Summary
Background
Asymmetric cell divisions generate daughter cells with distinct fates by polarizing fate determinants into separate cortical domains. Atypical protein kinase C (aPKC) is an evolutionarily conserved regulator of cell polarity. In Drosophila neuroblasts, apically restricted aPKC is required for segregation of neuronal differentiation factors such as Numb and Miranda to the basal cortical domain. While Numb is polarized by direct aPKC phosphorylation, Miranda asymmetry is thought to occur via a complicated cascade of repressive interactions (aPKC –| Lgl –| Myosin II –| Miranda).
Results
Here we provide biochemical, cellular, and genetic data to show that aPKC directly phosphorylates Miranda to exclude it from the cortex and Lgl antagonizes this activity. Miranda is phosphorylated by aPKC at several sites in its cortical localization domain and phosphorylation is necessary and sufficient for cortical displacement suggesting that the repressive cascade model is incorrect. In investigating key results that led to this model, we found that Y-27632, a Rho Kinase inhibitor used to implicate Myosin II, efficiently inhibits aPKC. Lgl3A, a non-phosphorylatable Lgl variant used to implicate Lgl in this process, inhibits the formation of apical aPKC crescents in neuroblasts. Furthermore, Lgl directly inhibits aPKC kinase activity.
Conclusions
Miranda polarization during neuroblast asymmetric cell division occurs by displacement from the apical cortex by direct aPKC phosphorylation. Lgl does not mediate Miranda cortical displacement but instead promotes Par-6/aPKC asymmetry by directly inhibiting aPKC. The role of Myosin II in this process, if any, is unknown.
Introduction
The presence of molecularly distinct cortical domains is a hallmark of many cell types [1, 2]. In epithelial cells, apical and basolateral domains mediate junction formation and transport [2]. Drosophila neuroblasts utilize polarized cortical domains during mitosis to generate progeny that assume different developmental fates [3, 4]. Early in mitosis, fate determinants that specify neuroblast fate form an apical cortical domain whereas those that specify differentiation form a basal domain and these domains are ultimately segregated into separate daughter cells [5]. In these cases and many others, a polarized cell cortex is essential for proper function. Although polarity is a fundamental characteristic of diverse cell types, many questions concerning its molecular origins remain.
The atypical Protein Kinase C (aPKC) is a key regulator of cell polarity [6]. In embryonic neuroblasts, aPKC localizes to the apical cortex where it restricts factors that specify basal cell fate, such as Miranda and Numb, to the basal cortex [5, 7, 8].
Recruitment of aPKC to the apical neuroblast cortex occurs through the combined action of Bazooka (Baz; aka Par-3), Par-6, and Cdc42 [7–9]. Apically localized Baz recruits GTP-bound Cdc42, which in turn binds the semi-CRIB domain of Par-6 to recruit aPKC [6].
Although aPKC apical recruitment is well understood, the mechanisms by which aPKC activity is translated into the polarization of downstream components are poorly defined. Although Numb has recently been identified as an aPKC substrate [10], and phosphorylation leads to its cortical displacement, the mechanism of Miranda polarization is thought to be much more complex. Apical aPKC phosphorylates the tumor suppressor Lgl [11, 12], a candidate Myosin II inhibitor [13]. Myosin II is thought to physically displace Miranda from the cortex, “pushing” it from the apical to basal cortex [14]. This leads to a complex “negative cascade” pathway in which aPKC phosphorylates Lgl, preventing its inhibition of Myosin II, ultimately removing Miranda from the cortex at sites of aPKC activity. However, the mechanisms by which Lgl might inhibit Myosin II and how Myosin II displaces Miranda from the cortex are entirely unknown. Furthermore, several key observations are inconsistent with this model, including the cortical association of Miranda in lgl aPKC mutants [5].
Here we have examined the mechanism by which aPKC polarizes Miranda and have found that Miranda is an aPKC substrate. Using a Miranda cortical displacement assay in cultured S2 cells we identify several Miranda phosphorylation sites that are responsible for its removal from the cortex. These results, along with expression of non-phosphorylatable Miranda mutants in neuroblasts, lead us to conclude that phosphorylation is necessary and sufficient for cortical displacement, inconsistent with the negative cascade model. By examined key pieces of evidence that supported this model, we found that a Rho kinase inhibitor used to implicate Myosin II [14, 15] is non-specific and efficiently inhibits aPKC at concentrations well below those used in previous studies. A non-phosphorylatable Lgl variant used to place Lgl between aPKC and Miranda [12] inhibits aPKC apical crescent formation. Furthermore, Lgl directly inhibits aPKC kinase activity. Based on these observations, we propose that aPKC directly phosphorylates Miranda to yield mutually exclusive cortical domains and that Lgl’s role in polarity is to restrict aPKC activity to the apical cortex.
