Abstract
WNT signaling is an important determinant of bone formation. The WNT co-receptor, Frizzled homolog 1 (FZD1), initiates WNT signal transduction. To study the influence of FZD1 genetic variation on measures of bone health, we first sequenced a 6.8-kb region surrounding FZD1 in 48 samples of African ancestry. We genotyped all common polymorphisms and performed association analysis with bone phenotypes in a larger sample. Only 3 of 35 SNPs identified were present in ≥5% of the sample and assayed further in 1084 men of African ancestry. Two of these SNPs were in the FZD1 promoter (rs2232157, rs2232158) and were associated with femoral neck areal BMD (p = 0.041 and 0.009, respectively). The minor alleles of these two SNPs were also associated with larger bone size at the radius (p < 0.05 for both), and rs2232158 was associated with greater strength-strain index, an indicator of bone's ability to withstand torsion. Functional experiments were completed to assess the influence of the rs2232158 promoter polymorphism on transcriptional regulation of FZD1. The minor C allele in rs2232158 creates a binding site for the transcription factor Egr1, has higher Egr1 binding affinity, and has greater FZD1 promoter activity in MG63 and SaOS-2 cells, providing a plausible molecular mechanism for the population associations. This study indicates that a cis-regulatory polymorphism in the FZD1 promoter region may have a functional role in determining bone structural geometry.
Key words: Frizzled-1, polymorphism, BMD, WNT, Egr1
INTRODUCTION
The wnt/β-catenin signaling pathway has recently emerged as an important regulator of early skeletal development and regeneration of tissue throughout the lifespan.(1–3) Control of bone formation, one of the diverse functions of the WNT/β-catenin pathway, is achieved by the regulation of osteoblast differentiation, proliferation, and apoptosis.(2–4) WNT signaling is transmitted to the nucleus when WNT molecules bind to the frizzled (FZD) and LRP5/6 co-receptor complex.(5,6) Mutations in the gene encoding the WNT co-receptor LRP5 were identified in both high and low bone mass conditions.(7,8) Additional studies, including a recent genome-wide association scan, have also observed associations between variation in LRP5 and normal variation in BMD in the general population.(9–12)
Members of the frizzled gene family are also logical candidate genes for bone mass. One family member, frizzled homolog 1 (FZD1), is a G-protein–coupled receptor capable of both transmission and repression of WNT signaling depending on the co-receptor bound to it,(13) and FZD1 is expressed in osteoblast-like cells.(14–16) To assess the influence of genetic variation in FZD1 on bone, we sequenced the FZD1 gene region to identify polymorphisms, conducted genetic association analyses with bone-related phenotypes, and performed in vitro functional analysis of an associated promoter variant. Our results suggest a novel role of genetic variation in the transcriptional regulation of FZD1 expression in osteoblast-like cells and an association with long bone size and biomechanical indices of bone strength.
MATERIALS AND METHODS
Population
The population sample was drawn from The Tobago Bone Health Study, an ongoing, population-based study of men >40 yr old from the Caribbean island of Tobago. In brief, 3300 men have been recruited since 1997, and this represents 62% of all age-eligible men on the island.(17,18) The ancestral makeup of this population is 94% African origin as determined by ancestry informative molecular markers.(19) Written informed consent was obtained from all participants, and the study was approved by both the Tobago Ministry of Health and Social Services and the University of Pittsburgh Institutional Review Boards.
Variant discovery
Common variation within the FZD1 gene region was poorly captured in public databases like the International HapMap project. Thus, to better characterize genetic variation in the FZD1 gene region, a 6.8-kb region including 2.1 kb upstream of the transcription start site, the 4.4-kb transcript, and 350 bp downstream of the transcript was sequenced in 48 genomic DNA samples collected from Afro-Caribbean men in the Tobago Bone Health Study. A sequencing project of this size should detect 99% of all SNPs with a minor allele frequency (MAF) of 5% and 87% of all SNPs with MAF of 1%.(20)
Sequencing was carried out by DNA Polymorphic (Alameda, CA, USA) on the ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Sequence analysis and SNP detection were completed with Sequencher 4.5 sequence analysis software (Genecodes, Ann Arbor, MI, USA). Polymorphisms present in more than one sequencing fragment were considered valid for these analyses.
Genotyping
Genomic DNA was isolated from either whole blood extracted by the salting out method or from blood clots collected in coagulation tubes and isolated by a Qiagen column procedure (Qiagen, Santa Clara, CA, USA). Common SNP variation identified by sequencing the FZD1 gene region (defined as MAF > 5%) was subsequently genotyped in 1084 men from the Tobago Bone Health Study who were of African ancestry. One polymorphism, rs2232163, was genotyped using TaqMan on the ABI Prism 7900HT (Applied Biostystems, Foster City, CA, USA). Two polymorphisms, rs2232157 and rs2232158, could not be genotyped by TaqMan and were genotyped using short read sequencing by SeqWright (Houston, TX, USA).
The success rates for genotyping were 99.6% for rs2232157, 98.2% for rs2232158, and 98.1% for rs2232163. Genotyping consensus was 100% in 46 samples that were assayed in duplicate.
Bone measurements
Areal BMD, BMC, and cross-sectional area (CSA) of the femoral neck were determined by DXA on a QDR 4500 scanner (Hologic, Bedford, MA, USA). Scans were analyzed with QDR software version 8.26a. Daily phantom scans were analyzed to ensure long-term scanner stability. CV was determined by repeating DXA measures on 12 participants (all CVs were ≤1.16%).
