Abstract
Overexpression of the ErbB2 receptor tyrosine kinase is prevalent in approximately 30% of human breast cancers and confers Taxol resistance. Our previous work has demonstrated that ErbB2 inhibits Taxol induced apoptosis in breast cancer cells by transcriptionally upregulating p21Cip1. However, the mechanism of ErbB2-mediated p21Cip1 upregulation is unclear. Here we show that ErbB2 upregulates p21Cip1 transcription through increased Src activity in ErbB2 overexpressing cells. Src activation further activated STAT3 that recognizes an SIE binding site on the p21Cip1 promoter required for ErbB2-mediated p21Cip1 transcriptional upregulation. Both Src and STAT3 inhibitors restored Taxol sensitivity in resistant ErbB2 overexpressing breast cancer cells. Our data suggest that ErbB2 overexpression can activate STAT3 through Src leading to transcriptional upregulation of p21Cip1 that confers Taxol resistance of breast cancer cells. Our study suggests a potential clinical application of Src and STAT3 inhibitors in Taxol sensitization of ErbB2 overexpressing breast cancers.
Keywords: ErB2, STAT3, Src, p21Cip1, breast cancer
Introduction
Overexpression of p185ErbB2 (ErbB2, HER2, neu) occurs in approximately 20−30% of breast cancer patients and results in increased incidence of metastasis and chemoresistance (1, 2). Previously, we and others have demonstrated that ErbB2 overexpression confers paclitaxel (Taxol) resistance in breast cancer cells (3, 4). Our further studies demonstrated that resistance occurs through ErbB2 direct phosphorylation of the cyclin dependent kinase p34Cdc2 in a kinase dependent manner and transcriptional upregulation of p21Cip1 in an ErbB2 kinase independent manner; however, the exact mechanism of how ErbB2 transcriptionally upregulates p21Cip1 is not yet defined (5, 6).
CDKN1A encodes a 21kDa protein, p21Cip1, known for its role as a cyclin dependent kinase inhibitor and p53 inducible gene (7-9). However, p21Cip1 upregulation can also occur through p53-independent mechanisms including growth factor stimulation (10). p21Cip1 partially governs the cell cycle at multiple checkpoints including the G1/S and the G2/M transitions (11). Increased levels of p21Cip1 at the G2/M transition result in a mitotic delay through the binding and inactivation of the Cdc2/Cyclin B complex (6). In addition, increased p21Cip1 levels may reduce Taxol sensitivity (12).
The p21Cip1 promoter contains multiple transcription factor binding sites including SIE sequences, which are consensus binding sites for signal transducer and activator of transcription (STAT) proteins (9, 13). STAT proteins are a family of transcription factors found latent in the cytoplasm. Upon activation by both receptor and non-receptor tyrosine kinases (RTK), STATs are phosphorylated and translocated to the nucleus, thereby activating downstream genes (14). Several STAT proteins are reported to have an oncogenic role (15). Particularly, STAT3 has been shown to be persistently activated in multiple human carcinomas, including breast cancers (15, 16).
We, along with others, have demonstrated that ErbB2 activates STATs and can signal through STAT responsive elements (17, 18). It has been previously reported that ErbB2 and STAT3 may not directly interact; however, ErbB2 mediated activation of STAT3 may occur through the participation of other non-RTKs (19). Our previous studies have shown that ErbB2 overexpression leads to a Src-ErbB2 constitutive association and elevated activated Src levels (20, 21). We therefore investigated whether ErbB2 mediated Src activation may regulate activation and downstream signaling of STAT3 in breast cancer cells and whether activation of this pathway is critical for ErbB2 transcriptional upregulation of p21Cip1 and ErbB2-mediated chemoresistance. In our study, we provide evidence to demonstrate that STAT3 activation, through ErbB2 and Src indeed led to transcriptional upregulation of p21Cip1 and targeting either STAT3 or Src kinase sensitized ErbB2 overexpressing breast cancer cells to Taxol.
Results
Hyper-activated STAT3 binds to the STAT- SIE on the p21Cip1 promoter in ErbB2 overexpressing cells
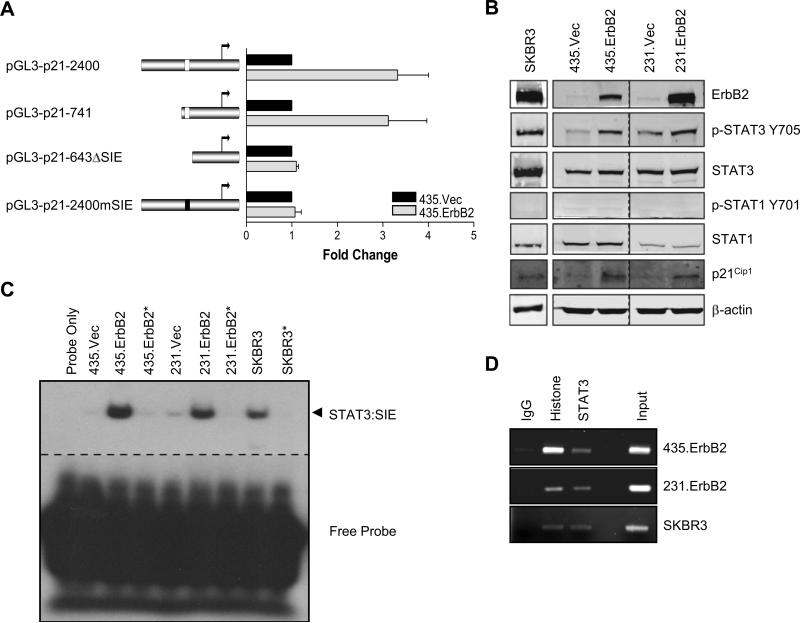
To investigate the mechanism of ErbB2-mediated p21Cip1 transcriptional upregulation, we examined the 2.4 kb promoter of p21Cip1. Among the transcription factor binding sites, we focused on the SIE1 sequence, previously identified as a STAT1/3 binding site, located at −679 to −669 base pairs, because we previously have shown that ErbB2 overexpression can lead to STAT3 activation (18). To directly test whether the STAT binding site on the p21Cip1 promoter was required for ErbB2 mediated transcriptional upregulation, we generated four different p21Cip1 promoter driven luciferase reporters with or without STAT binding site mutations: a full length 2.4 kb wild-type promoter (pGL3-p21−2400), a 5’ truncated promoter containing the SIE1 site (pGL3-p21−741), a 5’ truncated promoter with the SIE1 sequence deleted (pGL3-p21−643ΔSIE), and a full length promoter with SIE1 sequence mutations (pGL3-p21−2400mSIE). We transiently transfected each reporter construct into MDA-MB-435 human breast cancer cells stably overexpressing ErbB2 (435.ErbB2) or control vector (435.Vec) and measured the luciferase activity. Increased luciferase activities were detected in ErbB2 high-expressing 435.ErbB2 cells transfected with the pGL3-p21−2400 and pGL3-p21−741 promoters. In contrast, there was no increase in luciferase activity when 435.ErbB2 cells were transfected with pGL3-p21−643ΔSIE and pGL3-p21−2400mSIE (Fig. 1A), indicating that the SIE1 site is necessary for ErbB2-mediated p21 transcriptional upregulation. Next, to assure STAT proteins are activated by ErbB2 overexpression and contribute to p21 upregulation, we detected phosphorylated STAT3 and STAT1 in a panel of breast cancer cell lines (Fig. 1B). SKBR3, an endogenously overexpressing ErbB2 breast cancer cell line, had increased phosphorylation of STAT3 but not STAT1. Similarly, the ErbB2 transfectants 435.ErbB2 and 231.ErbB2 showed increased levels of phosphorylated STAT3 but not STAT1 compared to the ErbB2 low expressing 435.Vec and 231.Vec cells. Furthermore, increased STAT3 phosphorylation in ErbB2 overexpressing cells correlated with increased p21Cip1 protein levels (Fig. 1B). These data suggest that STAT3 is activated in ErbB2 overexpressing cells and activation of STAT3 may play a role in ErbB2 mediated p21Cip1 upregulation.
