Abstract
A diverse array of infectious agents, including prions and certain neurotropic viruses, bind to the laminin receptor (LR), and this determines tropism to the CNS. Bacterial meningitis in childhood is almost exclusively caused by the respiratory tract pathogens Streptococcus pneumoniae, Neisseria meningitidis, and Haemophilus influenzae, but the mechanism by which they initiate contact with the vascular endothelium of the blood brain barrier (BBB) is unknown. We hypothesized that an interaction with LR might underlie their CNS tropism. Using affinity chromatography, coimmunoprecipitation, retagging, and in vivo imaging approaches, we identified 37/67-kDa LR as a common receptor for all 3 bacteria on the surface of rodent and human brain microvascular endothelial cells. Mutagenesis studies indicated that the corresponding bacterial LR-binding adhesins were pneumococcal CbpA, meningococcal PilQ and PorA, and OmpP2 of H. influenzae. The results of competitive binding experiments suggest that a common adhesin recognition site is present in the carboxyl terminus of LR. Together, these findings suggest that disruption or modulation of the interaction of bacterial adhesins with LR might engender unexpectedly broad protection against bacterial meningitis and may provide a therapeutic target for the prevention and treatment of disease.
Introduction
The 37-kDa laminin receptor (LR) precursor protein is highly conserved in eukaryotes. The precursor form is thought to mature into one or more homo- or heterodimeric 67-kDa dimeric forms that are present on eukaryotic cell membranes and interact with extracellular matrix components (1–4). Binding studies and crystal structure determination indicate that different LR domains bind the basement membrane components laminin, collagen, and elastin (1, 5). It is also known that expression of LR correlates with tumor invasiveness and metastasis (6). Recent attention has been drawn to the role of LR as a cellular target for infectious agents. LR on microvascular endothelial cells interacts with neurotropic viruses, including Sindbis virus (7), Dengue virus (8), adeno-associated virus (9), tick-borne encephalitis virus, and Venezuelan equine encephalitis virus (10). Additionally, LR binds the cellular prion protein PrP (11, 12) and the secreted cytotoxic necrotizing factor toxin of E. coli (13, 14). Thus, LR participates in diverse cell-cell and cell-pathogen interactions.
Historically, Streptococcus pneumoniae (pneumococcus), Haemophilus influenzae, and Neisseria meningitidis (meningococcus) have been the predominant causes of bacterial meningitis. While many bacteria can replicate in the bloodstream, this trio is noted for also binding to the endothelium of the blood-brain barrier (BBB) and crossing this barrier to gain access to the cerebrospinal fluid of the subarachnoid space. As a consequence of limited immune defenses in the CNS, rapid bacterial multiplication leads to meningitis and devastating neuronal damage.
Bacterial seeding of the CNS from the bloodstream requires 2 steps: bacteria must first adhere to the cerebral vascular endothelium and then circumvent its continuous tight junctions to penetrate the barrier. It is now known that pneumococcus, H. influenzae, and meningococcus share a common strategy for the second step of traversing endothelial cells: they carry surface-exposed phosphorylcholine that mediates binding to the human platelet-activating factor receptor (PAFr) (15–17). Phosphorylcholine on the bacterial surface is believed to act as a molecular mimic of the chemokine PAF. Binding of bacterial phosphorylcholine to PAFr results in activation of β-arrestin–mediated uptake of the bacteria into BBB cells, a crucial step for the development of pneumococcal meningitis, which does not occur in mice deficient in PAFr (18).
The convergent use of PAFr by these pathogens suggested to us that they might also share a strategy to mediate initial adherence to the BBB. Here we show that the unrelated pathogens pneumococcus, meningococcus, and H. influenzae bind to the endothelium of the BBB via the 37/67-kDa LR. Using in vitro binding and in vivo imaging in mice, we identified 37/67-kDa LR as a common receptor for all 3 bacteria on rodent and human brain microvascular endothelial cells. Mutagenesis studies indicated that the corresponding LR-binding adhesins were pneumococcal CbpA, meningococcal PilQ and PorA, and OmpP2 of H. influenzae. Competitive binding of the bacteria, purified adhesins, LR peptides and antibodies suggested a common LR-carboxyterminal recognition site.