In addition, a subset of men (N = 769) also had pQCT measurements completed on an XCT 2000 scanner (Stratec; Medizintechnik, Pforzheim, Germany). Specifically, cortical and trabecular volumetric BMD (vBMD), total BMC, total CSA, and polar strength-strain index at the radius and tibia were measured. To standardize placement of the measurements, a scout scan view of the endplate of the radius and tibia was taken, and the endplates were used as anatomic landmarks. The length of the radius and tibia were measured, and scans were obtained 4% and 33% of the length of the radius and tibia away from the distal endplate. The nondominant arm was used for radius measurements, and tibia measurements were made on the left leg. Scans were acquired using a 0.5-mm voxel size, slice thickness of 2.5 mm, and at a speed of 20 mm/s. Image processing was performed by a single investigator using the Stratec software package (Version 5.5E). All 4% ultradistal radius and tibia scans were analyzed using identical parameters for contour finding and separation of trabecular and cortical bone (contour mode 2, T = 169 mg/cm3; peel mode 1, area = 45%) to determine the vBMD of the trabecular (mg/cm3) bone compartment. All 33% proximal radius and tibia shaft scans were analyzed using identical parameters for contour finding and separation of total and cortical bone (contour mode 2, T = 169 mg/cm3; cortmode 1, T = 710 mg/cm3) to determine the vBMD of the cortical (mg/cm3) bone compartment. To take the material properties of bone into consideration for assessing the structural properties, the polar strength strain index (SSI, mm3) was also calculated as the integrated product of the section modulus and cortical density (CoD; Eq. 1). Section modulus (mm3) is calculated as (a × d 2)/d max, where a is the cross-sectional area of a voxel (mm2), d is the distance of the voxel from the center of gravity (mm), and d max is the maximum distance (eccentricity) of one voxel to the center of gravity (mm). The ratio of CoD and ND (normal physiological density, 1200 mg/mm3) provides an estimate of the modulus of elasticity.(21) SSI has been shown to be an accurate and precise indicator of the structural properties of long bones tested in bending.(22) SSI is more strongly correlated with experimentally determined breaking force than either DXA measures of areal BMD, or CSMI or cortical volumetric BMD alone.(22)
CVs were determined for pQCT scans by replicating measurements on 15 subjects (CV ≤ 4.2% for all measures). Daily phantom scans were analyzed to ensure long-term scanner stability.
Other measures
Ethnicity was determined by self-report, and only those reporting Afro-Caribbean ancestry were included in this analysis. Body weight and height were determined, without shoes, using a balance-beam scale and wall-mounted stadiometer, respectively.
In silico functional analysis
We used Transcription Element Search Software (TESS; http://www.cbil.upenn.edu/tess) to assess the potential functional significance of the two promoter variations (rs2232157, rs2232158). Because rs2232158 was predicted to alter early growth response-1 (Egr1) binding in an allele-specific manner, we focused subsequent in vitro functional experiments on this SNP.
Cell culture
MG63 and SaOS-2 cell lines were cultured in RPMI medium 1640 with l-glutamine, 10% FBS (vol/vol), and penicillin/streptomycin (2 units/ml) at 37°C in a humidified chamber with 5% CO2 (Invitrogen, Carlsbad, CA, USA). In addition, MG63 and SaOS-2 cells were treated with ATP because previous studies showed that ATP induces Egr1 in these cell lines.(23) For ATP treatment, cells were grown to subconfluence (∼90%) under normal conditions. At 1 h before the treatment, culture media were replaced with fresh media. Subsequently, ATP was added to the media to obtain a concentration of 100 μm (Cell Signaling, Danvers, MA, USA). The cells were allowed to grow under normal conditions for 30 min before being harvested for nuclear protein extraction.
Western blot analysis
Ten micrograms of nuclear protein isolated from untreated cells and cells treated with ATP were resolved by SDS-PAGE, transferred to nitrocellulose, and analyzed by standard immunoblotting using horseradish peroxidase (HRP)-conjugated secondary antibody and chemiluminescence detection (Amersham). Rabbit polyclonal anti-Egr1 antibody was used (Sc-189; Santa-Cruz Biotechnology, Santa Cruz, CA, USA).
EMSA
Recombinant Egr1 protein (0.25–0.75 band forming units; Sigma-Aldrich, St Louis, MO, USA) was incubated with 32P (5′-end)-labeled double-stranded oligonucleotides (10 fmol) specific for the two alleles of rs2232158 (5′-CGACGGAGGGCACC(C/G)GCGCAGAGGTCTCC-3′). DNA binding reactions were performed in 10 mM Tris-HCl (pH. 7.9), 1 mM DTT, 1 mM EDTA, 60 mM NaCl, 10% glycerol, and 1 μg poly (dIdC; Pharmacia, Uppsala, Sweden). For supershift assays where Egr1 antibody (sc-110; Santa-Cruz Biotechnology) was used, 2 μl antibody was first mixed with nuclear extract for 30 min on ice before the binding reaction. A wildtype Egr1-specific probe (5′-GGATCCAGCGGGGGCGAGCGGGGGCGA-3′) was also used to identify the Egr1-specific complex. The DNA-protein complexes were separated by 5% PAGE and detected by autoradiography.
Transcription factor ELISA
Nuclear extracts for the transcription factor ELISA assays were prepared using the Panomics Nuclear Extraction Kit (Panomics, Fremont, CA, USA). Nuclear protein concentration was determined using Microplate BCA Protein Assay Kit-Reducing Agent Compatible according to the manufacturer's instructions (Pierce, Rockford, IL, USA). The Panomics Egr1 transcription factor ELISA assay was used according to manufacturer specifications, except that biotinylated oligonucleotide probes specific for the rs2232158 alleles described above were used instead of the consensus sequence probe provided by the company. Specifically, 1 μg of nuclear protein from MG63, SaOS-2, and SaOS-2 + ATP cells was incubated with 10 μM of allele-specific, oligonucleotide probe in the binding buffer for 30 min at room temperature. Each condition was run in triplicate. The binding mixture was incubated with a streptavidin-coated plate to allow the binding of biotinylated probe/protein complex. The Egr1-specific complex was captured by anti-Egr1 antibody followed by HRP-conjugated secondary antibody. The intensity of substrate development by the HRP was analyzed using a spectrophotometer (OD450) and used as a measure of Egr1 binding. A concentration curve (1, 2, and 3 μg) for the Egr1 binding was also obtained using nuclear protein from MG63 cells.
Generation of FZD1 promoter luciferase reporter constructs
Allele-specific FZD1 promoter region constructs spanning the rs2232158 site were generated using Accuprime PCR kit (Invitrogen) and genomic DNA from individuals with the genotypes of interest. Primer sets 5′-GCGAGCTCGCCACCACCACTACTTCCTC-3′ and 5′-GAGATCTGGCACAAAGTTCCCAGCTC-3′ were used to amplify a 726-bp fragment spanning −655 to +71 nucleotides relative to the translation start point. A SacI and BglII recognition site (underlined) was included in the 5′ end of the forward and reverse primer, respectively. The PCR products were cloned into the pGMTeasy vector (Promega, Madison, WI, USA). Direct DNA sequencing was used to verify the inserts before the subcloning into the pGL3 basic vector (Promega).