Results
Miranda is an aPKC substrate
Recently, aPKC was shown to phosphorylate the cell-fate determinant Numb [10]. To determine whether Miranda might also be an aPKC substrate, we expressed and purified several Miranda truncations and incubated them with recombinantly purified aPKC. Miranda contains an NH2-terminal cortical localization domain (residues 1–290) [16–18] that is responsible for cortical targeting in neuroblasts and we observed phosphorylation specific to this region (Figures 1A,B). Consistent with Miranda being an aPKC substrate, Miranda co-immunoprecipitates with aPKC from transfected S2 cells (Figure 1C) and Drosophila embryonic extracts (Figure 1D) indicating that the two proteins interact with one another in vivo.
Figure 1. aPKC binds and phosphorylates Miranda.
(A) Miranda domain architecture. (B) The Miranda cortical localization domain is specifically phosphorylated by aPKC. Phosphorimage and Coomassie stained SDS gel of GST and GST:Miranda fusion proteins in the presence of recombinant HIS:aPKC and 32P. GST:Miranda 1–290 is phosphorylated whereas all other constructs are not. Commassie stain of gel as loading control. (C) Miranda interacts with aPKC in transfected S2 cells. Immunoblot of S2 cell lysates from cells transfected with aPKC:myc and HA:Miranda immunoprecipitated with anti-aPKC, -HA, or -Pins antibodies. Protein G and anti-Pins antibody used as controls. (D) Miranda interacts with aPKC in Drosophila embryonic extracts. Immunoblot of embryonic lysate immunoprecipitated with anti-aPKC, -Par-6, -Miranda, or -Pins antibodies. Controls as in panel C. (E) Identification of residues within Miranda 1–290 phosphorylated by aPKC in vitro. Purple-highlighted residues indicate coverage by LC/MS/MS, red lettering indicates residues identified as phosphorylated, and stars indicate residues tested by point mutation as in Figure 2. Phosphorylation prediction programs predicted residues 141 and 143 to be highly likely for phosphorylation by aPKC.
We identified the sites within Miranda that are phosphorylated by aPKC using mass spectrometry. Tandem mass spectrometry of recombinantly purified Miranda 1–290 that had been incubated with recombinant aPKC followed by trypsin digestion yielded nearly complete coverage of the cortical localization domain (Figure 1E). Analysis of these tryptic fragments yielded several groups of phosphorylated residues within the Miranda cortical localization domain (Figure 1E).
Miranda phosphorylation by aPKC is necessary and sufficient for cortical displacement
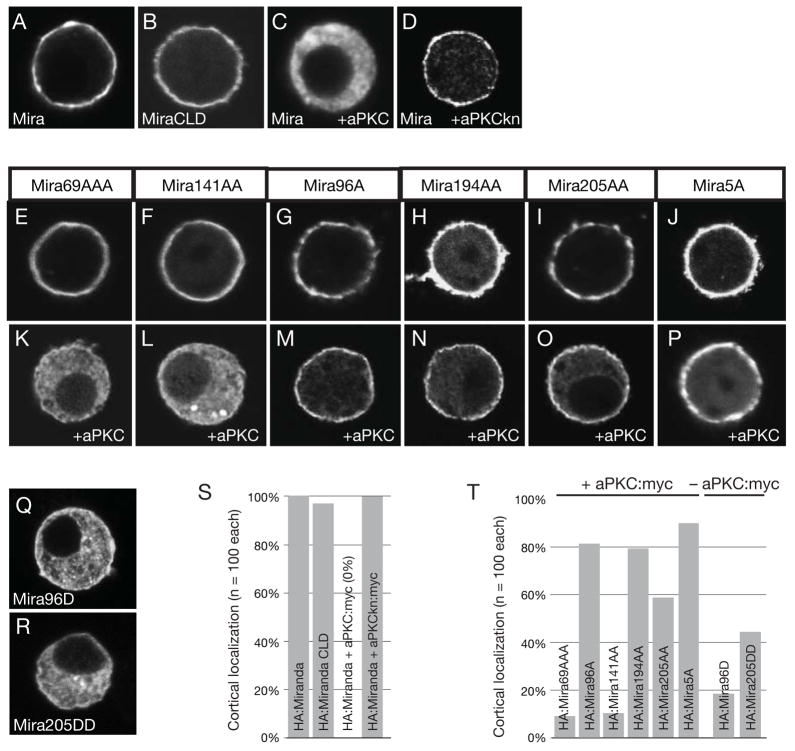
To rapidly determine which phosphorylation sites within Miranda may be responsible for cortical displacement, we developed a cortical displacement assay in cultured Drosophila S2 cells. We quantitatively assayed cortical-to-cytoplasmic signal ratios of transiently transfected Miranda, which is heavily enriched at the cortex of S2 cells (Figures 2A,S; Figure S1), similar to the basal enrichment of Miranda at the cortex of metaphase neuroblasts. The Miranda neuroblast cortical localization domain is sufficient for cortical localization in S2 cells (Figures 2B,S) suggesting that the association mechanism in neuroblasts is also utilized in S2 cells. Expression of aPKC with Miranda leads to loss of cortical staining and an increase in cytoplasmic signal (Figures 2C,S). Kinase activity is required for this effect, as a kinase-dead aPKC variant does not displace Miranda (Figures 2D,S). We conclude that aPKC removes Miranda from the S2 cell cortex in a manner similar to neuroblasts, and that this system can be used as a model for investigating aPKC-dependent Miranda cortical displacement.