Results
Interaction of bacterial meningeal pathogens with LR.
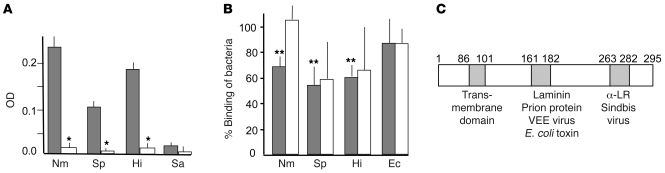
To determine whether pneumococcus, meningococcus, and H. influenzae bind LR, we first measured bacterial binding to immobilized recombinant LR (rLR) by ELISA. All 3 bacteria bound immobilized LR; in each case, binding was drastically reduced in the presence of soluble rLR (P < 0.001; Figure 1A). In contrast, Staphylococcus aureus, a species that does not seed the CNS from the bloodstream, failed to bind rLR (Figure 1A). Binding to LR was conserved among clinical isolates: 18 of 20 pneumococcal strains, 66 of 68 meningococcal strains, and all 39 H. influenzae strains (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI36759DS1).
Figure 1. Binding of pneumococcus, meningococcus, and H. influenzae to LR.
(A) Adhesion (OD405) of digoxigenin-labeled N. meningitidis MC58 (Nm), S. pneumoniae T4 (Sp), H. influenzae Rd (Hi), S. aureus NRS193 (Sa) to rLR (solid-phase antigen) after preincubation with BSA (gray bars) or soluble rLR (white bars). *P < 0.001 compared with adhesion without preincubation with soluble rLR; Student’s t test. Error bars indicate SD. (B) rBCEC6 cells were incubated for 30 minutes with 107 bacteria as indicated, and binding was quantified (100% value was set for each bacteria as the following CFU/ml bound: S. pneumoniae, 1.5 × 105; H. influenzae, 5 × 103; N. meningitidis, 5 × 103; E. coli [Ec] 1.9 × 104). Competitive inhibition of binding by anti-LR antibody (gray bars) or laminin (white bars) was quantified as a percentage of no treatment control (mean ± SD, n = 5). **P < 0.01 compared with wild-type; 1-way ANOVA. Results for a control irrelevant protein (albumin) and isotype-matched, irrelevant antibody (anti-vimentin) were not statistically different from those for no treatment control (data not shown). (C) Schematic representation of LR ligand binding domains. α-LR, anti-LR antibody; VEE, Venezuelan equine encephalitis virus; E. coli toxin, cytotoxic necrotizing factor–1. Numbers indicate amino acid residues.
Adherence of the 3 pathogens was then modeled using cerebral endothelial cells. We detected LR on the surface of mouse, rat, and human cerebral endothelial cells (Supplemental Figure 1A); because rodent cells lack endothelial CD46 targeted by meningococcus, the rat brain endothelial cell line rBCEC6 afforded a simpler system than human cells for studying adherence to LR. LR expression in vitro (Supplemental Figure 1B) and in vivo (Supplemental Figure 1C) was upregulated by TNF-α or infection, as observed in other model systems (19), and was associated with consistently increased bacterial adherence (>200%). Binding of all 3 species to rBCEC6 cells was reduced effectively by antibody against the LR C terminus (Figure 1B), while laminin (which binds to another domain of LR as shown in Figure 1C) was an inconsistent inhibitor (Figure 1B). Neither LR nor antibody against LR affected binding of E. coli (Figure 1B). The C-terminal LR peptide 263–282 (mapping to the anti-LR antibody–binding domain [ref. 20]; Figure 1C) decreased bacterial adherence to human BBB cells as compared with scrambled 263–282 or laminin-binding domain peptide 161–182 (Supplemental Figure 1D).