FZD1 promoter activity analysis
The MG63 and SaOS-2 cells were seeded in 48-well culture plates 36 h before the transfection to obtain subconfluence (∼70%) at the time of transfection. Transfection was performed using Lipofectamine LTX (Invitrogen). The cells were harvested at 30–36 h after transfection, and firefly luciferase activities were analyzed using the luciferase assay system (Promega). Six replicates for each experimental condition were included.
Statistical analysis
All analyses were conducted using SAS 9.1.3 (SAS Institute, Cary, NC, USA). Hardy-Weinberg equilibrium was assessed by χ2 analysis. Linkage disequilibrium was assessed by pairwise D′ and r 2 for the genotyped polymorphisms.
Polymorphisms were tested for association with BMD and bone structural geometry parameters. Specifically, for rs2232157 and rs2232158, linear regression was used to test for an additive association between the number of copies of the minor allele and the trait. Because relatively few men were homozygous for the minor allele of rs2232163, a dominant model was tested where those with the AG or AA genotype were combined into one group and compared with the common, GG, genotype group. All analyses were adjusted for age, and age-adjusted, genotype-specific means and SEs are presented. To account for multiple testing, permutation testing was carried out using the statistical package R (version 2.5.1). Specifically, an empirical distribution was obtained for each bone trait by regressing each SNP against the phenotype and randomly permuting the phenotype 1000 times. The minimum p value across all FZD1 SNPs for each permutation was used to create the new empirical distribution, and the value observed in the single SNP analysis (referred to as unadj.) was corrected (referred to as corrected).
The Wilcoxon rank-sum test was used to assess the significance of the increased Egr1 binding of the rs2212158 C allele in the transcription factor ELISA experiment and to assess the increased promoter activity in the transfection experiment.
RESULTS
FZD1 sequence analysis showed 35 SNPs, 3 insertion deletion polymorphisms, and a proline repeat variant within the 6.8-kb gene region (Table 1). Our sequence analysis confirmed 12 of the 21 variants listed in dbSNP Build 128 for the sequenced gene region. To our knowledge, 24 SNPs, 2 insertion deletion polymorphisms, and the proline repeat variant that we identified have not been previously described. Three of the SNPs identified were nonsynonymous coding SNPs but were relatively uncommon in our population and thus not genotyped in the larger sample. Three SNPs (rs2232157, rs2232158, rs2232163) with a MAF ≥5% were genotyped in a larger sample (N = 1084) from the Tobago Bone Health Study.
Table 1.
FZD1 Polymorphism Discovery by DNA Sequence Analysis of 48 Afro-Caribbean Individuals
| Location | Relative position* | Position on chromosome 7† | Reference SNP ID |
Alleles |
Codon | Amino Acid change |
Genotype frequency |
Minor allele frequency | ||
| 1/2‡ | 1/1‡ | 1/2‡ | 2/2‡ | |||||||
| 5′ flanking region | −2408 to −2409 | 90,728,420–90,728,421 | rsl0654396 | AG/− | 47 | 1 | 0 | 0.010 | ||
| −2030 | 90,730,102 | C/T | 47 | 1 | 0 | 0.010 | ||||
| −1337 | 90,730,795 | C/A | 45 | 3 | 0 | 0.031 | ||||
| −1297 | 90,730,835 | G/A | 41 | 1 | 0 | 0.012 | ||||
| −1296 | 90,730,836 | C/T | 41 | 1 | 0 | 0.012 | ||||
| −1285 | 90,730,847 | C/T | 40 | 1 | 1 | 0.036 | ||||
| −1275 | 90,730,857 | C/T | 39 | 3 | 0 | 0.036 | ||||
| −1272 | 90,730,860 | C/T | 41 | 1 | 0 | 0.012 | ||||
| −1269 | 90,730,863 | G/T | 41 | 1 | 0 | 0.012 | ||||
| −1252 | 90,730,880 | C/T | 41 | 1 | 0 | 0.012 | ||||
| −1248 | 90,730,884 | C/T | 41 | 1 | 0 | 0.012 | ||||
| −652 | 90,731,480 | rs2232151 | A/G | 44 | 3 | 0 | 0.032 | |||
| −612 | 90,731,520 | G/A | 46 | 1 | 0 | 0.011 | ||||
| −611 | 90,731,521 | C/T | 46 | 1 | 0 | 0.011 | ||||
| −601 | 90,731,531 | G/A | 46 | 1 | 0 | 0.011 | ||||
| −587 | 90,731,545 | rs2232152 | T/A | 46 | 1 | 0 | 0.011 | |||
| −580 | 90,731,552 | rs2232154 | G/T | 46 | 1 | 0 | 0.011 | |||
| −570 | 90,731,562 | G/A | 44 | 1 | 0 | 0.011 | ||||
| −561 | 90,731,571 | G/A | 46 | 1 | 0 | 0.011 | ||||
| −449 | 90,731,683 | C/T | 47 | 1 | 0 | 0.010 | ||||
| −415 | 90,731,717 | C/T | 47 | 1 | 0 | 0.010 | ||||
| 5′ UTR | −225 | 90,731,907 | rs2232156 | C/T | 47 | 1 | 0 | 0.010 | ||
| −224 | 90,731,908 | rs2232157 | G/T | 29 | 11 | 3 | 0.198§ | |||
| −94 | 90,732,038 | rs2232158 | G/C | 21 | 19 | 6 | 0.337§ | |||
| Exon 1 | 190 | 90,732, 322 | G/A | 64 | Ala/Thr | 47 | 1 | 0 | 0.010 | |
| 265–281 | 90,732,396–90,732,412 | 4 repeats/5 repeats | 89–92 | CCG (Pro) | 13 | 22 | 9 | 0.455 | ||
| 408 | 90,732,539 | C/T | 136 | Asn/Asn | 45 | 1 | 0 | 0.011 | ||
| 540 | 90,732,671 | A/G | 180 | Leu/Leu | 42 | 4 | 0 | 0.043 | ||
| 1168 | 90,733,299 | G/A | 390 | Ala/Thr | 47 | 1 | 0 | 0.010 | ||
| 1368 | 90,733,499 | C/T | 456 | Ala/Ala | 47 | 1 | 0 | 0.010 | ||
| 1490 | 90,733,621 | T/G | 497 | Val/Gly | 47 | 1 | 0 | 0.010 | ||
| 1749 | 90,733,880 | rs2232161 | C/T | 583 | His/His | 45 | 1 | 1 | 0.032 | |
| 2040 | 90,734,171 | rs2232163 | G/A | 680 | Arg/Arg | 43 | 5 | 0 | 0.052§ | |
| 3′ UTR | 2636 | 90,734,767 | rs3750145 | T/C | 46 | 2 | 0 | 0.021 | ||
| 2841 | 90,734,972 | C/T | 47 | 1 | 0 | 0.010 | ||||
| 3536 | 90,735,667 | rsl052015 | A/C | 45 | 3 | 0 | 0.031 | |||
| 3687 | 90,735,818 | A/- | 46 | 2 | 0 | 0.021 | ||||
| 3770 | 90,735,901 | rsl0282617 | A/G | 47 | 1 | 0 | 0.010 | |||
| 3′ flanking region | 4095–4096 | 90,736,226-90,736,227 | AA/− | 40 | 6 | 0 | 0.065 | |||
* Base pair location is reported using the translation start site as a reference.