Figure 2. aPKC is necessary and sufficient to displace Miranda from the S2 cell cortex.
Expression of the Miranda and aPKC in fixed S2 cells and stained by indicated markers. All quantifications use n=100 cells. (A) Miranda localizes to the S2 cell cortex (100% - scored as in Figure S1), (B) The Miranda neuroblast cortical localization domain is also sufficient for S2 cortical localization (96%). (C) Miranda is displaced into the cytoplasm when expressed with aPKC (100%), (D) but not with a variant that lacks kinase activity, aPKCkn (100%). (E–J) All Miranda variants with alanine mutations localized to the cortex in the absence of aPKC. (K) Miranda with alanine mutations in residues 69, 70, and 71 (Mira69AAA) localizes predominantly to the cytoplasm in the presence of aPKC:myc (92%), (L) as well as a construct with alanine mutations in residues 141 and 143 (Mira141AA; 90%). (M) Miranda with alanine mutations in residues 96 (Mira96A) is predominantly cortical in the presence of aPKC:myc (82%), (N) as well as constructs with alanine mutations in residues 194 and 195 (Mira194AA; 79%) or (O) in residues 205 and 206 (Mira205AA; 58%). (P) Miranda with alanine mutations in all five residues showing substantial effects (Mira5A; residues 96, 194, 195, 205, and 206) localizes predominantly to the cortex in the presence of aPKC:myc (90%). (Q and R) Miranda with aspartic acid mutations at residues 96 (Mira96D; 82%) or residues 205 and 206 (Mira205DD; 55%) are predominantly cytoplasmic in the absence of aPKC:myc. (S and T) Quantification of Miranda constructs and point mutants in the presence, or absence, of aPKC:myc.
To identify phosphorylation sites that might be coupled to Miranda cortical association, we examined the localization behavior of Miranda variants in which the in vitro phosphorylation sites had been changed to alanine and tested if these proteins could be displaced into the cytoplasm by aPKC. All Miranda variants localized to the cortex in the absence of aPKC (Figures 2E–J). Mutation of residues 69, 70, and 71 (Mira69AA) or 141 and 143 (Mira141AA) did not have any appreciable effect on Mira cortical exclusion by aPKC (Figures 2K,L,T). However, mutation of residue 96 (Mira96A), residues 194 and 195 (Mira194AA), or residues 205 and 206 (Mira205AA) strongly prevented Miranda displacement by aPKC (Figures 2M-O,T). We combined the mutated residues that reduced aPKC’s ability to displace Miranda from the cortex into one protein (Mira5A) and assayed its localization in the presence of aPKC. Mira5A remains cortical in the presence of aPKC (90% cortical) indicating that these sites regulate cortical exclusion of Miranda (Figures 4P,T). We conclude that aPKC phosphorylation of Miranda is necessary for excluding Miranda from the cell cortex of S2 cells.
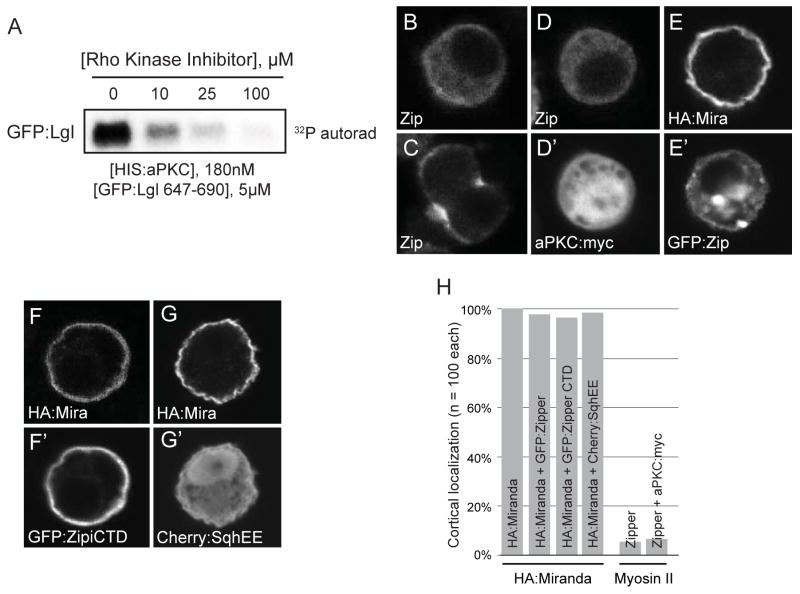
Figure 4. The Rho kinase inhibitor Y-27632 inhibits aPKC and Myosin II does not affect Miranda cortical localization in S2 cells.
