted in the cartoon. The deletion of domain II (HCVΔII) is represented by a dotted line.
An additional clue to the mechanism of mRNA loading came from the discovery that a subunit of eIF3, eIF3j, only binds stably to 40S subunits in the absence of mRNA9,18. Directed hydroxyl radical probing showed that the C-terminus of eIF3j lies in the 40S mRNA entry channel, where it presumably disfavors mRNA binding in the absence of other initiation factors19. Upon initiator tRNA recruitment, as part of the ternary complex, eIF3j is displaced and mRNA binding is enhanced. How viral IRESs trigger this critical switch, leading to proper positioning of the viral mRNA on the 40S subunit, is not known.
To address this question, we used directed hydroxyl radical probing and ribosome toeprinting of reconstituted translation initiation complexes to determine the steps required for HCV IRES-mediated mRNA positioning in the 40S decoding channel. We established that mRNA and the C-terminus of eIF3j do not bind simultaneously in the ribosome entry channel, and that eIF3j displacement signals mRNA entry and binding in the channel. Using this as an assay for initiation complex formation, we show that the 40S conformational change induced by the IRES domain II is necessary but not sufficient for mRNA entry into the decoding channel. In addition, both the mRNA strand downstream of the AUG codon and the ternary complex are required for the mRNA to displace eIF3j. Importantly, this effect does not require AUG recognition by the initiator tRNA, implying that correct mRNA positioning is a function of ribosome conformation rather than mRNA-tRNA base pairing. Our results support an ordered pathway for HCV IRES-mediated translation initiation in which an eIF3-and IRES-stabilized 40S subunit conformational state favors viral mRNA entry into the decoding channel, but only after ternary complex binding.
RESULTS
The HCV IRES domain II promotes mRNA entry into the binding channel
Previous studies showed that initiation factors eIF1, −1A, −2 and −3 are necessary for stable mRNA positioning in the ribosomal decoding channel as monitored by toeprinting and reduced eIF3j affinity to the 40S subunit19,20. To test whether the HCV IRES similarly favors mRNA binding and eIF3j displacement from the mRNA binding channel, we monitored 18S rRNA cleavages induced by bromoacetamidobenzyl-EDTA-Fe (BABE-Fe) modified eIF3j proteins19 (Fig. 1c). As shown previously, eIF3j proteins containing a single BABE-Fe moiety at several positions near the C-terminus lead to site-specific rRNA cleavages in the mRNA entry channel (helix 34) in complexes containing only the 40S subunit and eIF3j. Addition of the wild type HCV IRES mRNA to this complex largely prevented these cleavages (Figs. 1d and Supplementary Fig. 1b). This observation could indicate that the C-terminus of eIF3j dissociates from the 40S mRNA binding cleft upon HCV mRNA association. Consistent with this idea, we observed no cleavage of the mRNA segment of the HCV IRES RNA in IRES-40S-eIF3j complexes, suggesting that eIF3j is no longer in the channel (data not shown).
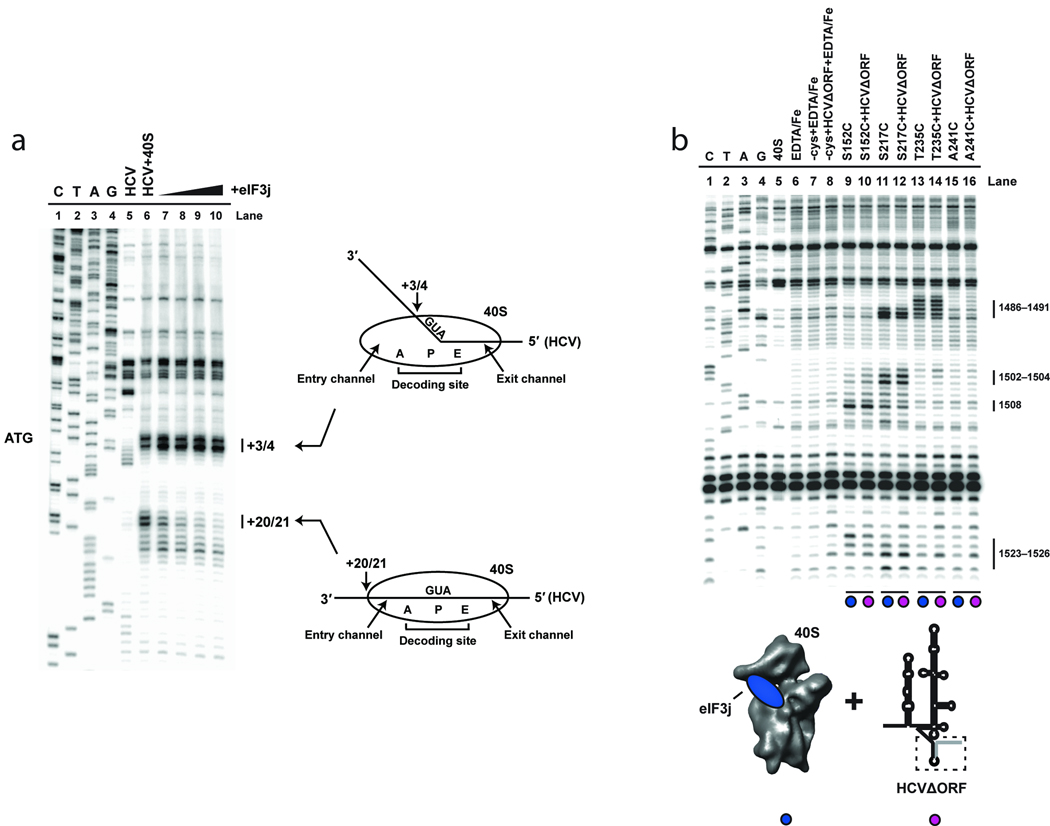
To distinguish between competitive binding of eIF3j and HCV mRNA and HCV mRNA-induced protection of 18S rRNA cleavage sites, the 40S subunit position on the IRES mRNA was mapped by toeprinting21–23. Consistent with previous results7,15, the association of the HCV mRNA with the 40S subunit induces a toeprint at nucleotides +20 and +21 downstream of the AUG codon, reflecting the leading edge of the 40S subunit on the mRNA (Fig. 2a). This toeprint was inhibited upon addition of increasing concentrations of eIF3j, indicating that the association of mRNA with the entry channel requires eIF3j displacement. Notably, the toeprint located at nucleotides +3 and +4 is still evident, even at a very large molar excess of eIF3j (5–40 µM), suggesting that eIF3j influences only mRNA association with the entry channel and not HCV mRNA association with the rest of the 40S subunit. This effect likely explains our previous data indicating that a short mRNA is able to bind concurrently with eIF3j19.