† Based on March 2006 human reference sequence (NCBI build 36.1).
‡ 1, major allele; 2, minor allele.
§ Common variation (minor allele frequency > 0.05) discovered and selected for genotyping in the population sample.
In the larger population sample, the observed MAFs were 21% for rs2232157, 29% for rs2232158, and 9% for rs2232163, which was similar to that observed in the sequencing panel. All three polymorphisms met the expectations of Hardy-Weinberg equilibrium. Linkage disequilibrium was assessed between rs2232157 and rs2232158 (r 2 = 0.668; D′ = 0.993), rs2232157 and rs2232163 (r 2 = 0.024; D′ = 0.932), and rs2232158 and rs2232163 (r 2 = 0.038; D′ = 0.959).
On average, the 1084 genotyped men were 60 yr old (range, 40–88 yr), 174 cm tall (range, 156–192 cm), and weighed 83 kg (range, 45–127 kg). There was no association between any of the three polymorphisms genotyped and height or body weight and therefore height and weight were not adjusted for in subsequent analyses.
There was a nominally significant association with femoral neck BMD and the two common promoter variants, rs2232157 and rs2232158 (Table 2). Men with the less common genotypes had 3–4% lower femoral neck BMD. This result was significant for rs2232158 but not rs223157 after correction for multiple testing (corrected p = 0.024 and p = 0.111, respectively). Further study of the two determinants of areal BMD (i.e., BMC and CSA) indicated that the association with areal BMD for both promoter SNPs seems to be driven by a nonsignificant decrease in BMC among those with the less common genotype and a borderline significant increase in CSA among carriers of the minor allele. The coding SNP, rs2232163, was not associated with femoral neck BMD, BMC, or CSA.
Table 2.
Association of Common Variation in FZD1 with DXA Measures at the Total Hip and Femoral Neck in Afro-Caribbean Men
| rs2232157 |
Adjusted mean* (SE) |
p† |
|||
| GG (N = 670) | GT (N = 355) | TT (N = 55) | Additive | Dominant | |
| Total hip BMD (g/cm2) | 1.15 (0.006) | 1.14 (0.008) | 1.16 (0.019) | 0.522 | |
| Femoral neck BMD (g/cm2) | 0.99 (0.005) | 0.97 (0.007) | 0.96 (0.018) | 0.041 | |
| BMC (g) | 5.29 (0.03) | 5.28 (0.042) | 5.20 (0.106) | 0.629 | |
| CSA (cm2) | 5.37 (0.014) | 5.43 (0.019) | 5.39 (0.047) | 0.046 | |
| rs2232158 | GG (N = 543) | GC (N = 427) | CC (N = 94) | ||
| Total hip BMD (g/cm2) | 1.15 (0.006) | 1.15 (0.007) | 1.13 (0.015) | 0.254 | |
| Femoral neck BMD (g/cm2) | 0.99 (0.006) | 0.98 (0.007) | 0.95 (0.014) | 0.009 | |
| BMC (g) | 5.30 (0.033) | 5.31 (0.038) | 5.12 (0.081) | 0.168 | |
| CSA (cm2) | 5.37 (0.015) | 5.40 (0.017) | 5.44 (0.037) | 0.057 |
| rs2232163 | GG (N = 878) | GA and AA (N = 185) | |||||||
| Total hip BMD (g/cm2) | 1.15 (0.005) | 1.14 (0.010) | 0.380 | ||||||
| Femoral neck BMD (g/cm2) | 0.98 (0.005) | 0.97 (0.010) | 0.322 | ||||||
| BMC (g) | 5.29 (0.026) | 5.24 (0.057) | 0.399 | ||||||
| CSA (cm2) | 5.39 (0.012) | 5.39 (0.026) | 0.876 |
* Adjusted for age.
† p values are unadjusted for multiple comparisons.
CSA, cross-sectional area.
The association of these variants with DXA measures at the femoral neck prompted further study of these polymorphisms with more refined phenotypic measures of bone structural geometry made with pQCT. At the proximal radius, bone structural measures, but neither cortical nor trabecular vBMD, differed by genotype (Table 3). Compared with the GG genotype, the presence of the T allele for rs2232157 was associated with greater total BMC and larger CSA (unadj. p = 0.019 and 0.010; corrected p = 0.048 and 0.022, respectively). Similarly, those with the less common genotype for the rs2232158 polymorphism had a higher CSA and BMC than those with the more common genotype (unadj. p = 0.003 and p = 0.013; corrected p = 0.006 and p = 0.036, respectively). This displacement of bone mass outward resulted in a 2.1% greater SSI for those with the CC compared with the GG genotype at rs2232158 (unadj. p = 0.029), but this association did not persist after correction for multiple comparisons (corrected p = 0.070). In contrast to the findings at the radius, none of the polymorphisms were associated with pQCT traits at the tibia (Table 4).
Table 3.