FIGURE 1.
Constitutive activation of STAT3 in ErbB2 overexpressing cells results in p21Cip1 transcriptional upregulation. A. left, schematic diagram of the 5’ promoter region of the p21 gene. White boxes represent STAT SIE location on promoter; black box represents mutated SIE. right, Luciferase activity of extracts prepared from 435.Vec and 435.ErbB2 cells transfected with the indicated p21Cip1 promoter reporter constructs. Bars indicate luciferase activity standardized to vector control for each construct. Error bars represent standard deviation (S.D.) B. Lysates from SKBR3, along with MDA-MB--435 and MDA-MB-231 breast cancer cells stably transfected with either control vector or wild-type ErbB2 were analyzed by Western blotting with the indicated antibodies. C. Nuclear extracts were collected from ErbB2 stable transfectants and vector control cells and SKBR3 cells for Electrophoretic Mobility Shift Assay (EMSA) analysis. Arrow indicates STAT protein:DNA complexes. Asterisk indicates addition of non-radiolabeled competitor probe. D. Chromatin Immunoprecipitation (ChIP assay) for SIE p21Cip1 promoter sequence using antibodies against histone and STAT3 in ErbB2 overexpressing cells. Histone and IgG immunoprecipitation served as positive and negative controls, respectively. Input represents 10% of the total.
STAT proteins (STAT1 and STAT3) have been shown to bind to the SIE in the p21 promoter (13, 22). To determine whether activated STAT3 in ErbB2 high-expressing cells may have increased binding to the SIE1 region on the p21Cip1 promoter, we performed an electrophoretic mobility shift assay (EMSA). Compared to ErbB2 low expressing cells with low STAT3 activity, elevated STAT:SIE1 DNA binding to the −685 to −665 bp region of the p21Cip1 promoter was detected in the ErbB2 overexpressing breast cancer cell lines (435.ErbB2, 231.ErbB2, and SKBR3) with activated STAT3 (Fig. 1C). To determine whether STAT3 bound to the p21Cip1 promoter in these cells in vivo, we performed a chromatin immunoprecipitation (ChIP) assay. Chromatin fragments were immunoprecipitated from ErbB2 high-expressing cell lines using anti-rabbit IgG, anti-histone, or anti-STAT3 antibodies. PCR of co-precipitated DNA using primers corresponding to the SIE1 site of p21Cip1 promoter detected signals from the anti-histone positive control and anti-STAT3 precipitated DNA, but not from anti-rabbit IgG (Fig. 1D). These data indicated that high ErbB2 expression results in increased STAT3:SIE1 DNA binding activity, which correlated with increased p21Cip1 protein expression.
STAT3 is required for ErbB2-mediated p21Cip1 transcriptional upregulation
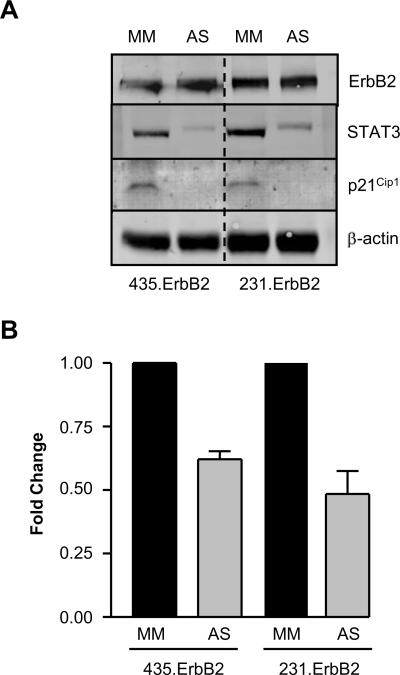
To evaluate whether STAT3 was required for p21Cip1 upregulation, we transfected ErbB2 high-expressing 435.ErbB2 and 231.ErbB2 cells with STAT3 Antisense (AS) or control Mismatch (MM) oligonucleotides. STAT3 protein levels were decreased in AS treated cells, which corresponded to decreased p21Cip1 protein levels (Fig. 2A). To further investigate whether inhibition of STAT3 expression led to reduced p21Cip1 expression at the transcriptional level, we treated ErbB2 high-expressing cells with STAT3 AS or MM oligonucleotides and co-transfected with the 2.4kb wild type p21 promoter reporter gene. Luciferase assays showed decreased p21Cip1 promoter activity by STAT3 AS treatment compared with MM treated control cells (Fig. 2B). These experiments revealed that both the STAT3 protein and the SIE1 binding site on the p21Cip1 promoter are required for ErbB2-mediated p21Cip1 transcriptional upregulation in breast cancer cells.
FIGURE 2.
p21Cip1 transcriptional activation by ErbB2 is dependant on STAT3 protein. A. 435.ErbB2 and 231.ErbB2 cells treated with 300 nM of Mismatch (MM) or STAT3 Antisense (AS) oligonucleotides. Cells were harvested 5 days post-transfection and analyzed by Western blot with the indicated antibodies. B. Standardized firefly luciferase activity of the wild type p21 promoter (pGL3-p21−2400) reporter plasmid in 435.ErbB2 and 231.ErbB2 cells treated with either STAT3 MM or AS oligonucleotides. Fold reduction was determined by standardizing to Mismatch control treated sample for each cell line. Error bars represent S.D.