(A) aPKC activity is inhibited by the Rho kinase inhibitor. An autoradiograph of a GFP:fusion of the Lgl phosphorylation site after incubation with aPKC and increasing concentrations of Y-27632 is shown. (B–H) Expression of indicated constructs in fixed S2 cells and stained by indicated markers. All quantifications use n=100 cells. (B and C) Myosin II is mostly cytoplasmic and enriched at the cleavage furrow in the absence of aPKC:myc (96%) and (D) remains cytoplasmic in the presence of aPKC:myc (94%). (E) HA:Miranda remains cortical in the presence of GFP:Zipper (97%) and (F) in the presence of the interphase cortical targeting domain [22] of GFP:Zipper (94%).(G) Expression of an activated form of sqh (Cherry:SqhEE) does not affect cortical HA:Miranda (98%). (H) Quantification of Miranda and Myosin II localization in the presence of the indicated transiently transfected proteins.
We next tested whether phosphorylation by itself can lead to Miranda displacement from the S2 cell cortex. We generated aspartic acid mutations at sites of aPKC phosphorylation and examined the localization of the phosphomimetic protein in the absence of aPKC. Mutation of residue 96 (Mira96D) or residues 205 and 206 (Mira205DD) caused Miranda to become cytoplasmic even in the absence of aPKC indicating phosphorylation is sufficient for cortical exclusion (Figures 2Q,R,O). Thus, based on the results of both alanine and aspartic acid mutations we conclude that phosphorylation is necessary and sufficient for excluding Miranda from the cortex of S2 cells.
We verified that Miranda phosphorylation leads to cortical displacement in neuroblasts using transgenic flies expressing Mira5A. We expressed wild-type Miranda and Mira5A in larval neuroblasts using the prospero-GAL4 driver and assayed for Mira5A localization and neuroblast polarity. Mira5A was unpolarized in mitotic larval neuroblasts in stark contrast to the basal enrichment in wild-type neuroblasts (Figures 3A–C,G). To determine whether non-phosphorylatable Miranda is cortically enriched throughout development, we expressed Mira5A in embryonic neuroblasts using the worniu-GAL4 driver and found similar uniform cortical localization (Figure 3D,G). In fact, Mira5A localized uniformly cortical throughout mitosis (Figures 3E,F). The presence of Mira5A had no effect on apical complex formation as Par-6 and aPKC correctly localized to the apical cortex (Figures 3B–D,G). Mira5A is functional, retaining the ability to bind the cell-fate determinant Prospero, as indicated by the uniformly cortical localization of Prospero in neuroblasts expressing Mira5A (Figure 3B″). In addition, Mira5A expression was not able to rescue embryonic lethality of mirandaZZ176 animals [16], although a wild-type form is able to do so [19] indicating phosphorylation and proper polarization of Miranda by aPKC is necessary for normal development (data not shown). We conclude that Miranda phosphorylation by aPKC is necessary and sufficient for Miranda cortical displacement.
Figure 3. Phosphorylation of Miranda is necessary for exclusion from the neuroblast apical cortex at metaphase.
(A–F) An unphosphorylatable Miranda is depolarized in embryonic and larval neuroblasts. Expression of UAS transgenes by prospero-GAL4 in brains at 96 hALH, or worniu-GAL4 in embryos at stages 11–13, and stained by the indicated markers. (A) Wild-type Flag:Miranda was used as a control and localized to the basal cortex (100%; n=47), whereas aPKC and Par-6 localized to the apical cortex (100%; n=47). (B–D) HA:Mira5A was not restricted to the basal cortex and localized uniformly cortical in larval nerublasts (100%; n=71) and in embryonic neuroblasts (100%, n=52) during metaphase, (E and F) and throughout mitosis. (B–D) aPKC (larval- 100%, n=71; embryonic- 96%, n=52) and Par-6 (larval- 100%, n=64) remained wild-type. (B″) Prospero was mislocalized uniformly cortical in Mira5A expressing neuroblasts. (G) Quantification of neuroblast metaphase localization.
The Rho Kinase Inhibitor Y-27632 inhibits aPKC
The observation that phosphorylation of Miranda by aPKC is necessary and sufficient for cortical displacement leads to a dramatic simplification of the negative cascade model: Miranda polarity occurs by a mechanism in which cortical regions containing aPKC activity displace Miranda by direct phosphorylation. Is it possible to explain the previous observations that gave rise to the negative cascade model in terms of this simpler mechanism?
Myosin II was thought to “push” Miranda from the apical cortex of neuroblasts leading to basal enrichment [14]. Much of the evidence for Myosin II involvement in aPKC-mediated cortical displacement of Miranda in the negative cascade model resides on the observation that Miranda polarity is lost in neuroblasts exposed to the Rho kinase inhibitor Y-27632 [14, 15]. Rho kinase phosphorylates the Myosin II RLC, which induces filament assembly and motor activity [20]. As the uniform cortical Miranda localization in Y-27632 treated neuroblasts is identical to apkc neuroblasts and kinase inhibitors are often not entirely specific, we hypothesized that the inhibitor phenotype results from inhibition of aPKC (in fact, inhibition of canonical PKC was noted in the original description of Y-27632 [21]). Using an in vitro kinase assay with purified, recombinant aPKC, we find that Y-27632 efficiently inhibits aPKCwith an IC 50 < 10 μM (Figure 4A). The concentration of this compound used to examine its effect on embryonic neuroblast polarity (~50 mM Y-27632 [14, 15]) vastly exceeds the IC50 of aPKC and Rho kinase (~1μM [21]) indicating that aPKC was likely to have been inhibited in these experiments. Given the cross-reactivity of Y-27632, an alternative explanation that aPKC is inhibited in drug-treated embryos displaying loss of Miranda polarity is consistent with our direct phosphorylation model.