Association of Common Variation in FZD1 With pQCT Measures at the Radius in Afro-Caribbean Men
| rs2232157 |
Adjusted mean* (SE) |
p† |
|||
| GG (N = 471) | GT (N = 258) | TT (N = 40) | Additive | Dominant | |
| Proximal site | |||||
| Cortical BMD (mg/cm3) | 1211 (1.1) | 1209 (1.4) | 1214 (3.6) | 0.770 | |
| Total BMC (mg/mm) | 147 (0.8) | 151 (1.1) | 150 (2.8) | 0.019 | |
| Total CSA (mm2) | 152 (1.0) | 158 (1.4) | 154 (3.5) | 0.010 | |
| Polar SSI (mm3) | 420 (3.6) | 438 (4.9) | 418 (12.3) | 0.062 | |
| Distal site | |||||
| Trabecular BMD (mg/cm3) | 208 (2.3) | 204 (3.1) | 207 (7.9) | 0.379 | |
| rs2232158 | GG (N = 380) | GC (N = 311) | CC (N = 62) | ||
| Proximal site | |||||
| Cortical BMD (mg/cm3) | 1211 (1.2) | 1209 (1.3) | 1212 (2.9) | 0.740 | |
| Total BMC (mg/mm) | 147 (0.9) | 151 (1.0) | 150 (2.2) | 0.013 | |
| Total CSA (mm2) | 151 (1.1) | 157 (1.2) | 156 (2.8) | 0.003 | |
| Polar SSI (mm3) | 419 (4.0) | 436 (4.4) | 428 (9.9) | 0.029 | |
| Distal site | |||||
| Trabecular BMD (mg/cm3) | 207 (2.6) | 207 (2.8) | 207 (6.3) | 0.998 | |
| rs2232163 | GG (N = 628) | GA and AA (N = 132) | |||||||
| Proximal site | |||||||||
| Cortical BMD (mg/cm3) | 1210 (0.9) | 1209 (2.0) | 0.670 | ||||||
| Total BMC (mg/mm) | 149 (0.7) | 145 (1.5) | 0.020 | ||||||
| Total CSA (mm2) | 155 (0.9) | 150 (1.9) | 0.036 | ||||||
| Polar SSI (mm3) | 429 (3.1) | 415 (6.8) | 0.062 | ||||||
| Distal site | |||||||||
| Trabecular BMD (mg/cm3) | 206 (2.0) | 207 (4.3) | 0.922 | ||||||
* Adjusted for age.
† p values are unadjusted for multiple comparisons.
CSA, cross-sectional area; SSI, strength-strain index.
Table 4.
Association of Common Variation in FZD1 With pQCT Measures at the Tibia in Afro-Caribbean Men
| rs2232157 |
Adjusted mean* (SE) |
p† |
|||
| GG (N = 471) | GT (N = 258) | TT (N = 40) | Additive | Dominant | |
| Proximal site | |||||
| Cortical BMD (mg/cm3) | 1177 (1.1) | 1177 (1.5) | 1177 (4.0) | 0.780 | |
| Total BMC (mg/mm) | 416 (2.5) | 416 (3.4) | 419 (8.8) | 0.999 | |
| Total CSA (mm2) | 507 (3.1) | 509 (4.2) | 514 (11.0) | 0.540 | |
| Polar SSI (mm3) | 2453 (19.4) | 2486 (26.1) | 2502 (68.2) | 0.262 | |
| Distal site | |||||
| Trabecular BMD (mg/cm3) | 233 (1.8) | 232 (2.4) | 234 (6.2) | 0.860 | |
| rs2232158 | GG (N = 380) | GC (N = 311) | CC (N = 62) | ||
| Proximal site | |||||
| Cortical BMD (mg/cm3) | 1177 (1.3) | 1178 (1.4) | 1176 (3.2) | 0.950 | |
| Total BMC (mg/mm) | 417 (2.8) | 417 (3.0) | 410 (7.0) | 0.586 | |
| Total CSA (mm2) | 508 (3.5) | 511 (3.8) | 504 (8.8) | 0.875 | |
| Polar SSI (mm3) | 2448 (21.4) | 2501 (23.5) | 2427 (54.2) | 0.467 | |
| Distal site | |||||
| Trabecular BMD (mg/cm3) | 233 (2.0) | 233 (2.2) | 232 (5.0) | 0.813 | |
| rs2232163 | GG (N = 628) | GA and AA (N = 132) | |||||||
| Proximal site | |||||||||
| Cortical BMD (mg/cm3) | 1177 (1.0) | 1174 (2.2) | 0.172 | ||||||
| Total BMC (mg/mm) | 418 (2.2) | 408 (4.7) | 0.064 | ||||||
| Total CSA (mm2) | 509 (2.7) | 504 (5.9) | 0.421 | ||||||
| Polar SSI (mm3) | 2474 (16.7) | 2408 (36.5) | 0.100 | ||||||
| Distal site | |||||||||
| Trabecular BMD (mg/cm3) | 233 (1.5) | 231 (3.4) | 0.549 | ||||||
* Adjusted for age.
† p values are unadjusted for multiple comparisons.
CSA, cross-sectional area; SSI, strength-strain index.
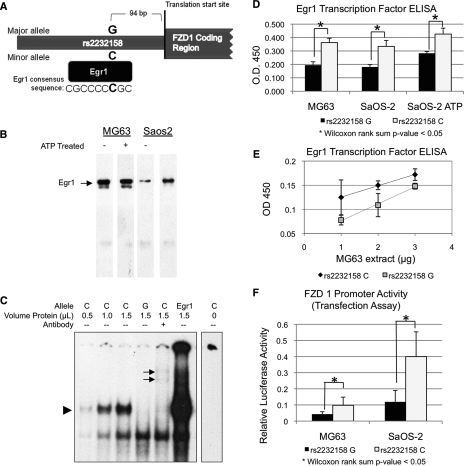
In silico prediction of transcription factor binding was completed using TESS for the sequence surrounding the two promoter polymorphisms (rs2232157 and rs2232158). This analysis showed that the Egr1 transcription factor is predicted to bind at the rs2232158 site only when the minor C allele is present. The sequence containing the G allele at rs2232158 is not predicted to bind Egr1 (Fig. 1A). The presence of Egr1 in the osteosarcoma cell lines, MG63 and SaOS-2, was next confirmed by Western blot (Fig. 1B). Although MG63 highly expressed Egr1, SaOS-2 expressed a lower level of Egr1 unless treated with ATP in agreement with Pines et al.(23)
FIG. 1.