ErbB2, Src, and STAT3 form a complex in ErbB2 overexpressing cells
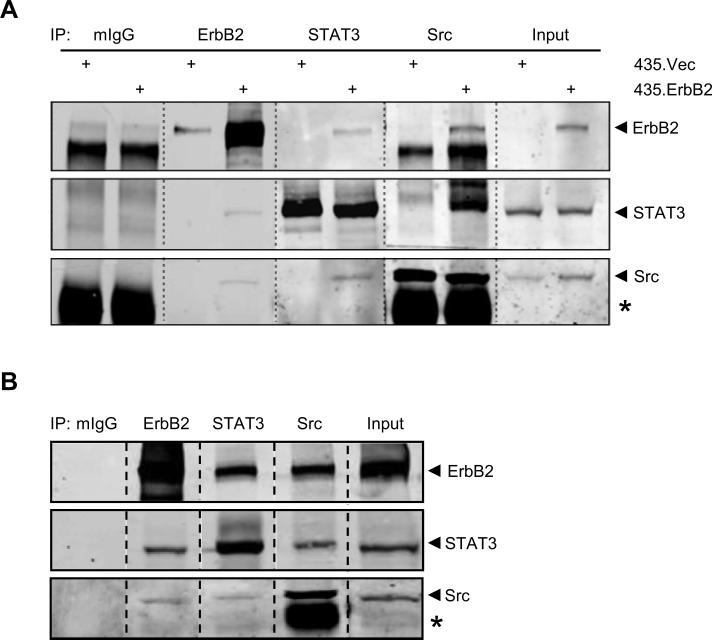
Our laboratory has shown that Src is activated and constitutively associated with the ErbB2 receptor in ErbB2 overexpressing breast cancer cells (20). Src has been implicated to activate STATs in mammary epithelial cells (23, 24). To investigate whether the activated Src in ErbB2 overexpressing breast cancer cells is involved in STAT3 activation, we first examined whether ErbB2, STAT3, and Src may co-exist in a heterocomplex. Immunoprecipitation of ErbB2, STAT3, or Src in 435.ErbB2 cells, followed by Western blotting of ErbB2, STAT3, and Src detected a heterocomplex containing the three proteins (Fig. 3A). A similar result was also seen in SKBR3 cells (Fig. 3B). The data suggests that formation of the signaling complex consisting of ErbB2, Src, and STAT3 in ErbB2 overexpressing cells may contribute to ErbB2 mediated upregulation of p21Cip1.
FIGURE 3.
ErbB2, Src, and STAT3 form a complex in ErbB2 overexpressing cells. Whole cell lysates from A. 435.ErbB2 or B. SKBR3 cells were immunoprecipitated (IP) with the indicated antibodies followed by immunoblot analysis for ErbB2, STAT3, and Src. Input lanes represent 10% of total protein immunoprecipitated. Asterisk indicates IgG heavy chain.
Src is required for p21Cip1 upregulation in ErbB2 overexpressing cells
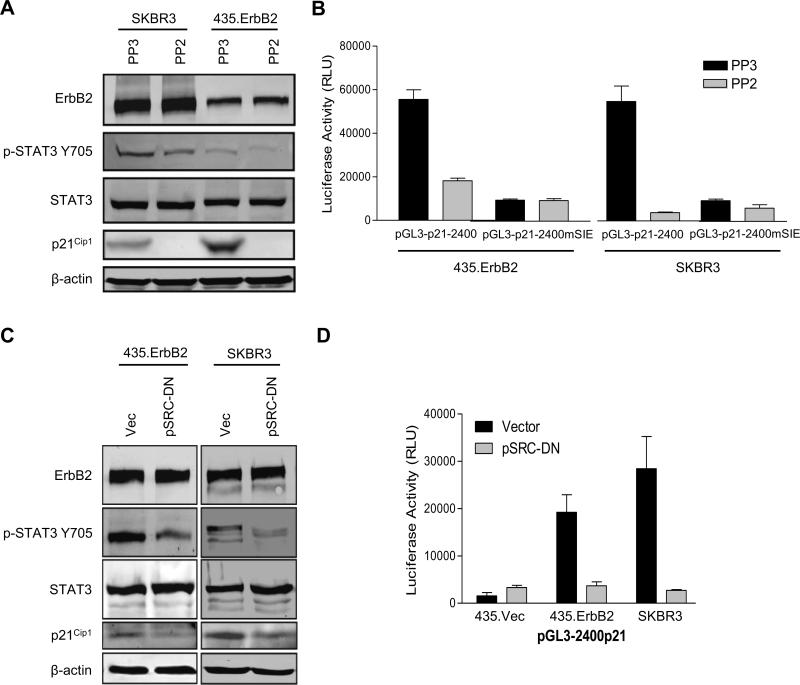
To determine whether ErbB2-mediated Src activation was required for ErbB2-mediated STAT3 activation and subsequent p21Cip1 upregulation, we treated ErbB2 overexpressing cells (SKBR3 and 435.ErbB2) with the Src family kinase inhibitor PP2 and examined STAT3 phosphorylation and p21Cip1 expression. Treatment of both SKBR3 and 435.ErbB2 cells with PP2 resulted in a decrease of STAT3 phosphorylation and reduction in p21Cip1 protein levels when compared to PP3 control treated cells (Fig. 4A). To determine whether PP2 treatment reduced p21 expression at the transcriptional level via inhibition of STAT3, we co-transfected the wild-type 2.4 kb p21 promoter (pGL3-p21−2400) or the SIE STAT3 binding site mutant p21 promoter (pGL3-p21−2400mSIE) driven luciferase reporters into 435.ErbB2 and SKBR3 cells that were treated with the Src inhibitor, PP2, or the control chemical, PP3. We found that PP2 repressed pGL3-p21−2400 promoter activity whereas PP2 had no effect on pGL3-p21−2400mSIE promoter activity (Fig. 4B). In addition to chemical inhibitors, we co-transfected 435.ErbB2 and SKBR3 cells with a dominant negative Src mutant (pSRC-DN) or empty vector (Vec) as control and repeated the assays described in Fig. 4A and B. Similar to PP2 treatment, pSRC-DN resulted in decreased phosphorylated STAT3, decreased total p21Cip1 protein levels, and reduced p21Cip1 promoter-driven luciferase activities as compared to vector control transfection in ErbB2 overexpressing cells (Fig. 4C and D).
FIGURE 4.