Bacterial ligands for LR.
Pneumococcus is known to require choline-binding protein A (CbpA) to adhere to the BBB in vitro (21) and to cause meningitis in vivo (22). Using CbpA as bait, we identified 37/67-kDa LR as a ligand from an rBCEC6 lysate by affinity chromatography with matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) analysis. This finding was confirmed by reactivity of the band with anti-LR antibody in immunoblot experiments (data not shown) and by coimmunoprecipitation of endothelial LR with rCbpA (Supplemental Figure 2A).
Using retagging, a contact-dependent cross-linking method (23), in conjunction with MALDI-TOF, we identified the pilus secretin protein PilQ (NMB1812) and the major outer membrane porin PorA (NMB1429) as meningococcal LR-binding surface proteins (Supplemental Figure 2B), and the porin OmpP2 was identified for H. influenzae (Supplemental Figure 2C). PilQ and PorA were shown to bind LR specifically and independently, as PorA was detected in lysates of the ΔpilQ mutant (Supplemental Figure 2B, lane 2) and PilQ was detected in lysates of the ΔporA mutant (data not shown; the protein complex was too large to enter the resolving gel). The retagging approach independently confirmed the CbpA-LR interaction (Supplemental Figure 2D) and the absence of LR-interacting ligands for E. coli. Binding for all 3 species was further confirmed by using biotin-labeled cross-linked proteins purified with magnetic beads (data not shown).
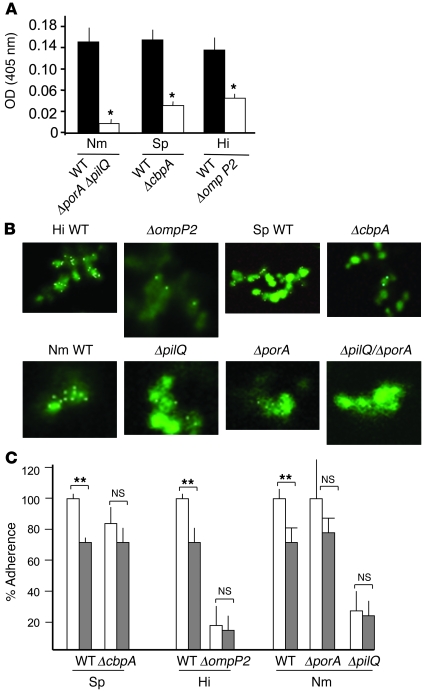
Mutational analysis was undertaken to further support the proposed LR-binding activity of CbpA, PilQ, PorA, and OmpP2. Wild-type S. pneumoniae and H. influenzae bound to rLR-coated ELISA plates, whereas the ΔcbpA and ΔompP2 deletion mutants showed dramatically less adherence (P < 0.005) (Figure 2A). Complementation of ΔcbpA with wild-type cbpA on a plasmid restored binding to 93% ± 1% (OD, 0.15 ± 0.001) of the wild-type parent (P < 0.005, Student’s t test). When porA and pilQ of meningococcus were inactivated, LR binding was reduced to less than 7% of wild-type in ELISA (P < 0.005) (Figure 2A); single mutants bound LR at almost wild-type levels (data not shown). In order to confirm these findings, we reconstructed the same genotype in a second clinical isolate with a high LR-binding activity: strain Z4181. Binding of Z4181ΔporAΔpilQ (OD, 0.13 ± 0.01) was 54% of the level in the isogenic wild-type strain (OD, 0.27 ± 0.03), a decrease similar to the 59% seen in the MC58 background. N. meningitidis porA is known to be highly variable. To investigate whether this could account for the observed differences in LR-binding activity of clinical isolates, we transferred the porA gene of strain MC58 to a clinical isolate with a low LR-binding activity: strain Z4682. Strain Z4682 binds rLR at 50% of the level of MC58 (OD, 0.09 ± 0.006 vs. 0.18 ± 0.004); when the MC58 porA gene was introduced into strain Z4682, replacing its native gene, rLR binding was increased to 94% of the level of strain MC58 (OD, 0.17 ± 0.007), confirming that the porA variant determines the rLR binding levels. Further confirmation of LR binding by the candidate adhesins from the 3 bacteria was obtained by visualizing binding of Cy5-LR to the bacteria. Cy5-LR clearly labeled wild-type bacteria, but not ΔcbpA pneumococci or ΔompP2 Haemophilus (Figure 2B). Cy5-LR bound strongly to wild-type meningococcus, while ΔpilQ and ΔporA mutants were labeled to a lesser degree, and the double mutant was virtually unlabeled (Figure 2B).