Functional characterization of the FZD1 rs2232158 promoter polymorphism. (A) Schematic of the FZD1 5′ UTR region. Egr1 consensus sequence and location of the C allele of rs2232158 is shown. (B) Western blot analysis of Egr1 protein in MG63 and SaOS-2 cells. The cells were treated with or without ATP. The Egr1-specific band is indicated by an arrow. (C) Allelic-specific binding to the rs2232158-specific probes with recombinant Egr 1 (E5777; Sigma) using EMSA. Increasing Egr1-specific complex (see arrowhead) was detected for the C allele with increasing Egr1 recombinant protein concentration. The Egr1-specific binding was abolished with Egr1 antibody treatment, and super shift bands were observed (see arrows). Additionally, radiolabeled probe for the Egr1 consensus binding region was incubated with Egr1 recombinant protein and a strong Egr1-specific complex was observed. A faster migrating complex was also observed for all of the probes; however, no alteration in the band was observed with Egr1 antibody, indicating that this was a non–Egr1-specific complex. (D) Binding affinity for Egr1 to rs2232158 alleles. Transcription factor ELISA (Panomics) and biotinylated probes designed for each allele of rs2232158 (G allele in black and C allele in white) were used. Differences in binding were tested using the Wilcoxon rank sum test (*p < 0.05). (E) Binding affinity for Egr1 to rs2232158 alleles at various concentrations of nuclear extract (G allele in black and C allele in gray). (F) Activity of the rs2232158-specific FZD1 promoter. The promoter activity for different rs2232158 alleles were assessed by transfecting MG63 and SaOS-2 cell lines with luciferase reporter plasmids (G allele in black and C allele in white). Differences in activity were tested using the Wilcoxon rank sum test (*p < 0.05).
To assess whether the rs2232158 G-to-C polymorphism alters Egr1 binding affinity, an EMSA was completed using recombinant Egr1 protein (Fig. 1C). As the Egr1 protein increased (lanes 1–3), more Egr1-specific complex was detected for the rs2232158C probe. No Egr1-specific complex was detected for the G-specific probe (lane 4). The specificity of the Egr1 complex was confirmed by using a wildtype Egr1 probe (lane 6) and supershift with an Egr1 specific antibody (lane 5).
To investigate further the differential allele-specific Egr1 binding with rs2232158, a transcription factor ELISA was completed (Fig. 1D). This experiment showed higher Egr1 binding for the C allele compared with the G allele (p = 0.04). Specifically, a 1.9-, 1.8-, and 1.5-fold increase in Egr1 binding with the C allele was shown when using nuclear extract from the MG63-, SaOS-2-, and ATP-treated SaOS-2 cells, respectively. In addition, different concentrations of nuclear protein from MG63 cells were assessed for allele-specific binding of Egr1 to ensure specificity of the assay. The C allele had higher Egr1 binding for all concentrations of nuclear protein used (p = 0.04 for each condition). Specifically a 1.6-, 1.4-, and 1.2-fold higher binding was observed for the 1-, 2-, and 3-μg conditions, respectively (Fig. 1E).
Promoter activity was next assessed by transient expression of allele-specific FZD1 promoter reporter plasmids in MG63 and SaOS-2 cells (Fig. 1F). This luciferase reporter experiment showed a 2.3-fold higher FZD1 activity for the C allele construct relative to the G allele construct in MG63 cells and an ∼3.4-fold increase in untreated SaOS-2 cells. This finding was statistically significant in MG63 and SaOS-2 cells (p = 0.012 and p = 0.003, respectively).
DISCUSSION
We sequenced a 6.8-kb region encompassing the FZD1 gene and identified 39 polymorphisms including 3 common SNPs with minor allele frequency ≥0.05. Genetic association analysis of these common SNPs suggests that FZD1 promoter variation is associated with areal BMD at the femoral neck and cortical bone geometry at the proximal radius. Although there was no difference in volumetric BMD at the proximal radius, the difference in bone structural geometry at this skeletal site may impact long bone strength as reflected in the polar SSI. To our knowledge, our results are the first to show an association between FZD1 polymorphisms and bone-related phenotypes and further implicate the WNT signaling pathway in the regulation of bone health.
In silico functional analysis of rs2232158 suggested that the C allele creates an Egr1 transcription factor binding site, whereas the G allele does not. Consistent with the in silico analysis, the Egr1-specific complex was only detected in the C allele–specific probe and not the G probe in EMSA. The TF-ELISA experiments in osteoblast-like MG63 and SaOS-2 cells in vitro further confirmed the differential binding of the rs2232158 alleles to Egr1. FZD1 promoter activity in these two cell lines was also significantly increased for the C allele. Some binding was observed for the G allele–specific probe in the ELISA experiment, and this may have been because of an interaction of the Egr1 protein to other transcription factor(s) that directly bind to the probe. Further experiments will be necessary to test this hypothesis directly and to identify the other factors involved. Nonetheless, our results suggest a novel role of a FZD1 promoter variant in the Egr1-dependent regulation of FZD1 expression, long bone size, and biomechanical indices of bone strength.
To our knowledge, an influence of Egr1 on FZD1 expression has not been previously shown. The observation that the C allele of rs2232158 altered the Egr1-dependent transactivation of the FZD1 promoter is particularly interesting because of the known role of Egr1 in bone development. High Egr1 expression is detected in developing mouse bone, including the periosteal region of the developing long bones.(24) Moreover, Egr1 knockout mice have 10% lower areal BMD than wildtype mice with the same genetic background.(25) Egr1 is a transcription factor that is induced in osteoblasts by mechanical stress and ATP.(23) Mechanical stimulation causes ATP to be produced and released by osteoblast cells, and this ATP release may be a mechanism underlying the effect of skeletal loading on osteogenesis.(23,26) Mechanical loading also increases Wnt signaling in osteoblasts and increases the expression of several Wnt pathway components.(27) Therefore, it is plausible that Egr1 could mediate part of the previously described effect of skeletal loading on WNT pathway activation by upregulating Frizzled-1, the transmembrane co-receptor for WNT.