Src is required for ErbB2 mediated STAT3 activation and p21Cip1 upregulation. A. ErbB2 overexpressing cells were treated with 5 μM control (PP3) or Src inhibitor (PP2). Cell lysates were collected and analyzed by Western blotting. B. 435.ErbB2 and SKBR3 cells were transfected with the p21Cip1 reporter plasmids wild type (pGL3-p21−2400) or SIE mutant (pGL3-p21−2400mSIE) and treated with 5 μM PP3 or PP2 for 24 hours. Cell lysates were collected and luciferase activities were measured and standardized by transfection efficiency using renilla values. Error bars represent S.D. C. Indicated cells were transfected with either vector control (Vec) or dominant negative Src mutant (pSRC-DN) for 24 hours. Lysates were analyzed by Western blotting. D. Cells were co-transfected with vector control or pSrc-DN plasmids and the pGL3-p21−2400 reporter plasmid. Luciferase activities were measured as in (C).
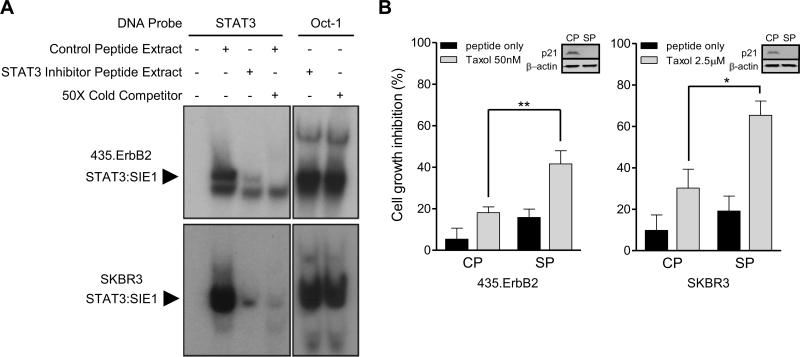
STAT3 inhibition restores Taxol sensitivity in ErbB2 overexpressing cells
Our results indicated that ErbB2 overexpressing breast cancer cells upregulate p21Cip1 through activation of STAT3. We have shown p21Cip1 upregulation is important for ErbB2 mediated Taxol resistance (6). Therefore, we examined whether inhibiting STAT3 DNA binding by a STAT3 inhibitory peptide would sensitize ErbB2 overexpressing breast cancer cells to Taxol treatment. To validate the efficacy of the STAT3 peptide inhibitor, we performed an EMSA using a STAT3 binding consensus sequence as a DNA probe and nuclear lysates collected from STAT3 inhibitor treated and control treated 435.ErbB2 (Fig. 5A, upper panel) and SKBR3 (Fig. 5A, lower panel) cells. We observed a dramatic decrease in STAT3:DNA binding in STAT3 inhibitor peptide treated cells, indicating that the STAT3 inhibitor effectively inhibited STAT3 binding to DNA (Fig. 5A). Next, ErbB2 overexpressing breast cancer cells were treated with either control peptide (CP) or STAT3 inhibitor peptide (SP). The treatment resulted in a decrease in p21Cip1 protein levels (Fig. 5B, inset). Both 435.ErbB2 and SKBR3 cell lines treated with STAT3 inhibitor peptide plus Taxol showed more effective inhibition of cell growth when compared to control peptide plus Taxol treated cells (Fig. 5B). Therefore, inhibition of STAT3 can sensitize ErbB2 overexpressing breast cancer cells to Taxol treatment.
FIGURE 5.
Inhibition of STAT3 sensitized ErbB2 overexpressing breast cancer cells to Taxol. A. 435.ErbB2 and SKBR3 cells were treated with 50 μM of STAT3 inhibitory peptide or control peptide for 24 hours. Nuclear extracts were then subjected to EMSA. Band shifts indicated a STAT3 protein:DNA complex. Oct-1 protein:DNA binding serves as an loading control. B. 435.ErbB2 and SKBR3 cells were treated with control (CP) or STAT3 inhibitor peptide (SP) (100 μM for 435.ErbB2 and 50 μM for SKBR3) for 12 hours. Cells were then treated with different concentrations of Taxol for 24 hours. Cell growth inhibition was determined by MTS assay. Error bars represent S.D. Inset: Representative Western blot for p21Cip1 and β-actin (loading control) from cells treated with either control (CP) or STAT3 inhibitor peptide (SP) treatment (** p = 0.004; * p = 0.016; t-test).
Src inhibition restores Taxol sensitivity in ErbB2 overexpressing cells
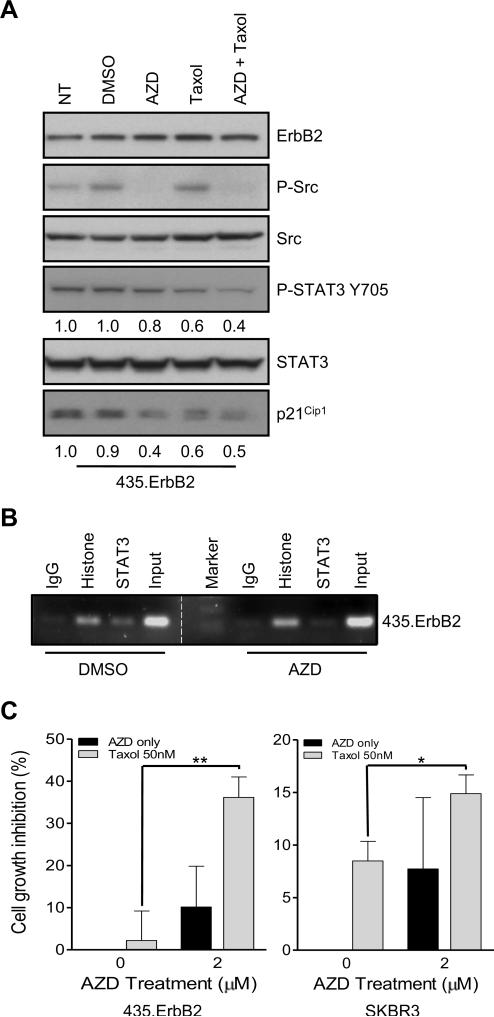
Since STAT3 is activated in ErbB2 overexpressing cells through Src activation, we investigated whether inhibiting Src activation may also sensitize ErbB2 overexpressing breast cancer cells to Taxol. 435.ErbB2 cells were treated with a clinically applicable Src inhibitor, AZD0530, or with DMSO solvent as a control (25). AZD0530 effectively inhibited Src phosphorylation without altering Src protein expression level (Fig. 6A). Furthermore, STAT3 phosphorylation was also reduced by AZD0530 treatment. Moreover, AZD0530 treatment reduced p21Cip1 expression (Fig. 6A). Similar results were also seen in SKBR3 cells (data not shown). To confirm Src inhibition by AZD0530 disrupted STAT3 binding to the p21Cip1 promoter, we performed a ChIP assay. PCR of STAT3 immunoprecipitated chromatin fragments indicated that AZD0530 treatment dramatically reduced STAT3 binding to the SIE site of the p21Cip1 promoter compared to DMSO treated cells (Fig. 6B). We then treated the cells with AZD0530, Taxol, or AZD0530 plus Taxol. We found that AZD0530 sensitized the cells to Taxol treatment in both 435.ErbB2 and SKBR3 cells (Fig. 6C). Thus, Src inhibition in combination with Taxol treatment can more effectively inhibit ErbB2 overexpressing breast cancer cell proliferation.