Figure 2. Bacterial ligands mediating binding to LR and endothelial cells.
(A) Specific binding of digoxigenin-labeled wild-type (black bars) or isogenic mutant (white bars) bacteria to rLR-coated ELISA plates (mean ± SD, n = 4) was determined by subtracting the absorbance in BSA-coated wells from that in rLR-coated wells. *P < 0.005 compared with wild-type; 1-way ANOVA. (B) Pneumococcal CbpA–dependent, meningococcal PorA– and PilQ–dependent, and H. influenzae OmpP2–dependent binding to LR. Wild-type and isogenic mutants were labeled with CFSE and incubated with Cy5-conjugated LR. After washing, colocalization (white) was detected by confocal microscopy (original magnification, ×100). (C) LR siRNA (gray bars) reduced adherence of wild-type pneumococcus (Sp), H. influenzae, and N. meningitidis to rBCEC6 cells but did not affect mutants lacking CbpA, OmpP2, or PilQ, respectively. MAPK siRNA (white bars) was used as a negative control. **P < 0.005; Student’s t test; 100% value was set for each bacteria as the following CFU/ml bound: Sp, 104; Hi, 105; Nm, 4.3 × 105 (MAPK siRNA adherence values were not statistically different from untransfected control values for any of the 3 bacteria). Error bars indicate SD.
When LR expression was reduced by transfection of rBCEC6 cells with LR siRNA (Supplemental Figure 3), wild-type pneumococcal, Haemophilus, and meningococcal binding were significantly reduced compared with that of control transfected cells (Figure 2C); the binding of ΔcbpA, ΔompP2, ΔporA, and ΔpilQ mutants was not affected by LR siRNA (Figure 2C).
In vivo BBB tropism.
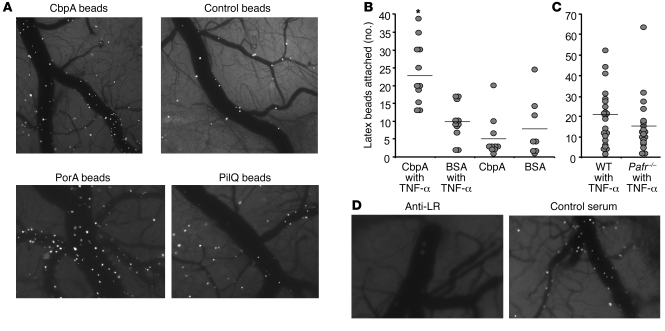
To study tropism in vivo, we first established that fluorescent microspheres coated with LR-binding adhesins and injected intravascularly into mice could be visualized binding to cerebral endothelium through a cranial window. Beads coated with CbpA, PilQ, and PorA adhered rapidly and firmly to the endothelial lining of the cerebral vasculature, whereas those coated with control proteins failed to do so (Figure 3, A and B). This interaction with the brain microvasculature was amplified by pretreatment of mice with TNF-α (Figure 3B). Binding of CbpA-coated beads was determined to be independent of the known BBB bacterial uptake receptor PAFr, as adherence was equally high in Pafr–/– mice and their wild-type littermates (Figure 3C). Furthermore, CbpA bead adherence was inhibited by pretreatment of mice with anti-LR antibody (43 ± 35 beads/×4 field) compared with control antiserum (131 ± 32 beads/×4 field) (Figure 3D; P = 0.006, Student’s t test).