The two promoter variants, rs2232157 and rs2232158, are in strong linkage disequilibrium and consequently show similar patterns of association with bone phenotypes. It is possible that rs2232157 also influences FZD1 promoter activity, and further experiments are needed to assess this possibility. The synonymous SNP, rs2232163, was associated with cortical BMD and bone area but not with BMD at the femoral neck. Although this SNP is not predicted to be functional by in silico analysis, this has not been experimentally validated.
Although both DXA and pQCT indicated that the less common allele of the promoter variants is associated with larger bone size, there were differences in results between these measurement techniques for BMC and BMD. There are several possible explanations for the differences between the areal and volumetric BMD results. The different results for areal and volumetric BMD may be a consequence of the different measurement techniques. Areal BMD is likely to be underestimated in those with smaller bones, whereas QCT measures of vBMD are not influenced by bone size.(28) Indeed, our analysis suggests that the FZD1 promoter variants are associated with the external dimensions of cortical bone and bone CSA and not volumetric BMD at the proximal radius. The femoral neck and proximal radius are fundamentally different structures that are subjected to different weight-bearing and mechanical loading. Moreover, these skeletal sites have different amounts of cortical and cancellous bone. We found little evidence for association between FZD1 variants and trabecular BMD in this analysis. Thus, it is plausible that the effect of FZD1 may be different at the femoral neck and proximal radius skeletal sites.
This analysis was limited to Afro-Caribbean men, and our findings may not be generalizable to other populations. Furthermore, the technology to determine volumetric BMD measures at the femoral neck was not available in Tobago, so the differences between areal and volumetric findings cannot be examined at the same skeletal site. Future association studies of FZD1 variation and both areal and volumetric measurements of the central skeleton would be of interest. Furthermore, our study was designed to investigate common variation in FZD1 and bone phenotypes, and we cannot exclude the possibility that less common variants within FZD1 are also associated with bone phenotypes. Finally, we examined a narrow region encompassing the FZD1 transcript and the 5′ and 3′ regulatory regions and thus cannot exclude the likelihood that more distal regulatory variants are also associated with bone phenotypes.
Our study also has a number of methodological strengths. Both volumetric and areal BMD were measured. Few genetic association studies to date have included QCT measures of bone-related phenotypes. In addition, this analysis provided a comprehensive examination of the role of common genetic variation in FZD1 and bone phenotypes by identifying all common variants by sequence analysis, assessing association with refined skeletal measures, and assessing the potential molecular mechanisms underlying SNP associations of interest in an in vitro osteoblast model. This strategy of using in vitro models has previously been used to study the role of an Sp1 binding site polymorphism in the COLIA1 gene.(29)
In conclusion, WNT signaling is important in skeletal development and bone metabolism, and several genes in this pathway have been implicated in BMD regulation. The current genetic association analysis provides evidence that, in addition to LRP5, another member of the WNT pathway, FZD1, may contribute to the genetic regulation of bone mass and geometry. Moreover, our functional analysis also supports a role of promoter variation in the regulation of FZD1 activity in osteoblast-like cells, possibly through increased Egr1 transcription factor binding.
ACKNOWLEDGMENTS
This study was supported, in part, by Grant R01-AR049747 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases and funding from the National Osteoporosis Foundation and Western Pennsylvania Arthritis Foundation. SPM and LMY were supported by National Institute of Aging Grant T32-AG00181.
Footnotes
The authors state that they have no conflicts of interest.
REFERENCES
- 1.Baron R, Rawadi G, Roman-Roman S. Wnt signaling: A key regulator of bone mass. Curr Top Dev Biol. 2006;76:103–127. doi: 10.1016/S0070-2153(06)76004-5. [DOI] [PubMed] [Google Scholar]
- 2.Hartmann C. A Wnt canon orchestrating osteoblastogenesis. Trends Cell Biol. 2006;16:151–158. doi: 10.1016/j.tcb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: Arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 6.Umbhauer M, Djiane A, Goisset C, Penzo-Mendez A, Riou JF, Boucaut JC, Shi DL. The C-terminal cytoplasmic Lys-thr-X-X-X-Trp motif in frizzled receptors mediates Wnt/beta-catenin signalling. EMBO J. 2000;19:4944–4954. doi: 10.1093/emboj/19.18.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao SC, Eustace B, Lappe MM, Spitzer L, Zweier S, Braunschweiger K, Benchekroun Y, Hu X, Adair R, Chee L, FitzGerald MG, Tulig C, Caruso A, Tzellas N, Bawa A, Franklin B, McGuire S, Nogues X, Gong G, Allen KM, Anisowicz A, Morales AJ, Lomedico PT, Recker SM, Van Eerdewegh P, Recker RR, Johnson ML. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, Arslan-Kirchner M, Batch JA, Beighton P, Black GC, Boles RG, Boon LM, Borrone C, Brunner HG, Carle GF, Dallapiccola B, De Paepe A, Floege B, Halfhide ML, Hall B, Hennekam RC, Hirose T, Jans A, Juppner H, Kim CA, Keppler-Noreuil K, Kohlschuetter A, LaCombe D, Lambert M, Lemyre E, Letteboer T, Peltonen L, Ramesar RS, Romanengo M, Somer H, Steichen-Gersdorf E, Steinmann B, Sullivan B, Superti-Furga A, Swoboda W, van den Boogaard MJ, Van Hul W, Vikkula M, Votruba M, Zabel B, Garcia T, Baron R, Olsen BR, Warman ML. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 9.Ferrari SL, Deutsch S, Choudhury U, Chevalley T, Bonjour JP, Dermitzakis ET, Rizzoli R, Antonarakis SE. Polymorphisms in the low-density lipoprotein receptor-related protein 5 (LRP5) gene are associated with variation in vertebral bone mass, vertebral bone size, and stature in whites. Am J Hum Genet. 2004;74:866–875. doi: 10.1086/420771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koller DL, Ichikawa S, Johnson ML, Lai D, Xuei X, Edenberg HJ, Conneally PM, Hui SL, Johnston CC, Peacock M, Foroud T, Econs MJ. Contribution of the LRP5 gene to normal variation in peak BMD in women. J Bone Miner Res. 2005;20:75–80. doi: 10.1359/JBMR.041019. [DOI] [PubMed] [Google Scholar]