Figure 2.
Toeprinting analysis of the 40S-HCV-eIF3j complexes. (a) Lanes C, T, A, and G depict sequencing lanes corresponding to HCV mRNA, with the AUG codon indicated. Toeprinting reactions of HCV mRNA in the absence (lane 5) or presence (lane 6) of 40S subunits is shown. Additional reactions including 40S subunits in the presence of 5 µM (lane7), 10 µM (lane 8), 20 µM (lane 9), or 40 µM (lane 10) eIF3j are indicated. The positions of toeprints that correspond to 40S-HCV complexes are indicated (+3/4 and +20/21). Numbering is from the A (+1) of the AUG codon. Cartoons depicting the 40S-HCV complexes formed are also indicated. (b) Primer extension analysis of 18S rRNA cleaved by BABE-Fe-modified eIF3j in the absence or presence of HCV IRES RNA truncated after the AUG codon (HCVΔORF). Sequencing and control lanes are as indicated as described in Fig. 1d). The lanes corresponding to eIF3j in the absence (lanes 9, 11, 13, and 15) or presence (lanes 10, 12, 14, and 16) of HCVΔORF are indicated. As described in Fig. 1d, the colored circles correspond to the components added in each reaction, as indicated by the cartoons.
Since the conformational changes induced by domain II of the HCV IRES have been suggested to facilitate mRNA entry into the binding channel, we tested an HCV mRNA lacking this domain in our cleavage assay (HCVΔII in figures). Previous studies showed that this truncated form of the HCV IRES binds with similar affinity to the 40S subunit but fails to induce efficient translation initiation7,24. We found that the cleavages induced at nucleotides 1486–1491 in helix 34 by BABE-Fe-modified C-terminal positions on eIF3j were restored (Fig. 1d), suggesting that domain II is necessary for eIF3j displacement from, and mRNA binding to, the decoding channel. However, it is not possible from these data to determine the order in which these events occur. We next tested an otherwise wild-type version of the HCV IRES mRNA truncated after the AUG codon to determine if the rRNA cleavage sites induced by BABE-Fe modified eIF3j are generated when the mRNA strand does not extend into the eIF3j binding site on the 40S subunit. Although this shortened mutant associates with the 40S subunit (data not shown), the rRNA cleavage sites were essentially unchanged relative to those observed for 40S-eIF3j complexes (Fig. 2b). This suggests that in addition to the conformational changes induced by domain II, the presence of mRNA in the entry channel and A site of the 40S subunit is required to promote eIF3j displacement. It should be noted that domain II does not stably associate with the 40S subunit in the absence of other domains of the HCV IRES.
The eIF3 complex regulates HCV mRNA entry into the binding channel
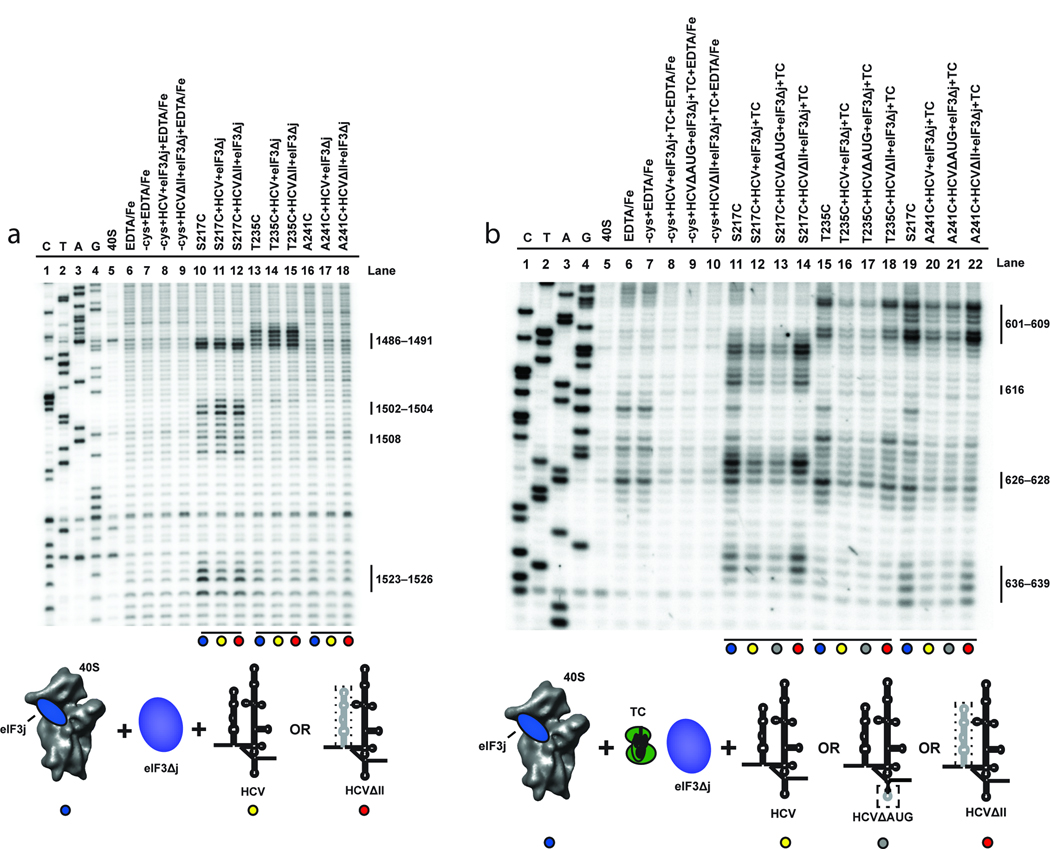
While the C-terminal half of eIF3j is located in the mRNA binding channel of the 40S subunit, its N-terminus interacts with the eIF3b subunit of eIF325. If maintained in the presence of the 40S-eIF3-HCV IRES mRNA complex, this interaction should increase the local concentration of eIF3j on the 40S subunit and thus enhance eIF3j’s ability to compete for binding to the entry channel with mRNA. To test this possibility, we analyzed BABE-Fe-modified eIF3j-induced cleavage of 18S rRNA in reconstituted 40S-eIF3-HCV IRES mRNA complexes. This was achieved by the addition of BABE-Fe modified eIF3j together with an eIF3 complex purified without endogenous eIF3j attached (eIF3Δj). In contrast to complexes containing the eIF3j subunit alone, those containing intact eIF3 yielded similar cleavage patterns to those observed in the absence of the HCV IRES mRNA (Fig. 3a). Similar