Figure 3. Bacterial adhesins binding to cerebral endothelial LR in vivo.
(A) Cranial window image (original magnification, ×4) 10 minutes after intravascular injection of a mouse with fluorescent beads bearing CbpA, PilQ, PorA, or control pneumococcal protein Grr (intracellular response regulator). Images are representative of 10 mice per group, each imaged over 6 hours. (B) Quantitation of binding of fluorescent beads bearing CbpA or BSA (control) to mouse cerebral endothelium (beads/×40 field) as shown in A. Mice (1 symbol per animal; combined from 3 experiments; bars are the mean of the group) were injected i.v. with saline or TNF-α 1 hour prior to beads. *P < 0.01 versus BSA beads; 1-way ANOVA. (C) A comparison, similar to that in B, of the binding of CbpA beads in wild-type versus Pafr-null mice (NS, Student’s t test). (D) As in A, mice were pretreated with antibody against LR or control antiserum followed by infusion of CbpA beads. Images at 10 minutes after infusion are shown; images are representative of 3 mice per group.
CbpA contains 2 nearly identical cell-binding domains, R1 and R2, each formed by 3 anti-parallel helices (Figure 4A) (24). The RNYPT sequence of R1 and R2 is exposed at the turn between the first 2 helices (Figure 4A) and is known to bind to the epithelial polymeric immunoglobulin receptor (pIgR) (25, 26). This region did not appear to bind LR, because beads bearing a CbpA:RNGPT mutant protein that fails to bind pIgR bound as well as native CbpA to rBCEC6 cells (105% ± 20%). The sequence EPRNEEK is exposed at the second turn in the anti-parallel helices (Figure 4A) and is broadly conserved in the R2 domain across strains (P, 67%; R, 97%; N, 83%; EEK, 100% of 30 clinical isolates) but less so in the R1 domain (P, 37%; R, 92%; N, 32%; E, 90%; E, 42%; K, 75% of 40 clinical isolates). Its biological importance was tested using recombinant proteins bearing single amino acid substitutions in this sequence expressed in trans in ΔcbpA pneumococcus. Bacteria expressing CbpAP392G or CbpAR393G bound poorly to endothelial cells, while the other mutants bound as well as bacteria with wild-type CbpA (Figure 4B). We therefore focused on the double mutant CbpAP392G-R393G and compared it with wild-type CbpA, both of which were expressed in trans in the cbpA deletion mutant background (ΔcbpA pneumococci were transformed with plasmids bearing either cbpAWT or cbpAP392G-R393G). These strains grew with the same kinetics and expressed CbpA equally well (Supplemental Figure 4, A and B); binding to A549 lung epithelial cells and Factor H, which involve regions of CbpA distinct from that which binds endothelial cells, remained intact (Supplemental Figure 4, C and D). However, the double mutant bound to endothelial cells at only 42% ± 5% of the wild-type level (P < 0.001, Students t test). Recombinant CbpAP392G-R393G protein also bound to LR at 26% ± 11% of wild-type binding values in ELISA (P < 0.04, Student’s t test).
Figure 4. CbpA-LR binding determines development of pneumococcal meningitis in vivo.
(A) Schematic representation of the anti-parallel helices that form each of the 2 nearly identical R domains of CbpA (R2 is shown) (24). Square: binding site for pIgR showing amino acids at the turn; circle: putative binding site for LR showing amino acids at the turn. (B) Adherence to rBCEC6 cells of ΔcbpA pneumococci expressing the indicated wild-type or mutant ΔcbpA (100% adherence, 107 ± 14 bacteria/well). *P < 0.001 compared with wild-type; 1-way ANOVA. Error bars indicate SD. CpbA–, strain not expressing CbpA protein. (C) Cranial window images (original magnification, ×4) showing adherence of beads bearing wild-type R12 or mutant CbpAP392G-R393G. (D) BALB/c mice were inoculated i.v. with 1 × 106 to 4 × 106 CFU/ml T4 pneumococci. Bacteria were counted in blood and cerebrospinal fluid (CSF) at 18 hours. Circles, wild-type CbpA (n = 10; open circle, mean); X, CbpAP392G-R393G (n = 9; asterisk indicates the mean). Symbols in the gray area were at or below the lower limit of detection.