- 11.van Meurs JB, Trikalinos TA, Ralston SH, Balcells S, Brandi ML, Brixen K, Kiel DP, Langdahl BL, Lips P, Ljunggren O, Lorenc R, Obermayer-Pietsch B, Ohlsson C, Pettersson U, Reid DM, Rousseau F, Scollen S, Van Hul W, Agueda L, Akesson K, Benevolenskaya LI, Ferrari SL, Hallmans G, Hofman A, Husted LB, Kruk M, Kaptoge S, Karasik D, Karlsson MK, Lorentzon M, Masi L, McGuigan FE, Mellstrom D, Mosekilde L, Nogues X, Pols HA, Reeve J, Renner W, Rivadeneira F, van Schoor NM, Weber K, Ioannidis JP, Uitterlinden AG. Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. JAMA. 2008;299:1277–1290. doi: 10.1001/jama.299.11.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, Wilson SG, Andrew T, Falchi M, Gwilliam R, Ahmadi KR, Valdes AM, Arp P, Whittaker P, Verlaan DJ, Jhamai M, Kumanduri V, Moorhouse M, van Meurs JB, Hofman A, Pols HA, Hart D, Zhai G, Kato BS, Mullin BH, Zhang F, Deloukas P, Uitterlinden AG, Spector TD. Bone mineral density, osteoporosis, and osteoporotic fractures: A genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zilberberg A, Yaniv A, Gazit A. The low density lipoprotein receptor-1, LRP1, interacts with the human frizzled-1 (HFz1) and down-regulates the canonical Wnt signaling pathway. J Biol Chem. 2004;279:17535–17542. doi: 10.1074/jbc.M311292200. [DOI] [PubMed] [Google Scholar]
- 14.Malbon CC. Frizzleds: New members of the superfamily of G-protein-coupled receptors. Front Biosci. 2004;9:1048–1058. doi: 10.2741/1308. [DOI] [PubMed] [Google Scholar]
- 15.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoang BH, Kubo T, Healey JH, Sowers R, Mazza B, Yang R, Huvos AG, Meyers PA, Gorlick R. Expression of LDL receptor-related protein 5 (LRP5) as a novel marker for disease progression in high-grade osteosarcoma. Int J Cancer. 2004;109:106–111. doi: 10.1002/ijc.11677. [DOI] [PubMed] [Google Scholar]
- 17.Hill DD, Cauley JA, Sheu Y, Bunker CH, Patrick AL, Baker CE, Beckles GL, Wheeler VW, Zmuda JM. Correlates of bone mineral density in men of African ancestry: The Tobago Bone Health Study. Osteoporos Int. 2008;19:227–234. doi: 10.1007/s00198-007-0450-9. [DOI] [PubMed] [Google Scholar]
- 18.Bunker CH, Zmuda JM, Patrick AL, Wheeler VW, Weissfeld JL, Kuller LH, Cauley JA. High bone density is associated with prostate cancer in older Afro-Caribbean men: Tobago prostate survey. Cancer Causes Control. 2006;17:1083–1089. doi: 10.1007/s10552-006-0047-1. [DOI] [PubMed] [Google Scholar]
- 19.Miljkovic-Gacic I, Ferrell RE, Patrick AL, Kammerer CM, Bunker CH. Estimates of African, European and Native American ancestry in Afro-Caribbean men on the island of Tobago. Hum Hered. 2005;60:129–133. doi: 10.1159/000089553. [DOI] [PubMed] [Google Scholar]
- 20.Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 21.Schiessl H, Ferretti J, Tysarczyk-Niemeyer G, Willnecker J. Noninvasive bone strength index as analyzed by peripheral quantitative computed tomography (pQCT) In: Schoneau E, editor. Paediatric Osteology: New Developments in Diagnostics and Therapy. New York, NY, USA: Elsevier; 1996. pp. 141–146. [Google Scholar]
- 22.Ferretti JL, Capozza RF, Zanchetta JR. Mechanical validation of a tomographic (pQCT) index for noninvasive estimation of rat femur bending strength. Bone. 1996;18:97–102. doi: 10.1016/8756-3282(95)00438-6. [DOI] [PubMed] [Google Scholar]
- 23.Pines A, Romanello M, Cesaratto L, Damante G, Moro L, D'Andrea P, Tell G. Extracellular ATP stimulates the early growth response protein 1 (Egr-1) via a protein kinase C-dependent pathway in the human osteoblastic HOBIT cell line. Biochem J. 2003;373:815–824. doi: 10.1042/BJ20030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahon AP, Champion JE, McMahon JA, Sukhatme VP. Developmental expression of the putative transcription factor Egr-1 suggests that Egr-1 and c-fos are coregulated in some tissues. Development. 1990;108:281–287. doi: 10.1242/dev.108.2.281. [DOI] [PubMed] [Google Scholar]
- 25.Cenci S, Weitzmann MN, Gentile MA, Aisa MC, Pacifici R. M-CSF neutralization and egr-1 deficiency prevent ovariectomy-induced bone loss. J Clin Invest. 2000;105:1279–1287. doi: 10.1172/JCI8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romanello M, Pani B, Bicego M, D'Andrea P. Mechanically induced ATP release from human osteoblastic cells. Biochem Biophys Res Commun. 2001;289:1275–1281. doi: 10.1006/bbrc.2001.6124. [DOI] [PubMed] [Google Scholar]
- 27.Hens JR, Wilson KM, Dann P, Chen X, Horowitz MC, Wysolmerski JJ. TOPGAL mice show that the canonical Wnt signaling pathway is active during bone development and growth and is activated by mechanical loading in vitro. J Bone Miner Res. 2005;20:1103–1113. doi: 10.1359/JBMR.050210. [DOI] [PubMed] [Google Scholar]
- 28.Carter DR, Bouxsein ML, Marcus R. New approaches for interpreting projected bone densitometry data. J Bone Miner Res. 1992;7:137–145. doi: 10.1002/jbmr.5650070204. [DOI] [PubMed] [Google Scholar]
- 29.Grant SF, Reid DM, Blake G, Herd R, Fogelman I, Ralston SH. Reduced bone density and osteoporosis associated with a polymorphic Sp1 binding site in the collagen type I alpha 1 gene. Nat Genet. 1996;14:203–205. doi: 10.1038/ng1096-203. [DOI] [PubMed] [Google Scholar]