experiments using HCVΔII produced small, but reproducibly enhanced cleavage intensities in rRNA helix 34 relative to wild type HCV IRES mRNA (Fig. 3a). These results suggest that the eIF3 complex enables eIF3j to compete more effectively with HCV IRES mRNA for binding to the mRNA entry channel, particularly in the absence of the IRES domain II-induced 40S conformational change. These data raise the question of how the HCV mRNA is able to efficiently associate with the mRNA entry channel in the presence of eIF3j and the eIF3 complex during the process of translation initiation.
Figure 3.
Effects of eIF3 and eIF2-Met-tRNAi on directed hydroxyl radical probing of 18S rRNA with BABE-Fe-eIF3j. (a) Lanes include 40S subunits in the absence or presence of EDTA/Fe and mock-derivatized eIF3j (−cys+EDTA-Fe) in the absence or presence of HCV constructs and eIF3 complex without endogenous eIF3j (eIF3Δj). Lanes corresponding to eIF3j derivatized with BABE-Fe at the positions indicated in the absence (lanes 10, 13, and 16), or presence of eIF3Δj and wild type HCV IRES (HCV; lanes 11, 14, and 17), or domain III of the HCV IRES (HCVΔII; lanes 12, 15, and 18) are indicated. (b) Lanes include 40S subunits in the absence or presence of EDTA/Fe and mock-derivatized eIF3j (−cys+EDTA-Fe) in the absence or presence of HCV constructs and other initiation factors as indicated. Lanes corresponding to BABE-Fe-modified eIF3j at the positions indicated in the absence (lanes 11, 15, and 19) or presence of eIF3Δj, eIF2-Met-tRNAi (Ternary complex; TC) and HCV (lanes 12, 16, and 20), HCV\AUG (lanes 13, 17 and 21), or HCVΔII (lanes 14, 18, and 22) are indicated. For each gel, sequencing lanes (C, T, A, and G) and cleavage nucleotide positions in the 18S rRNA are indicated. Colored circles correspond to the components added, as depicted in the cartoons. Relevant mutations in each HCV IRES construct are represented by a dotted line.
Previously, we showed that ternary complex association with the 40S subunit enhances the affinity of a short unstructured mRNA for the 40S subunit in the presence of eIF3j (ref. 19), hinting at a role for the ternary complex in displacing eIF3j during mRNA loading. To test this possibility, we determined the effect of ternary complex on HCV IRES mRNA association in the 40S subunit mRNA entry channel in the presence of eIF3j and intact eIF3Δj using site-directed hydroxyl radical probing. Unfortunately, it was not possible to utilize the cleavage site that we have described in helix 34 for this purpose since association of the ternary complex alone protects helix 34 from eIF3j induced cleavage (Supplementary Fig. S2a). This occurs even though eIF3j is still present in the mRNA entry channel, indicated by the unchanged cleavage intensity of helix 18 by BABE-Fe modified eIF3j in the presence of ternary complex (Supplementary Fig. S2b). Since cleavage of helix 18 by BABE-Fe modified eIF3j is unchanged by the association of eIF3Δj with the 40S subunit in the absence (Supplementary Fig. S2c), or presence (Supplementary Fig. S2d) of the HCV mRNA this allows visualization of this region to determine any ternary complex specific changes during HCV mRNA association. Upon recruitment of the ternary complex to the 40S subunit in the presence of HCV mRNA and eIF3Δj, cleavage intensities at all nucleotide positions in helix 18 in the mRNA entry channel are diminished (Fig. 3b). This observation implies that once ternary complex associates, even in the presence of intact eIF3, HCV IRES mRNA enters the entry channel and eIF3j is displaced. Notably, this effect is not seen in complexes containing HCVΔII (Fig. 3b).