FIGURE 6.
Inhibition of Src kinase sensitized ErbB2 overexpressing breast cancer cells to Taxol. A. 435.ErbB2 cells were treated as indicated for 18 hours. Cell lysates were collected and analyzed by western blot analysis for the indicated antibodies. Numbers represent relative intensity of each band compared to the non-treated (NT) control. Total STAT3 was used as the loading control for both P-STAT3-Y705 and p21. B. 435.ErbB2 cells were treated with DMSO (control) or AZD0530 for 18 hours. Chromatin Immunoprecipitation (ChIP assay) for SIE p21Cip1 promoter sequence was performed using antibodies against histone and STAT3. IgG and Histone immunoprecipitation served as negative and positive controls, respectively. Input represents 10% of the total. C. 435.ErbB2 and SKBR3 cells were treated with DMSO only or AZD0530 (2 μM) for 12 hours. After 12 hours, cell media was replaced with medium containing either Taxol (50 nM) only, AZD0530 only or combination of Taxol and AZD0530. Cell growth inhibition was determined by MTS assay at 36 hours. Results were normalized to DMSO controls (** p = 0.002; * p = 0.013; t-test).
Discussion
Our data demonstrated that ErbB2 increases p21Cip1 levels by transcriptional upregulation. ErbB2 mediated transcriptional upregulation of target genes can occur through the increased activation of transcription factors, such as STATs (26-29). In our model system, high-expression of ErbB2 led to increased activated phosphorylation of STAT3 mediated by Src kinase down-stream of ErbB2. Mutation of the SIE1 STAT3 binding site in the p21Cip1 promoter decreased p21Cip1 promoter activity. Likewise, inhibition of STAT3 or Src kinase activity reduced p21 expression at the transcriptional level and sensitized ErbB2 overexpressing breast cancer cells to Taxol treatment. Our data demonstrated the requirement for STAT3 and Src activity in ErbB2-mediated p21Cip1 upregulation and therapeutic resistance.
Previous studies have shown that STAT3 activation by Src is required for Src-mediated transformation (30-32). Constitutive activation of STAT3 by overexpression of ErbB2 has also been reported (19, 29, 33-35); however, the role of ErbB2-mediated Src activation leading to STAT3 activation and subsequent p21Cip1 upregulation is unique to our study. Our data demonstrated that ErbB2, Src, and STAT3 are associated in a complex in ErbB2 overexpressing cells. Src association with STAT3 provides a previously unknown link between ErbB2 and STAT3 activation. Our data support a model whereby formation of this signaling complex consisting of ErbB2, Src, and STAT3 may drive constitutive expression of downstream targets.
Although the SIE1 site on the p21Cip1 promoter had originally been identified as a STAT1 binding site (13), others have reported that the SIE1 site on the p21Cip1 promoter may bind STAT3 (36-39). These differences could be a result of different cell backgrounds. In our system, both ChIP and Western blot experiments indicated that STAT3 bound the p21Cip1 promoter at the SIE1 site in ErbB2 overexpressing breast cancer cells. Furthermore, when STAT3 protein was decreased or inhibited, p21Cip1 expression and promoter activity was reduced. This indicated that STAT3 and the SIE1-STAT binding site are both required to drive transcription of the p21Cip1 gene, in response to ErbB2 overexpression.
Increased activation of STAT3 has been shown to lead to chemotherapeutic resistance due to STAT3 upregulation of Bcl-2, which resulted in apoptotic resistance of Taxol treated cells (40). The essential role of STAT3 in oncogenesis supports the use of STAT-targeting inhibitors in combination with conventional chemotherapy; however, clinically relevant STAT inhibitors are not available. An alternative target in this pathway is Src kinase, which has been well documented to be associated with increased phophosphorylation and activation of Stat3. Interestingly, in our model system, we observed a modest decrease in STAT3 phosphorylation after Src inhibition by AZD0530 treatment. A recent report demonstrated that another Src inhibitor, dasatinib, reduced STAT3 phosphorylation levels in only one of the lung cancer cell lines tested while phosphorylation of STAT3 was not affected in the other cell lines (41). It has also been shown that dasatinib treatment in multiple cancer cell lines led to reduced STAT3 phosphorylation levels initially; however, after long term treatment, STAT3 phosphorylation levels returned to basal levels even though Src was inhibited (42). This was explained by the activation of compensatory pathways (42). This may explain why AZD0530 as a single agent produced modest cell growth inhibition in our model system. Nevertheless, we have demonstrated AZD0530 treatment led to decreased STAT3 binding to the p21 promoter that corresponded with the sensitization to Taxol treatment. Together, our new findings showed that Src-mediated activation of STAT3 upregulated p21Cip1 and this pathway is involved in Taxol resistance of ErbB2 overexpressing cells. The findings further provide a strong rationale for using either STAT3 or Src inhibition to sensitize Taxol response, especially in ErbB2 overexpressing breast cancer cells.
In conclusion, our data provided evidence on the pivotal role of Src-mediated STAT3 activation and subsequent p21Cip1 upregulation in Taxol resistance of ErbB2 overexpressing breast cancer cells. Blockade of STAT3 DNA binding activity and inhibition of Src activation led to decreased p21Cip1 expression and restoration of Taxol sensitivity. Currently in the oncology field, combinational targeted therapy is an area of intense investigation (43-45). Since the Src inhibitor, AZD0530, is currently used in clinical trials (25), patients with Taxol resistant tumors due to deregulation of the ErbB2-Src-STAT3-p21Cip1 pathway may benefit from personalized cancer care using combination treatment of Taxol plus AZD0530 to target this pathway.