Small animal models of meningitis are well developed for pneumococcus but not for meningococcus or Haemophilus. Thus, the functional significance of bacteria-LR binding in the development of meningitis was tested in vivo in mice using the pneumococcal model. As observed in the cranial window (Figure 4C), substantially fewer beads bearing the double mutant CbpAP392G-R393G bound to cerebral endothelium than those bearing wild-type CbpA (Supplemental Videos 1 and 2). To test whether the interaction was important to the development of meningitis, we challenged mice with ΔcbpA pneumococcus expressing from a plasmid either wild-type CbpA or mutant CbpAP392G-R393G, inducing the sustained high-titer bacteremia (105 to 106 CFU/ml) that is required for the development of meningitis. Bacterial titers in the blood and cerebrospinal fluid were monitored. Animals challenged with bacteria expressing wild-type CbpA developed meningitis at blood bacteria titers 106 CFU/ml or less. In contrast, mice challenged with bacteria expressing CbpAP392G-R393G rarely developed meningitis despite blood bacterial titers as high as 107 CFU/ml (Figure 4D).
Discussion
We suggest that the family of LR-binding pathogens extends to the 3 most commonly neuroinvasive bacteria: pneumococcus, H. influenzae, and meningococcus. The interaction of LR with bacteria apparently differs from its interaction with most viruses and prions, in that bacteria appear to target the carboxyl terminus of LR. It is striking that such a broad range of medically significant pathogens use the same molecular target to initiate interaction with the BBB. This finding suggests that LR is a strong candidate as a therapeutic target for agents designed to block BBB penetration in the context of infection. This possibility is supported by the fact that pneumococci fail to cause meningitis in mice when they bear a substitution of only 2 amino acids in CbpA that are required for binding to LR.
The bacterial adhesins binding LR were identified as CbpA of pneumococcus, the outer membrane proteins PilQ and PorA of meningococcus, and the porin OmpP2 of H. influenzae. All of these are abundant, multifunctional proteins that contain surface-exposed loop structures; they include several eukaryotic receptors in their ligand repertoire and are protective antigens in models of sepsis (27, 28). CbpA acts as a bridging adhesin to human cells (28). The primary binding site for LR on CbpA was localized to a highly conserved, surface-exposed loop that is independent of that which binds the mucosal receptor pIgR (25, 26). Meningococcal PorA and H. influenzae OmpP2 are integral outer membrane porins (29, 30) that also play fundamental roles in adhesion and invasion of host cells and complement binding. Meningococcal PilQ is an essential component of the type IV pilus, which plays key roles in the binding of bacteria to epithelial and endothelial cells (31). The critical sequence enabling CbpA to bind to LR included the loop EPRNEEK, a region missing in the CbpA of the 2 clinical strains that failed to bind LR. While this sequence was not found in PilQ, PorA, or OmpP2, we identified the sequence NEENEEK within the domain of CNF1 toxin of E. coli that is believed to bind to LR (13).
We suggest that LR serves as a common receptor for bacterial pathogens that target the BBB. The interaction with LR is likely to complement the numerous other ligand-receptor interactions used by these bacteria and may act synergistically with them to promote adherence and invasion (32). Such an effect may involve localization of LR to lipid rafts (33), structures that have been implicated in invasion of host cells by a growing number of viral and bacterial pathogens (34). Our finding that upregulation of LR on the surface of eukaryotic cells is required for adherence is consistent with the need for increased expression of carcinoembryonic antigen–related cell adhesion molecules (CEACAMs) to demonstrate binding by Opa proteins (32). Inflammation during the bacteremic phase of disease prior to onset of meningitis is likely to be important for upregulating LR prior to its interaction with bacteria.