One explanation for these results is that AUG recognition by the initiator tRNA stabilizes mRNA on the 40S subunit. To test this, we mutated the AUG codon and surrounding nucleotides (ATCATG to AAAAAA) in domain IV of the HCV IRES and analyzed complexes containing this construct by site-directed hydroxyl radical probing. Surprisingly, in the absence of the AUG codon, ternary complex recruitment still promotes HCV mRNA binding in the 40S subunit entry channel, as determined by the dissociation of eIF3j (Fig. 3b). Taken together, these data show that in the presence of intact eIF3, association of ternary complex with the 40S subunit stabilizes HCV IRES mRNA in the entry channel in a process that is independent of AUG codon recognition by the initiator tRNA.
eIF3 binding triggers a structural rearrangement of the 40S subunit
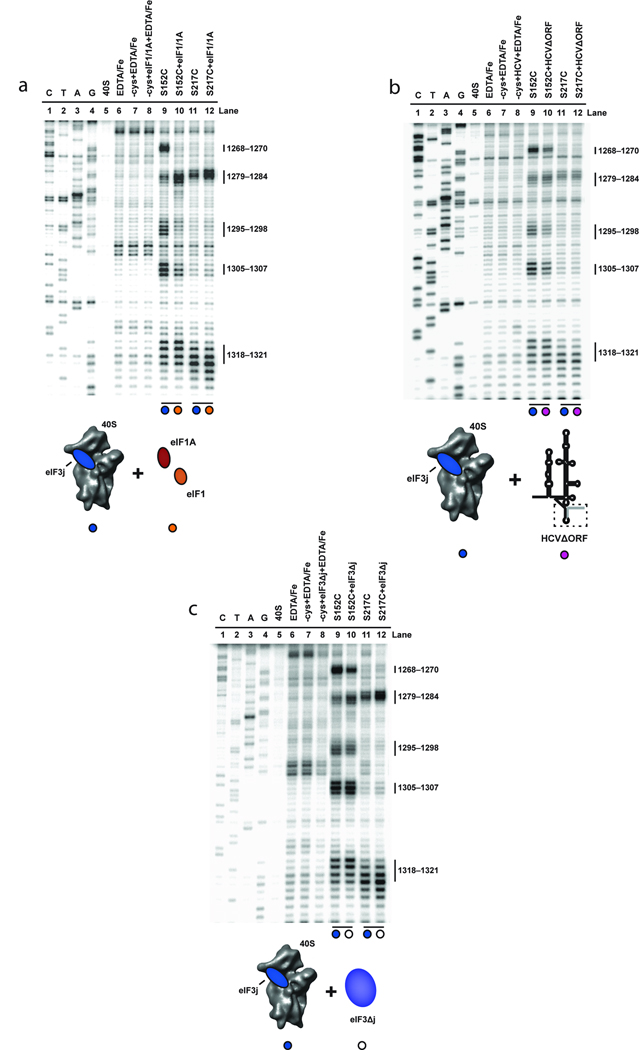
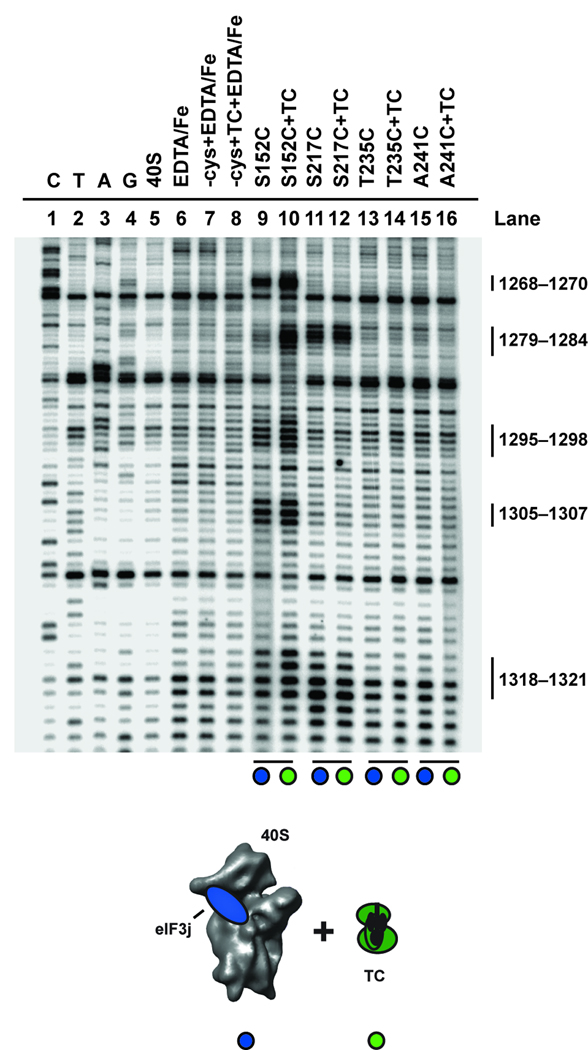
How do eIF3 and the ternary complex stabilize mRNA in the 40S subunit decoding channel irrespective of AUG codon recognition? An important clue to a possible mechanism came from observations of distinct conformational states of the 40S subunit by using cryo-EM to compare empty 40S subunits to 40S subunits complexed with viral IRES mRNA or initiation factors eIF1 and eIF1A13,14,17. These structures showed similar motions of the head relative to the body of the 40S subunit, altering the architecture of the mRNA binding channel, which could thereby affect access to the mRNA binding channel. To test whether these characterized structural changes are detectable using our site-directed hydroxyl radical probing assay, we formed each complex and determined the cleavage pattern from hydroxyl radicals generated from eIF3j. To visualize its association with the 40S subunit, the HCV mRNA was truncated at the AUG codon to avoid eIF3j dissociation, as discussed above. 18S rRNA cleavages resulting from BABE-Fe generated hydroxyl radicals from different sites on eIF3j were used to estimate changes in the three-dimensional structure of the 40S subunit, as previously validated for the bacterial ribosome26. Distance changes between the location of the BABE-Fe moiety and a specific nucleotide in the rRNA result in altered cleavage intensities, with more intense cleavages representing reduced distances between BABE-Fe and the affected sites. We compared the rRNA cleavage patterns generated in parallel experiments using two different BABE-Fe-modified eIF3j variants (Fig. 1c). Cleavage sites to be monitored were selected in the region of the 40S subunit beak (Fig. 1a), since the conformation of this structure changes significantly upon IRES or initiation factor binding13,14,17. Consistent with the cryo-EM structures, 18S rRNA cleavage patterns generated from BABE-Fe positions in eIF3j change upon the association of saturating amounts of eIF1 and eIF1A or the HCV mRNA with the 40S subunit (Fig. 4a, b). Specifically, the cleavage intensities at nucleotides 1268–1270, 1295–1298 and 1305–1307 in helices 32 and 33 generated from position 152 in eIF3j are reduced in both complexes (Fig. 4a, b, compare lanes 9 and 10). However, in contrast to the recruitment of HCV mRNA, eIF1 and eIF1A association with the 40S subunit results in increased cleavage intensities of nucleotides 1279–1284 generated from BABE-Fe tethered to amino acid positions 152 and 217 on eIF3j (Fig. 4a, b, compare lanes 9 and 