Materials and Methods
Cell Lines and Reagents
Human breast cancer cell lines MDA-MB-231 and SKBR3 were obtained from the American Type Culture Collection (Rockville, MD) and the MDA-MB-435 cell line was obtained from Dr. Janet Price, M. D. Anderson Cancer Center. There is evidence suggesting MDA-MB-435 is not a breast cancer derived cell line; however, we have shown that MDA-MB-435 is most likely a breast epithelial cell line that has undergone lineage infidelity (46, 47). The vector control and wild-type erbB2 stable transfectants, 435.Vec/435.ErbB2 were generated as previously described (48) and 231.Vec/231.ErbB2 was a generous gift from Dr. Patricia Steeg, NIH. All cells, except SKBR3, were grown in DMEM/F-12 50/50 medium and supplemented with 8% fetal bovine serum. SKBR3 cells were grown in McCoy's 5A medium supplemented with 10% fetal bovine serum and 5% L-Glutamine.
Inhibitors used include Src family kinase inhibitor 4-Amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine (PP2) or control 4-Amino-7-phenylpyrazol[3,4-d]pyrimidine (PP3) for a final concentration of 50 μM for 24 hours (Calbiochem, San Diego, CA).
The Src/Abl kinase inhibitor N-(5-Chloro-1,3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl)ethoxy]-5-(tetrahydro-2H-pyran-4-yloxy)quinazolin-4-amine (AZD0530) was a gift from AstraZeneca (Wilmington, DE).
Plasmid Constructs
The pGL3-p21−2400 plasmid was generated by digestion of the WWP-Luc plasmid (9, 49) to yield the 2.4 kb p21Cip1 promoter region that was cloned into pGL3-basic vector (Promega, Madison, WI). pGL3-p21−741 and pGL3-p21−643ΔSIE plasmid were made by PCR amplification of the pGL3-p21−2400 plasmid using primers 5'-AATTCTTCTGTTTCCCTGGAGATCA-3' and 5'-TTTTTGGCGTCTTCCATGGTGGCTTT-3', and 5'-CTTAAGTTCAGTGGACCTCAATTT-3' and 5'-TTTTTGGCGTCTTCCATGGTGGCTTT-3', respectively, and cloned into pGL3-basic vector. pGL3-p21−2400mSIE plasmid was constructed using the QuikChange Site-directed mutagenesis kit (Stratagene, La Jolla, CA) with primers 5’-CTCCAATTCCCTCCAAGCTTGAAGCATGTGACAATC-3’ and 5’-GATTGTCACATGCTTCAAGCTTGGAGGGAATTGGAG-3’. The plasmid pSrc-DN has been previously described (21).
Luciferase Assays
Cells were grown to 60% confluence in 6 well plates. Cells were then transfected with 0.5 μg of indicated plasmid and 0.05 μg of control renilla plasmid using Lipofectamine (Invitrogen, Carlsbad, CA). Luciferase assays were performed 24 hours post-transfection using the Dual-Luciferase Assay Reporter System (Promega, Madison, WI) according to manufactures protocol.
Western Blot and Immunoprecipitation Analysis
Total cell lysates were collected in IP lysis buffer (20 mM Tris pH 7.5, 150 mM, NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM sodium orthovanadate, protease inhibitor cocktail, 1 mM PMSF). Lysates were subjected to SDS-PAGE followed by transfer to nitrocellulose membranes. Membranes were incubated with the following antibodies: ErbB2 (Calbiochem); STAT1, STAT3, phospho-STAT3-pY705, anti-p21Cip1 (Cell Signaling Technology, Danvers, MA), anti-Src (Santa Cruz Technology, Santa Cruz, CA), β-actin (Sigma, St. Louis, MO). Secondary antibody detection utilized Alexa-Fluor secondary antibodies (Invitrogen Molecular Probes). Proteins were detected using the LI-COR Odyssey detection system (Lincoln, Nebraska).
For immunoprecipitation experiments, 500 μg of protein was pre-cleared with 30 μl Protein G-Agarose beads for 1 h at 4°C. Immunoprecipitation was performed using 5 μg of isotyped IgG control (Millipore, Billerica, MA) or the indicated primary antibody at 4°C overnight followed by incubation with 40 μl of Protein G-Agarose beads for 4 hours. Immune complexes were washed with lysis buffer and immunoprecipitates were separated by SDS-PAGE followed by Western blotting.
Electrophoretic Mobility Shift Assay
Cell membranes were lysed (10mM Hepes pH 7.9, 10mM KCl, 0.1mM EDTA, 0.4% NP-40) and nuclear extracts were prepared in nuclear extraction buffer (20mM Hepes pH 7.9, 0.4M NaCl, 1mM EDTA). Nuclear extracts (5 μg) were incubated in a reaction mixture containing a double stranded, 5’ end radiolabeled, oligonucleotide sequence derived from the SIE1 site on the p21Cip1 promoter, 5’-AGCTCCTTCCCGGAAGCATG-3’. Cold competitor reactions included 2 pmols of unlabeled double stranded probe that was incubated for 30 minutes at room temperature in a 50-fold molar excess to the radiolabeled probe. All reactions were then incubated for 15 minutes at 37°C. Reaction complexes were resolved on a 5 % nondenaturing polyacrylamide gel, dried, and exposed to film overnight at −80°C.
Chromatin Immunoprecipitation Assays (ChIP)
ChIP assays were performed based on the protocol from the ChIP assay kit (Millipore) with minor modifications. Briefly, soluble chromatin solutions from 1 % formaldehyde fixed and sonicated cells were pre-cleared and then immunoprecipitated with 5 μg isotyped IgG, Histone H1 (Santa Cruz Biotechnology) or STAT3 (Santa Cruz Biotechnology) antibodies overnight at 4°C. Immune complexes were precipitated with Protein Sepharose A beads (GE Healthcare, Pittsburgh, PA). Complexes were washed extensively, eluted, and proteins were removed.
Co-precipitated DNA (2 μL) was analyzed by PCR (30−35 cycles) using specific primers, 5’-AATTCTTCTGTTTCCCTGGAGATCA-3’ and 5’-AAATTGAGGTCCACTGAACTTAAG-3’, to the STAT3 binding region (SIE1) of the p21Cip1 promoter. PCR products were resolved on a 2 % TAE agarose gel stained with ethidium bromide.
STAT3 Antisense
Oligonucleotides against STAT3 were synthesized using phospho-isothiolate modifications (indicated by lowercase as follows: mismatch oligonucleotide 5’-aaaAAGAGGCCTGATTgccC-3’ and antisense oligonucleotide 5’-aaaAAGTGCCCAGATTgccC-3’. Cells were grown to 40 % confluence on day 1 and transfected twice (day 1 and 3) with 200 nM of either Mismatch or Antisense oligonucleotide in Serum-Free Opti-MEM medium using Lipofectamine Plus (Invitrogen). Cells were harvested in lysis buffer on day 5.