As the S2 cell system accurately recapitulates aPKC-mediated cortical displacement of Miranda, we also examined the effect of Myosin II on Miranda localization in S2 cells. Although we were unable to reduce Myosin II levels sufficiently using RNAi, we tested whether Myosin II behaves as predicted by the negative cascade model. This model predicts cells expressing sufficient aPKC to displace Miranda should alter Myosin II levels at the cortex [14]. Endogenous Myosin II is predominantly cytoplasmic in S2 cells and enriches at the cleavage furrow during mitosis (Figures 4B,C,H). However, we observe no appreciable change in cortical Myosin II levels in cells expressing aPKC (Figure 4D,H). The model also predicts Myosin II physically displaces Miranda at sites along the cortex, suggesting competition for the same cortical binding sites. We attempted to directly displace Miranda using Myosin II overexpression, but were unable to do so with full-length heavy chain (Figures 4E,H; Zipper in Drosophila) or a heavy chain fragment that localizes predominantly to the cortex (Figures 4F,H)[22] indicating the presence of Myosin II at the cortex is not sufficient to displace Miranda. As active Myosin II may be necessary to displace Miranda from the cortex, we expressed a phosphomimetic form of the regulatory light chain (RLC) spaghetti squash (sqhEE) with Miranda in S2 cells but again observed no change in cortical Miranda (Figure 4G,H). This protein also fails to have an effect in neuroblasts [14].
Lgl directly inhibits aPKC to depolarize Miranda
In the negative cascade model, aPKC phosphorylation of Lgl is required for Miranda cortical displacement. This aspect of the model arose from the observation that neuroblasts expressing Lgl3A, a non-phosphorylatable variant, exhibit Miranda polarization defects [12, 14], which was interpreted to result from a requirement for Lgl phosphorylation in Miranda cortical release. To explore the nature of the Lgl3A phenotype more closely, we examined neuroblasts expressing Lgl3A driven by worniu-GAL4. Consistent with previous observations, the majority of neuroblasts exhibited ectopic cortical Miranda compared to wild-type (Figures 5A–C). However, we also found that apical aPKC and Par-6 crescents are severely reduced compared to wild-type in neuroblasts with ectopic Miranda cortical localization (Figures 5B,C). We also examined the effect of Lgl3A on Miranda cortical displacement in S2 cells. Miranda and Lgl3A co-localize at the S2 cell cortex as expected (Figures 5D,F). However, when aPKC is also expressed, Lgl3A remains at the cortex but Miranda is displaced into the cytoplasm (Figures 5E,F), inconsistent with Lgl phosphorylation being a prerequisite for Miranda displacement. Thus, an alternative explanation of Lgl3A induced Miranda depolarization is that it results from compromised aPKC activity.
Figure 5. aPKC can displace Miranda from the cortex independently of Lgl.
(A–F), Wild-type and expression of the indicated UAS transgenes by worniu-GAL4 in brains at 96h after larval hatching (ALH) and labeled with the indicated markers. (A), Wild-type larval neuroblasts display normal cortical polarity, whereas (B,C) Lgl3A-expressing neuroblasts show ectopic cortical localization of aPKC (88%; n=16) and Par-6 (92%; n=13) and disrupted apical crescents whereas Miranda shows ectopic cortical localization (76%; n=29). (D–F) Expression of indicated constructs in fixed S2 cells and stained by indicated markers. All quantifications use n=100 cells. (D) Miranda is cortical in the presence of GFP:Lgl3A (94%) in S2 cells, (E) but is displaced into the cytoplasm in the presence of GFP:Lgl3A and aPKC:myc (98%). (F) Quantification of Miranda localization in Lgl3A-expressing background. (G) Expression of aPKCΔN, which is not efficiently inhibited by Lgl, can displace Miranda from the cortex of neuroblasts even in the presence of Lgl3A (96%; n=27). (H) Lgl directly inhibits aPKC. The phosphorylation of GST:Miranda 1–290 by HIS:aPKC detected by phosphorimaging is reduced upon addition of GFP:Lgl 647–690. (I) Model of aPKC-induced neuroblast polarity.
We sought to determine whether the Lgl3A-mediated Miranda phenotype results from excessive inhibition of aPKC by Lgl3A, or a requirement for Lgl phosphorylation in Miranda displacement. We examined Miranda localization in neuroblasts expressing Lgl3A along with an active, predominantly cytoplasmic aPKC (aPKCΔN) that no longer binds Lgl through Par-6 (N-terminal portion that binds Par-6 is missing [12]). If the Lgl3A phenotype results from inhibition of aPKC activity, aPKCΔN should overcome this inhibition and displace Miranda from the cortex, whereas if phosphorylation of Lgl is required for Miranda displacement, Miranda should remain cortical. We find that Miranda is efficiently driven into the cytoplasm of neuroblasts expressing Lgl3A and aPKCΔN (Figure 5G), further suggesting that Lgl phosphorylation is not required for aPKC-mediated Miranda cortical displacement. This observation is consistent with the cytoplasmic Miranda observed in neuroblasts expressing aPKC-CAAX (containing a lipid modification tag) and Lgl3A[5].