10). Therefore, although eIF1 and eIF1A, or the HCV mRNA induce similar structural changes in the 40S subunit, these data obtained in solution imply some differences in the way these components alter the 40S subunit structure, particularly surrounding the mRNA entry channel and A-site. It should be noted that the observed cleavage differences could also result partly or entirely from direct interactions between eIF3j with eIF1 and eIF1A. We have previously shown that the interaction between eIF3j and eIF1A is anti-cooperative19, which likely indicates altered conformations of eIF3j and eIF1A on the 40S subunit.
Figure 4.
Effects of eIF1, eIF1A, HCV and eIF3 and on directed hydroxyl radical probing of 18S rRNA from BABE-Fe-eIF3j. (a) Primer extension analysis of 18S rRNA cleaved by BABE-Fe-modified eIF3j in the absence (lanes 9 and 11) or presence of eIF1 and eIF1A (lanes 10 and 12). (b) Analysis of 18S rRNA cleaved by BABE-Fe-modified eIF3j in the absence (lanes 9 and 11) or presence of HCVΔORF (lanes 10 and 12). (c) Analysis of 18S rRNA cleavage by BABE-Fe-modified eIF3j in the absence (lanes 9 and 11) or presence of eIF3Δj (lanes 10 and 12). In each gel the sequencing lanes are indicated (C, T, A, and G). Other lanes include 40S subunits in the absence or presence of EDTA/Fe and mock-derivatized eIF3j (−cys+EDTA-Fe) in the absence or presence of HCV and other initiation factors as indicated. Cleavage nucleotide positions in the 18S rRNA are indicated and colored circles correspond to the components added in each reaction, as depicted in the cartoons.
We next tested whether the eIF3 complex alters the conformation of the 40S subunit upon binding. To this end, comparing 18S rRNA cleavages generated from BABE-Fe tethered to positions in eIF3j in the absence or presence of the eIF3 complex indicates that association of the eIF3 complex alters the cleavage pattern in a similar, but distinct manner to the different complexes described above (Fig. 4c). In particular, the cleavage intensities of 18S rRNA nucleotides 1279–1284 generated by BABE-Fe modified eIF3j at positions 152 and 217 are enhanced by the addition of eIF3 or eIF1 and eIF1A to the 40S subunit (Fig. 4a, c, compare lanes 9 and 10). However, in contrast to cleavage patterns observed upon addition of the HCV IRES or eIF1 and eIF1A, the cleavage intensities of nucleotides 1295–1298 and 1305–1307 by hydroxyl radicals generated from position 152 in eIF3j remain unchanged upon addition of eIF3 (Fig. 4a, c, compare lanes 11 and 12). Interestingly, the eIF3 complex, eIF1 and eIF1A, or the HCV IRES mRNA all reduce the cleavage intensities of nucleotides 1268–1270 in helix 32 that are generated from BABE-Fe modified eIF3j at position 152 (Fig. 4a–c, compare lanes 9 and 10). Thus, the eIF3 complex stabilizes a specific 40S subunit conformation that has some similarities to the structures previously reported for HCV mRNA or eIF1 and eIF1A bound to the 40S subunit. However, these conformational changes are not sufficient to favor mRNA binding to the decoding channel, as detected by eIF3j displacement or 40S toeprinting20, in the absence of additional initiation factors. Furthermore, we also observe differences in cleavage intensity changes between complexes containing eIF1 and eIF1A, or eIF3-j in helix 18 (Supplementary Fig. S2b), confirming the similar, but unique conformations that these initiation factors promote in the 40S subunit.