Taxol Sensitivity and Cell Proliferation Assay
SKBR3 and 435.ErbB2 cells were seeded in 96 well plates at a density of 5×103 cells/well in 100 μL culture medium. After 24 hours of adherence, medium was aspirated and replaced with medium containing control or STAT3 inhibitor peptide (Calbiochem) concentrations of 100 μM for 435.eB and 50 μM for SKBR3 continuing for additional 12 hours. Cells were then treated with different concentration of paclitaxel (Taxol) for 24 hours. For Src inhibition, cells were treated with 2PM of AZD0530 for 12 hours followed by Taxol treatment for 24 hours. Viable cells were determined by CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay kit according to the manufacturer's protocol (Promega).
Acknowledgements
We would like to thank Dr. Jun Yao for his valuable discussion and Nina T. Nguyen for her critical reading of the manuscript.
Footnotes
This work was supported by NIH Grants P30-CA 16672 (MDACC), PO1-CA0099031 project 4, 1RO1-CA109570, 1RO1CA119127, and P50 CA116199 project 4 (D. Yu). D. Yu is the Nylene Eckles Distinguished Professor in Breast Cancer Research.
References
- 1.Yu D, Hung MC. Role of erbB2 in breast cancer chemosensitivity. Bioessays. 2000;22(7):673–80. doi: 10.1002/1521-1878(200007)22:7<673::AID-BIES10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 2.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu protooncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 3.Witters LM, Santala SM, Engle L, Chinchilli V, Lipton A. Decreased response to paclitaxel versus docetaxel in HER-2/neu transfected human breast cancer cells. Am J Clin Oncol. 2003;26(1):50–4. doi: 10.1097/00000421-200302000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Yu D, Liu B, Jing T, et al. Overexpression of both p185c-erbB2 and p170mdr-1 renders breast cancer cells highly resistant to taxol. Oncogene. 1998;16(16):2087–94. doi: 10.1038/sj.onc.1201729. [DOI] [PubMed] [Google Scholar]
- 5.Tan M, Jing T, Lan KH, et al. Phosphorylation on tyrosine-15 of p34(Cdc2) by ErbB2 inhibits p34(Cdc2) activation and is involved in resistance to taxol-induced apoptosis. Mol Cell. 2002;9(5):993–1004. doi: 10.1016/s1097-2765(02)00510-5. [DOI] [PubMed] [Google Scholar]
- 6.Yu D, Jing T, Liu B, et al. Overexpression of ErbB2 blocks Taxol-induced apoptosis by upregulation of p21Cip1, which inhibits p34Cdc2 kinase. Mol Cell. 1998;2(5):581–91. doi: 10.1016/s1097-2765(00)80157-4. [DOI] [PubMed] [Google Scholar]
- 7.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–16. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Xiong Y, Beach D. Proliferating cell nuclear antigen and p21 are components of multiple cell cycle kinase complexes. Mol Biol Cell. 1993;4(9):897–906. doi: 10.1091/mbc.4.9.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Deiry WS, Harper JW, O'Connor PM, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54(5):1169–74. [PubMed] [Google Scholar]
- 10.Michieli P, Chedid M, Lin D, Pierce JH, Mercer WE, Givol D. Induction of WAF1/CIP1 by a p53-independent pathway. Cancer Res. 1994;54(13):3391–5. [PubMed] [Google Scholar]
- 11.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366(6456):701–4. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt M, Lu Y, Liu B, Fang M, Mendelsohn J, Fan Z. Differential modulation of paclitaxel-mediated apoptosis by p21Waf1 and p27Kip1. Oncogene. 2000;19(20):2423–9. doi: 10.1038/sj.onc.1203546. [DOI] [PubMed] [Google Scholar]
- 13.Chin YE, Kitagawa M, Su WC, You ZH, Iwamoto Y, Fu XY. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272(5262):719–22. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 14.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19(21):2468–73. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 15.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19(21):2474–88. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 16.Turkson J, Jove R. STAT proteins: novel molecular targets for cancer drug discovery. Oncogene. 2000;19(56):6613–26. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- 17.Karni R, Jove R, Levitzki A. Inhibition of pp60c-Src reduces Bcl-XL expression and reverses the transformed phenotype of cells overexpressing EGF and HER-2 receptors. Oncogene. 1999;18(33):4654–62. doi: 10.1038/sj.onc.1202835. [DOI] [PubMed] [Google Scholar]
- 18.Tan M, Lan KH, Yao J, et al. Selective inhibition of ErbB2-overexpressing breast cancer in vivo by a novel TAT-based ErbB2-targeting signal transducers and activators of transcription 3-blocking peptide. Cancer Res. 2006;66(7):3764–72. doi: 10.1158/0008-5472.CAN-05-2747. [DOI] [PubMed] [Google Scholar]
- 19.Ren Z, Schaefer TS. ErbB-2 activates Stat3 alpha in a Src- and JAK2-dependent manner. J Biol Chem. 2002;277(41):38486–93. doi: 10.1074/jbc.M112438200. [DOI] [PubMed] [Google Scholar]
- 20.Nagata Y, Lan KH, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6(2):117–27. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 21.Tan M, Li P, Klos KS, et al. ErbB2 promotes Src synthesis and stability: novel mechanisms of Src activation that confer breast cancer metastasis. Cancer Res. 2005;65(5):1858–67. doi: 10.1158/0008-5472.CAN-04-2353. [DOI] [PubMed] [Google Scholar]
- 22.Bellido T, O'Brien CA, Roberson PK, Manolagas SC. Transcriptional activation of the p21(WAF1,CIP1,SDI1) gene by interleukin-6 type cytokines. A prerequisite for their pro-differentiating and anti-apoptotic effects on human osteoblastic cells. J Biol Chem. 1998;273(33):21137–44. doi: 10.1074/jbc.273.33.21137. [DOI] [PubMed] [Google Scholar]
- 23.Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23(48):8017–23. doi: 10.1038/sj.onc.1208159. [DOI] [PubMed] [Google Scholar]
- 24.Olayioye MA, Beuvink I, Horsch K, Daly JM, Hynes NE. ErbB receptor-induced activation of stat transcription factors is mediated by Src tyrosine kinases. J Biol Chem. 1999;274(24):17209–18. doi: 10.1074/jbc.274.24.17209. [DOI] [PubMed] [Google Scholar]