Although Lgl has been shown to inhibit aPKC [23], the mechanism of inhibition has been unclear. To test if Lgl is capable of directly inhibiting aPKC in vitro, we incubated recombinantly purified aPKC with the cortical localization domain of Miranda and increasing concentrations of purified Lgl. Miranda is efficiently phosphorylated by aPKC in the absence of Lgl (Figure 5J). However, upon titration of Lgl, phosphorylation of Miranda was drastically reduced indicating that Lgl can directly inhibit aPKC activity. As Lgl is an aPKC substrate, it is possible that it competitively inhibits aPKC activity, although Baz is also an aPKC substrate and does not inhibit aPKC [23]. Thus, we conclude that Lgl directly regulates aPKC activity resulting in a simplified model for cell-fate determinant segregation where aPKC directly phosphorylates and displaces Miranda from the neuroblast cortex and Lgl antagonizes this activity (Figure 5L).
Discussion
We have examined the mechanism by which polarity is generated in Drosophila neuroblasts, a process required for the segregation of cell fate determinants during asymmetric cell division. This process utilizes aPKC, which is found in many polarized systems such as epithelia. Previously, polarization of the protein Miranda, which is normally restricted to the basal neuroblast cortex opposite aPKC, has been thought to occur by a complex cascade of repressive interactions involving the tumor suppressor Lgl and the motor protein Myosin II [12, 14]. Our finding that aPKC phosphorylation displaces Miranda from the cortex of neuroblasts and S2 cells led us to suspect that the negative cascade model might not accurately describe Miranda displacement. This prompted us to reexamine key results supporting the repressive cascade model.
Based on our investigation of previous results we propose that studies suggesting that Myosin II is involved in aPKC-mediated cortical displacement of Miranda are an artifact of inhibition of aPKC by the Rho kinase inhibitor Y-27632. Although we cannot exclude that the Miranda polarity defects observed in Y-27632 treated embryos are indeed the result of Myosin II inhibition, the fact that this phenotype is identical to that exhibited by apkc mutants, the efficient inhibition of aPKC, and the high concentrations of this compound used in previous reports (~50 mM compared to the IC50 <10 μM for aPKC and 1 μM for Rho kinase) indicate that the simplest interpretation of the Y-27632 phenotype is direct inhibition of aPKC activity. The role of Myosin II in Miranda cortical displacement, if any, is unclear.
We have also examined the central result that led to the placement of Lgl between aPKC and Miranda. Expression of a form of Lgl in which the aPKC phosphorylation sites have been inactivated results in uniformly cortical Miranda in neuroblasts [12]. This result can be interpreted in one of two ways: Lgl mediates Miranda cortical targeting and phosphorylation of Lgl represses this activity, or Lgl inhibits aPKC and this inhibition is repressed by aPKC phosphorylation (i.e. feedback). The key distinction between these two models is whether or not aPKC is repressed when Lgl3A is expressed. Several recent studies indicate that Lgl is a potent inhibitor of aPKC activity [5, 23]. Consistent with this, we find that Lgl3A expression dramatically reduces the localization of aPKC to the neuroblast apical cortex. Furthermore, we find that a form of aPKC that is not efficiently repressed by Lgl can overcome the effects of Lgl3A and drive Miranda into the cytoplasm, which is only consistent with Lgl phosphorylation not being a requirement for Miranda cortical displacement. In addition, we show that Lgl alone is sufficient for inhibition of aPKC activity. Thus, we conclude that Lgl can directly inhibit aPKC, but that Lgl is not required for Miranda cortical targeting.
We favor a simpler mechanism than the repressive cascade model for Miranda polarization by aPKC: aPKC phosphorylates Miranda causing it to be displaced from the cortex. The identification of Miranda as a direct aPKC substrate, the requirement of these phosphorylation events for cortical displacement both in S2 cells and neuroblasts, and the necessity of these phosphorylation events for normal development and viability support this model. The sufficiency of phosphorylation (phosphomimetic Miranda is cytoplasmic in the absence of aPKC) indicates that other phosphorylation events (such as phosphorylation of Lgl in the repressive cascade model) are not required for Miranda cortical displacement. We believe this new model dramatically simplifies our understanding of how asymmetric aPKC activity, as generated by Baz and Cdc42 (Figure 5I), is translated into the segregation of cell fate determinants. Thus, for three components downstream of aPKC, Miranda (this work), Numb [10], and Lgl [12] polarization appears to occur by direct aPKC phosphorylation. Further work will be required to determine if this mechanism is utilized by all factors that are polarized by aPKC.