Ternary complex binding induces a conformational change in the 40S
A reasonable explanation for ternary complex induced stability of mRNA irrespective of AUG recognition may be via additional conformational changes in the 40S subunit induced by the ternary complex itself. To test this we again compared rRNA cleavage patterns generated from BABE-Fe modified eIF3j in the location of the 40S subunit beak upon recruitment of the ternary complex to the 40S subunit in the absence of other initiation factors. Changes in the cleavage intensities from hydroxyl radicals generated from BABE-Fe tethered to position 152 in eIF3j are apparent (Fig. 5, compare lanes 9 and 10). Cleavage intensities dramatically increase at nucleotide positions 1268–1270 (helix 32) and 1279–1284 (helix 33), and increase slightly at nucleotides 1295–1298 (helix 33). These data suggest a conformational change upon ternary complex recruitment in which nucleotides in the 40S subunit beak move closer to the location of amino acid 152 in eIF3j (or into a more favorable environment or position for attack). Importantly, the cleavage intensities generated from eIF3j position 152 at nucleotides 1305–1307, or cleavage of nucleotides 1279–1284 generated from eIF3j position 217 in helix 33 do not change, suggesting that eIF3j does not move on the 40S subunit. Moreover, analysis of the cleavage pattern and intensity in helix 18 located inside the mRNA entry channel indicates that this region of the 40S subunit also does not change in relation to the location of eIF3j upon ternary complex recruitment (Supplementary Fig. S2b). These data reveal that the ternary complex promotes a structural rearrangement of the beak of the 40S subunit that would likely alter the conformation of the mRNA binding channel, especially in the A site of the 40S subunit. This change may contribute to the association of mRNA with the 40S subunit in a manner that is independent of AUG codon recognition by the initiator tRNA. Importantly, while a thermodynamically stable complex between the ternary complex and 40S subunit persists in the absence of any other initiation factors, it is expected that other initiation factors are required for accelerating the rate of complex formation.
Figure 5.
The effect of eIF2-Met-tRNAi on directed hydroxyl radical probing of 18S rRNA with BABE-Fe-eIF3j. Primer extension analysis of 18S rRNA cleaved by BABE-Fe-modified eIF3j. The sequencing lanes are indicated (C, T, A, and G). Other lanes include 40S subunits in the absence or presence of EDTA/Fe and mock-derivatized eIF3j (-cys+EDTA-Fe) in the absence (lane7) or presence of eIF2-Met-tRNAi (TC; lane 8). Lanes corresponding to eIF3j derivatized with BABE-Fe at the positions indicated in the absence (lanes 9, 11, 13, and 15), or presence of TC (lanes 10, 12, 14, and 16) are indicated. Nucleotide positions of cleavage sites in the 18S rRNA are indicated, and colored circles correspond to the components added in each reaction, as depicted in the cartoon.
Discussion
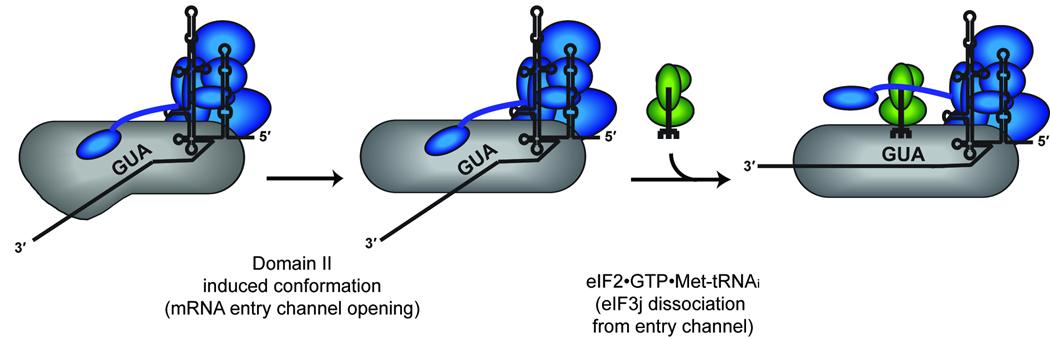
In this study, we investigated the streamlined initiation mechanism employed by the HCV IRES mRNA to examine the mechanism of mRNA positioning in the 40S mRNA binding channel. On the basis of site-directed hydroxyl radical probing and toeprinting we established that eIF3j and HCV IRES mRNA binding to the 40S subunit mRNA entry channel is mutually exclusive. This enabled us to use the dissociation of eIF3j from the mRNA entry channel as an indicator of mRNA binding to the 40S subunit entry channel. A proposed model for the pathway of HCV mRNA association with the 40S subunit is presented in Figure 6. In agreement with previous data7,13 the conformational change in the 40S subunit induced by domain II of the HCV IRES is required for mRNA positioning in the binding channel. This conformational change alone is not sufficient to allow the HCV mRNA to associate with the entry channel when both eIF3j and eIF3 are present. Instead, eIF3j displacement and consequent association of the HCV mRNA with the entry channel require ternary complex recruitment to the 40S subunit. It is worth noting here that the toeprinting competition experiment shown in Figure 2a requires a large molar excess of eIF3j to compete with HCV mRNA binding to the 40S subunit. This is likely required because the HCV mRNA is tethered to the 40S subunit through domains II and III, increasing its local concentration, while the absence of the interaction with the eIF3 complex reduces the local concentration of eIF3j. Although it is possible that the large excess of eIF3j used for toeprinting may bind non-specifically to the 40S subunit, this seems unlikely since the physiological local concentration of eIF3j on the 40S subunit would likely be high due to its interaction with the eIF3 complex.
Figure 6.