- 25.Hennequin LF, Allen J, Breed J, et al. N-(5-chloro-1,3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl)ethoxy]-5- (tetrahydro-2H-pyran-4-yloxy)quinazolin-4-amine, a novel, highly selective, orally available, dual-specific c-Src/Abl kinase inhibitor. J Med Chem. 2006;49(22):6465–88. doi: 10.1021/jm060434q. [DOI] [PubMed] [Google Scholar]
- 26.Loureiro RM, Maharaj AS, Dankort D, Muller WJ, D'Amore PA. ErbB2 overexpression in mammary cells upregulates VEGF through the core promoter. Biochem Biophys Res Commun. 2005;326(2):455–65. doi: 10.1016/j.bbrc.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 27.Mendoza-Gamboa E, Siwak DR, Tari AM. The HER2/Grb2/Akt pathway regulates the DNA binding activity of AP-1 in breast cancer cells. Oncol Rep. 2004;12(4):903–8. [PubMed] [Google Scholar]
- 28.Galang CK, Garcia-Ramirez J, Solski PA, et al. Oncogenic Neu/ErbB-2 increases ets, AP-1, and NF-kappaB-dependent gene expression, and inhibiting ets activation blocks Neu-mediated cellular transformation. J Biol Chem. 1996;271(14):7992–8. doi: 10.1074/jbc.271.14.7992. [DOI] [PubMed] [Google Scholar]
- 29.Fernandes A, Hamburger AW, Gerwin BI. ErbB-2 kinase is required for constitutive stat 3 activation in malignant human lung epithelial cells. Int J Cancer. 1999;83(4):564–70. doi: 10.1002/(sici)1097-0215(19991112)83:4<564::aid-ijc20>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 30.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18(5):2553–8. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odajima J, Matsumura I, Sonoyama J, et al. Full oncogenic activities of v-Src are mediated by multiple signaling pathways. Ras as an essential mediator for cell survival. J Biol Chem. 2000;275(31):24096–105. doi: 10.1074/jbc.m001606200. [DOI] [PubMed] [Google Scholar]
- 32.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18(5):2545–52. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carraway KL, 3rd, Sweeney C. Co-opted integrin signaling in ErbB2-induced mammary tumor progression. Cancer Cell. 2006;10(2):93–5. doi: 10.1016/j.ccr.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 34.DeArmond D, Brattain MG, Jessup JM, et al. Autocrine-mediated ErbB-2 kinase activation of STAT3 is required for growth factor independence of pancreatic cancer cell lines. Oncogene. 2003;22(49):7781–95. doi: 10.1038/sj.onc.1206966. [DOI] [PubMed] [Google Scholar]
- 35.Guo W, Pylayeva Y, Pepe A, et al. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell. 2006;126(3):489–502. doi: 10.1016/j.cell.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 36.Barre B, Avril S, Coqueret O. Opposite regulation of myc and p21waf1 transcription by STAT3 proteins. J Biol Chem. 2003;278(5):2990–6. doi: 10.1074/jbc.M210422200. [DOI] [PubMed] [Google Scholar]
- 37.Giraud S, Bienvenu F, Avril S, Gascan H, Heery DM, Coqueret O. Functional interaction of STAT3 transcription factor with the coactivator NcoA/SRC1a. J Biol Chem. 2002;277(10):8004–11. doi: 10.1074/jbc.M111486200. [DOI] [PubMed] [Google Scholar]
- 38.Lu S, Shen KC, Wang Y, Brooks SC, Wang YA. Impaired hepatocyte survival and liver regeneration in Atm-deficient mice. Hum Mol Genet. 2005;14(20):3019–25. doi: 10.1093/hmg/ddi333. [DOI] [PubMed] [Google Scholar]
- 39.Sinibaldi D, Wharton W, Turkson J, Bowman T, Pledger WJ, Jove R. Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: role of activated STAT3 signaling. Oncogene. 2000;19(48):5419–27. doi: 10.1038/sj.onc.1203947. [DOI] [PubMed] [Google Scholar]
- 40.Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21(50):7611–8. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- 41.Song L, Morris M, Bagui T, Lee FY, Jove R, Haura EB. Dasatinib (BMS-354825) selectively induces apoptosis in lung cancer cells dependent on epidermal growth factor receptor signaling for survival. Cancer Res. 2006;66(11):5542–8. doi: 10.1158/0008-5472.CAN-05-4620. [DOI] [PubMed] [Google Scholar]
- 42.Johnson FM, Saigal B, Tran H, Donato NJ. Abrogation of signal transducer and activator of transcription 3 reactivation after Src kinase inhibition results in synergistic antitumor effects. Clin Cancer Res. 2007;13(14):4233–44. doi: 10.1158/1078-0432.CCR-06-2981. [DOI] [PubMed] [Google Scholar]
- 43.Lu CH, Wyszomierski SL, Tseng LM, et al. Preclinical testing of clinically applicable strategies for overcoming trastuzumab resistance caused by PTEN deficiency. Clin Cancer Res. 2007;13(19):5883–8. doi: 10.1158/1078-0432.CCR-06-2837. [DOI] [PubMed] [Google Scholar]
- 44.Hiscox S, Morgan L, Green T, Nicholson RI. Src as a therapeutic target in anti-hormone/anti-growth factor-resistant breast cancer. Endocr Relat Cancer. 2006;13(Supplement1):S53–9. doi: 10.1677/erc.1.01297. [DOI] [PubMed] [Google Scholar]
- 45.Storniolo AM, Pegram MD, Overmoyer B, et al. Phase I Dose Escalation and Pharmacokinetic Study of Lapatinib in Combination With Trastuzumab in Patients With Advanced ErbB2-Positive Breast Cancer. J Clin Oncol. 2008;26(20):3317–23. doi: 10.1200/JCO.2007.13.5202. [DOI] [PubMed] [Google Scholar]
- 46.Sellappan S, Grijalva R, Zhou X, et al. Lineage infidelity of MDA-MB-435 cells: expression of melanocyte proteins in a breast cancer cell line. Cancer Res. 2004;64(10):3479–85. doi: 10.1158/0008-5472.CAN-3299-2. [DOI] [PubMed] [Google Scholar]
- 47.Ross DT, Scherf U, Eisen MB, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24(3):227–35. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 48.Yu D, Liu B, Tan M, Li J, Wang SS, Hung MC. Overexpression of c-erbB-2/neu in breast cancer cells confers increased resistance to Taxol via mdr-1-independent mechanisms. Oncogene. 1996;13(6):1359–65. [PubMed] [Google Scholar]
- 49.Baker SJ, Markowitz S, Fearon ER, Willson JK. Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249(4971):912–5. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]