Methods
S2 cell culture and quantification
S2 cells were cultured using Schneider’s medium (Sigma) containing 10% FBS. Constructs were cloned into pMT and transfected using Effectene (Qiagen). We generated alanine and aspartic acid point mutations by site-directed mutagenesis using pMT Miranda as a template. In order to quantify Mira localization, we analyzed 100 cells transfected with HA:Miranda in the presence, or absence, of aPKC:myc in ImageJ to generate histograms based on pixel intensity versus pixel distance. We compared the pixel intensity at the cortex to the pixel intensity in the cytoplasm (Figure S1A–C). Cells in which the ratio of cortex to cytoplasm staining was 2 or less were scored as cytoplasmic while cells with a ratio greater than 2 were scored as cortical.
Fly strains
Oregon R (wild type), lgl334(Bloomington), worniu-Gal4, pros-Gal4,UAS -HA:Mira5A, UAS-aPKCΔN [12], UAS-Flag:Mira (gift from C. Doe), UAS-lgl3A[12], miraZZ176. Stocks were balanced over CyO; CyO, actin::GFP; TM3, actin::GFP, Ser, e; or TM3, Sb. To produce Mira5A transgenic animals, we PCR amplified and subcloned the coding sequence into the pUAST vector downstream of a 5′ hemagglutinin (HA) tag and generated transformants using standard methods.
Antibodies and immunofluorescent staining
We fixed and stained larval brains and S2 cells as previously described [24]. Wild-type and lgl mutant larvae were aged at 25°C until 96 h after larval hatching (ALH). Flag:Miranda and HA:Mira5A expressing larvae were aged at 30°C until 96 h ALH (prospero-Gal4). HA:Mira5A expressing embryos were aged at 25°C until stages 11–13. Lgl3A and aPKCΔN mutant were aged at 30°C until 96 h ALH (worniu-Gal4). Rescue experiment was performed by expressing HA:Mira5A (worniu-Gal4) in zygotic mirandaZZ176 embryos. Primary antibodies: rabbit anti-PKCζ (C20; 1:1000; Santa Cruz Biotechnology Inc); rat anti-Par-6 (1:200) [8]; guinea pig anti-Mira (1:500); rat anti-Mira (1:500); rabbit anti-Phospho-Histone H3 (1:1000; Upstate); guinea pig anti-Baz (1:1000)[25]; mouse anti-HA (1:1000; Covance); mouse anti-Flag (1:100; Sigma); rabbit anti-zipper (1:2000)[22]; rabbit anti-GFP (1:1000; Torrey Pines); mouse anti-Prospero (1:100). Secondary antibodies were from Jackson ImmunoResearch Laboratories and Invitrogen. Confocal images were acquired on a Leica TCS SP2 microscope equipped with a 63×1.4 NA oil-immersion objective. Final figures were arranged using ImageJ, Adobe Photoshop, and Adobe Illustrator.
Protein purification, binding experiments, and mass spectrometry
All proteins were expressed and purified as previously described [7]. Drosophila embryonic lysate was prepared as previously described [7]. We immunoprecipitated proteins using approximately 5 μg rabbit anti-aPKC, rat anti-Par-6, rat anti-Pins [26], or mouse anti-HA with Protein G conjugated beads according to the manufacturer’s protocol (GE Healthcare). To determine which immunoprecipitates contained aPKC, we separated samples by SDS-PAGE and transferred to nitrocellulose followed by probing with anti-aPKC (1:2000) antibody.
For mass spectrometry, recombinantly purified HIS:Miranda 1–290 was incubated with purified HIS:aPKC as described in the following section. Phosphorylated Miranda 1–290 was purified by SDS-PAGE followed by coomassie staining. Gel slices containing protein were digested with trypsin and resuspended in formic acid. Samples were analyzed using OrbiTrap with neutral loss for +2, +3, +4 ions. Spectra were analyzed using MASCOT and X!Tandem to identify phosphorylated residues.
Kinase assay
We incubated HIS:aPKC purified from HEK cells [7]at 30°C for 15 min in reaction buffer (20mM HEPES pH 7.5, 10mM MgCl2, 1mM DTT, 10mM ATP) then added GST, GST:Miranda, or HIS:Miranda fragments (10μM final concentration) and 17 nM [γ-32P]-labeled ATP for 20 additional minutes. We quenched the reaction by addition of SDS loading buffer and heating at 95°C for 5 min and determined the extent of phosphorylation by SDS-PAGE and exposure onto a Phosphor screen (Molecular Dynamics) and detected using a STORM 860.
Supplementary Material
Supplemental Figure 1. Quantification of S2 cell signal intensity.
(A,B) HA:Miranda is cortical but is displaced into the cytoplasm with expressed with aPKC:myc. Pixel intensity profiles that correspond to the lines in each of the representative images are shown. (C) Histogram of cortical-to-cytoplasmic signals for cells expressing HA:Miranda (blue) or HA:Miranda and aPKC (red). Plotting pixel intensity at the cortex to pixel intensity in the cytoplasm generates a histogram of cortical-to-cytoplasmic signal ratios over 100 cells from each background. Cells with ratios 2 or below were scored as cytoplasmic whereas cells with ratios greater than 2 were scored as cortical. (D–F) Large fields of S2 cells transfected as shown.