A model for HCV IRES association with the mRNA binding channel of the 40S subunit. Following the association of the HCV IRES with the 40S subunit, domain II is required to promote an open conformation of the mRNA entry channel. The stable association of eIF3 with this complex places eIF3j in the mRNA entry channel, preventing the stable binding of HCV mRNA with the A-site and entry channel. The subsequent recruitment of eIF2-Met-tRNAi is necessary to shift the equilibrium to favor the stability of HCV mRNA in the entry channel over that of eIF3j.
Surprisingly, our data show that the stabilizing effect of the ternary complex on HCV mRNA association does not require base pairing between the AUG codon and the anti-codon of the initiator tRNA. This finding contradicts toeprinting data that indicated a strong requirement of the AUG codon for HCV mRNA association with the entry channel7. However, toeprinting only enables detection of the most thermodynamically stable complexes such as those enhanced by the codon-anticodon interaction. In addition, mutation of the authentic AUG codon only reduces protein synthesis by 50–60% in vitro27. The remaining activity may be due to the utilization of the ACG codon situated two codons downstream of the AUG codon. Since toeprinting is not possible with the mutant AUG construct7 we are not able to determine if our construct actually allows a codon/anticodon interaction to occur at the ACG codon. Therefore, while our data suggests that AUG recognition by the initiator tRNA is not required for eIF3j dissociation in our assay, we cannot rule out the weaker interaction between the initiator tRNA and the ACG codon that may be essential for mRNA stabilization and eIF3j dissociation. Interestingly, recent data indicate that eIF3j is able to dissociate mRNA from the 40S subunit in during ribosome recycling, but only in the presence of eIF120. Since eIF1 does not affect the eIF3j binding affinity to the 40S subunit19, this is likely due to the effect of eIF1 in dissociating the P site bound tRNA, which would subsequently shift the equilibrium towards eIF3j binding to the entry channel over that of mRNA. However, a detailed kinetic analysis is required before the role of eIF3j in mRNA dissociation can be made, as it is plausible that eIF3j only influences mRNA association.
In this study we also investigated the mRNA binding propensities of different 40S subunit conformations induced upon association of the HCV IRES or initiation factors. In agreement with cryo-EM studies13,14, our data provide evidence that the HCV IRES or eIF1 and eIF1A promote similar but distinct conformational changes in the 40S subunit. Importantly, we show that the eIF3 complex also induces a conformational change in the 40S subunit that has some similarities to those induced by the HCV mRNA or eIF1 and eIF1A. Such eIF3-mediated effects on the 40S subunit may provide an explanation for its involement in mRNA recruitment to the 40S subunit in addition to its role in recruiting the cap binding complex in vitro and in vivo8,9,28. A recent report also suggested a possible conformational change in the mRNA entry channel when eIF3 associates with the 40S subunit29. However, those experiments were completed in the absence of eIF3j, but instead contained poly(U) to provide affinity for the eIF3 complex to the 40S subunit. Therefore, it is not possible to determine from those experiments what relative effects eIF3 and poly(U) have on the structure of the 40S subunit.
Ternary complex association with the 40S subunit without other components induces a conformational change in the head of the 40S subunit, which may play a role in promoting eIF3j dissociation and/or mRNA stability with the entry channel. Importantly, this conformational change is not sufficient to promote HCV mRNA association with the entry channel since the ternary complex is unable to stabilize HCV mRNA in the entry channel in the absence of domain II. Therefore, since eIF1, eIF1A, eIF3 and the ternary complex enable AUG recognition on an unstructured mRNA30, it is tempting to speculate that eIF1 and eIF1A likely provide the necessary conformational change in the 40S subunit to allow mRNA positioning in the binding channel, as proposed previously14. Based on these findings, we propose that either the HCV IRES, or eIF1 and eIF1A, can induce a similar conformational state of the 40S subunit that favors mRNA loading into the decoding channel with consequent displacement of eIF3j upon ternary complex binding. This conclusion explains how the HCV IRES functionally replaces eIF1 and eIF1A during translation initiation on the human ribosome.
Methods
Sample purification and modification
We purified human eIF1, −1A, −2, −3j, Δ3j and 40S ribosomal subunits were as described previously19,31. Initiator tRNA was transcribed in vitro, purified and charged in vitro using a purified tRNA synthetase, as previously described19,32. Conjugation of bromoacetamidobenzyl-EDTA-Fe (BABE-Fe; Dojindo Molecular Technologies) to single-cysteine eIF3j proteins was carried out according to a published protocol19,33. All HCV mRNA numbering is according to a previous publication34. Wild type HCV mRNA (40–372) and HCVΔII (120–372) used in probing experiments were prepared as described previously34. A derivative HCV construct (40–344; HCVΔORF) was generated using QuikChange mutagenesis (Stratagene). The HCV construct used for toeprinting experiments included the firefly luciferase open reading frame cloned into the wild type HCV mRNA between the BamHI and HindIII restriction sites located in the wild type HCV construct34. This resulted in a short nucleotide linker (GGATCCTC) following nucleotide 372 of the original construct before the ATG of the luciferase gene. The HCV construct containing the mutated AUG codon was generated using QuikChange mutagenesis of the HCV construct including the firefly luciferase open reading frame. The resulting construct mutates the initiation codon ATCATG to AAAAAA. All constructs were verified by sequencing and HCV RNA was produced by in vitro transcription and purified by denaturing acrylamide gel electrophoresis as described34.
Directed hydroxyl radical probing
Complexes containing either mock-derivatized eIF3j (-Cys) or BABE-Fe-eIF3j bound to salt-washed 40S subunits were formed and radical probing was carried out as described19,35,36. Specifically, each probing reaction was carried out in 50 µl incubations in buffer A (50 mM Hepes, pH 7.5, 50 mM KCl, 2 mM Mg acetate). Reactions contained 16 pmols (320 nM) 40S subunits, 72 pmols (1.44 µM) eIF1 and eIF1A, 35 pmols (700 nM) eIF3j, 23 pmols eIF3Δj (460 nM) and 72 pmols HCV mRNA (1.44 µM), as indicated in the figure legends. For reactions containing eIF2 and charged initiator tRNA, eIF2 was first incubated with 1 mM GMP-PNP (Sigma-Aldrich) in Buffer A for 5 minutes at 30 degrees. A two-fold excess of charged initiator tRNA was then added in Buffer A supplemented with a final free concentration of 1 mM Mg acetate and incubated at 30 degrees for 10 minutes. Subsequent probing reactions included 30 pmols eIF2-GMP-PNP and 60 pmols initiator tRNA. Detection of 18S rRNA cleavage by BABE-Fe-eIF3j was carried out using reverse transcription and denaturing gel electrophoresis as described19,35,36.
Toeprinting
The position of 40S subunits on the HCV mRNA was determined by a primer extension inhibition assay (toeprinting) as described previously21–23,37 with minor modifications. 40S subunits (16 pmols; 400 nM) HCV-luciferase mRNA (6 pmols; 150 nM) and 10 pmols (250 nM) of a 5’ end labeled 32P-labeled primer (5’-GCGCCGGGCCTTTCTTTATG-3’) complementary to nucleotides 18–37 of firefly luciferase were incubated in 40 µl reactions in Buffer A supplemented with 1 mM DTT. Reactions were incubated at 37 degrees for 10 minutes and then placed on ice for 5 minutes. 4µl of 10X extension mix (80 mM Mg acetate, 10 mM DTT, 10 mM dNTPs, 5 U SuperScript III reverse transcriptase (Invitrogen)) was added and the reaction was incubated at 30 degrees for 15 minutes. The reaction was then cooled on ice, followed by RNA extraction and analysis by denaturing gel electrophoresis as described previously19.
Supplementary Material
Acknowledgments
We thank David King at UC Berkeley for expert mass spectrometry analysis of modified proteins. We gratefully acknowledge members of the Doudna laboratory for discussions and comments on the manuscript. In particular we would like to thank Richard Spanggord for advice on hydroxyl radical probing and Fai Siu for advice on RNA transcription protocols. This work was supported in part by a grant from the NIH to J.A.D. and J.W.B.H.
References
- 1.Pestova TV, Lorsch JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- 2.Fraser CS, Doudna JA. Quantitative studies of ribosome conformational dynamics. Q Rev Biophys. 2007;40:163–189. doi: 10.1017/S0033583507004647. [DOI] [PubMed] [Google Scholar]
- 3.Doudna JA, Sarnow P. Translation Initiation by Viral Internal Ribosome Entry Sites. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 129–153. [Google Scholar]
- 4.Elroy-Stein O, Merrick WC. Translation Initiation via Cellular Internal Ribosome Entry Sites. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 155–172. [Google Scholar]
- 5.Pisarev AV, Shirokikh NE, Hellen CU. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C R Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nature reviews. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 7.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trachsel H, Erni B, Schreier MH, Staehelin T. Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J Mol Biol. 1977;116:755–767. doi: 10.1016/0022-2836(77)90269-8. [DOI] [PubMed] [Google Scholar]
- 9.Benne R, Hershey JW. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 10.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci U S A. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spahn CM, et al. Structure of the 80S ribosome from Saccharomyces cerevisiae--tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 12.Taylor DJ, Frank J, Kinzy TG. Structure and Function of the Eukaryotic Ribosome and Elongation Fractors. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 59–85. [Google Scholar]
- 13.Spahn CM, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 14.Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Molecular cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 17.Spahn CM, et al. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser CS, Berry KE, Hershey JW, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Molecular cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 20.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 22.Anthony DD, Merrick WC. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- 23.Kozak M. Primer extension analysis of eukaryotic ribosome-mRNA complexes. Nucleic Acids Res. 1998;26:4853–4859. doi: 10.1093/nar/26.21.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. Rna. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ElAntak L, Tzakos AG, Locker N, Lukavsky PJ. Structure of eIF3b RNA recognition motif and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. J Biol Chem. 2007;282:8165–8174. doi: 10.1074/jbc.M610860200. [DOI] [PubMed] [Google Scholar]
- 26.Joseph S, Noller HF. Directed hydroxyl radical probing using iron(II) tethered to RNA. Methods Enzymol. 2000;318:175–190. doi: 10.1016/s0076-6879(00)18052-8. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds JE, et al. Unique features of internal initiation of hepatitis C virus RNA translation. Embo J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jivotovskaya AV, Valasek L, Hinnebusch AG, Nielsen KH. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol Cell Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraser CS, et al. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J Biol Chem. 2004;279:8946–8956. doi: 10.1074/jbc.M312745200. [DOI] [PubMed] [Google Scholar]
- 32.Pestova TV, Hellen CU. Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation in vitro. Rna. 2001;7:1496–1505. doi: 10.1017/s135583820101038x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spanggord RJ, Siu F, Ke A, Doudna JA. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nature structural & molecular biology. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- 34.Kieft JS, et al. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 35.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Culver GM, Noller HF. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol. 2000;318:461–475. doi: 10.1016/s0076-6879(00)18070-x. [DOI] [PubMed] [Google Scholar]
- 37.Pisarev AV, Unbehaun A, Hellen CU, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.