Acknowledgments
We would like to thank J. Knoblich for providing the Lgl3A transgenic line and C. Doe, R. Anderson, R. Newman, and J. Boone for helpful discussions. This work was supported by a NIH Developmental Biology Training Grant 5-T32-HD07348 (SXA), and NIH grant GM068032 (KEP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bryant DM, Mostov KE. From cells to organs: building polarized tissue. Nature reviews. 2008;9:887–901. doi: 10.1038/nrm2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mellman I, Nelson WJ. Coordinated protein sorting, targeting and distribution in polarized cells. Nature reviews. 2008;9:833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 4.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 7.Atwood SX, Chabu C, Penkert RR, Doe CQ, Prehoda KE. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J Cell Sci. 2007;120:3200–3206. doi: 10.1242/jcs.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol. 2003;163:1089–1098. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petronczki M, Knoblich JA. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat Cell Biol. 2001;3:43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- 10.Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, Berry DM, Bonneil E, Thibault P, Schweisguth F, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. Embo J. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betschinger J, Eisenhaber F, Knoblich JA. Phosphorylation-induced autoinhibition regulates the cytoskeletal protein Lethal (2) giant larvae. Curr Biol. 2005;15:276–282. doi: 10.1016/j.cub.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Betschinger J, Mechtler K, Knoblich JA. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- 13.Kalmes A, Merdes G, Neumann B, Strand D, Mechler BM. A serine-kinase associated with the p127-l(2)gl tumour suppressor of Drosophila may regulate the binding of p127 to nonmuscle myosin II heavy chain and the attachment of p127 to the plasma membrane. J Cell Sci. 1996;109(Pt 6):1359–1368. doi: 10.1242/jcs.109.6.1359. [DOI] [PubMed] [Google Scholar]
- 14.Barros CS, Phelps CB, Brand AH. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev Cell. 2003;5:829–840. doi: 10.1016/s1534-5807(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 15.Erben V, Waldhuber M, Langer D, Fetka I, Jansen RP, Petritsch C. Asymmetric localization of the adaptor protein Miranda in neuroblasts is achieved by diffusion and sequential interaction of Myosin II and VI. J Cell Sci. 2008;121:1403–1414. doi: 10.1242/jcs.020024. [DOI] [PubMed] [Google Scholar]
- 16.Fuerstenberg S, Peng CY, Alvarez-Ortiz P, Hor T, Doe CQ. Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding, and cortical release. Molecular and cellular neurosciences. 1998;12:325–339. doi: 10.1006/mcne.1998.0724. [DOI] [PubMed] [Google Scholar]
- 17.Matsuzaki F, Ohshiro T, Ikeshima-Kataoka H, Izumi H. miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development. 1998;125:4089–4098. doi: 10.1242/dev.125.20.4089. [DOI] [PubMed] [Google Scholar]
- 18.Shen CP, Knoblich JA, Chan YM, Jiang MM, Jan LY, Jan YN. Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes Dev. 1998;12:1837–1846. doi: 10.1101/gad.12.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- 20.Moussavi RS, Kelley CA, Adelstein RS. Phosphorylation of vertebrate nonmuscle and smooth muscle myosin heavy chains and light chains. Molecular and cellular biochemistry. 1993:127–128. 219–227. doi: 10.1007/BF01076773. [DOI] [PubMed] [Google Scholar]
- 21.Uehata M, Ishizaki T, Satoh H, Ono T, Kawahara T, Morishita T, Tamakawa H, Yamagami K, Inui J, Maekawa M, et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 22.Liu SL, Fewkes N, Ricketson D, Penkert RR, Prehoda KE. Filament-dependent and -independent localization modes of Drosophila non-muscle myosin II. J Biol Chem. 2008;283:380–387. doi: 10.1074/jbc.M703924200. [DOI] [PubMed] [Google Scholar]
- 23.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siegrist SE, Doe CQ. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development. 2006;133:529–536. doi: 10.1242/dev.02211. [DOI] [PubMed] [Google Scholar]
- 25.Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- 26.Nipper RW, Siller KH, Smith NR, Doe CQ, Prehoda KE. Galphai generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts; Proceedings of the National Academy of Sciences of the United States of America; 2007. pp. 14306–14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Quantification of S2 cell signal intensity.
(A,B) HA:Miranda is cortical but is displaced into the cytoplasm with expressed with aPKC:myc. Pixel intensity profiles that correspond to the lines in each of the representative images are shown. (C) Histogram of cortical-to-cytoplasmic signals for cells expressing HA:Miranda (blue) or HA:Miranda and aPKC (red). Plotting pixel intensity at the cortex to pixel intensity in the cytoplasm generates a histogram of cortical-to-cytoplasmic signal ratios over 100 cells from each background. Cells with ratios 2 or below were scored as cytoplasmic whereas cells with ratios greater than 2 were scored as cortical. (D–F) Large fields of S2 cells transfected as shown.