The CNS is an immunologically privileged site and is protected from infiltration by noxious agents, including pathogens, by the elaboration of tight junctions surrounding the cells of the vascular endothelium. Nonetheless, a limited number of pathogens, including select viruses, bacteria, and prions, are capable of invading this compartment with devastating results. We suggest that the shared ability to bind LR on the BBB is an important contributor to this capability. Because LR is present on many cell types at many body sites, this interaction may also play a role in the spread of infection elsewhere in the body. Conversely, other determinants are also likely to be operative at the BBB. However, LR binding appears to play a role in bringing circulating bacterial meningeal pathogens into intimate contact with BBB cells. This contact could then facilitate bacterial uptake into human cells via PAFr-mediated endocytosis. These 3 pathogens share the necessary surface phosphorylcholine ligand for PAFr. Our work suggests that disruption, or modulation, of the bacterial interaction with LR might engender unexpectedly broad protection against progression of disease to meningitis — the most serious consequence of infection with these agents. While effective vaccines have been developed against a subset of these pathogens, most of the more than 90 serotypes of pneumococcus, all non–type b H. influenzae, and the highly prevalent serogroup-B meningococcus are not covered by these vaccines. Broader therapeutic and/or preventive strategies based on common elements of pathogenesis rather than serotype could decrease global morbidity and mortality and prevent the emergence of replacement serotypes.
Methods
Bacterial strains and growth conditions.
E. coli strains XL10-Gold, BL21(DE3), TOP10Fι (both Invitrogen), and JM109 (Promega) and their derivatives containing plasmids were grown at 37°C in Luria-Bertani (LB) broth with agitation or on LB agar supplemented, when appropriate, with ampicillin (100 μg/ml), spectinomycin, and streptomycin (100 μg/ml each) or kanamycin (50 μg/ml). S. pneumoniae serotype 4 strain TIGR4; the unencapsulated TIGR4 derivative T4R; 19 clinical isolates (collection of E.I. Tuomanen); and Staphylococcus aureus NRS193 (provided by Jon McCullers, St. Jude Children’s Research Hospital) were grown on tryptic soy agar plates supplemented with 3% defibrinated sheep blood or in defined semisynthetic casein liquid medium supplemented with 0.5% yeast extract (35, 36). The ΔcbpA derivatives of unencapsulated T4R and wild-type T4 were created as described previously (28). N. meningitidis strains MC58, Z4181 (C: P1.5,2-1: F5-4: ST-11 [cc11]), Z4682 (B: P1.7-1,1: F5-8: ST-12 [–]), their mutant derivatives, and 68 additional clinical isolates (collection of D.A.A. Ala’Aldeen) and H. influenzae Rd and ATCC 10211, their derivatives, and 38 clinical isolates (collection of D.A.A. Ala’Aldeen) were cultured on chocolate agar at 37°C in 5% CO2. For selection of mutants, meningococcal cells were cultured on Mueller-Hinton agar plates supplemented with Vitox (Oxoid) and, where appropriate, streptomycin and spectinomycin (each 100 μg/ml) or kanamycin (100 μg/ml). All mutants retained wild-type growth rates. Liquid cultures of N. meningitidis were grown in Mueller-Hinton broth supplemented with Vitox at 37°C with agitation. The ΔompP2 mutant of H. influenzae Rd was a gift from Joachim Reidl, Biozentrum der Universität Würzburg, Am Hubland, Germany (29).
DNA manipulation.
Chromosomal DNA was prepared by using a DNeasy tissue kit (QIAGEN) and the protocol for bacterial cells recommended by the manufacturer. Plasmid DNA was prepared by using a QIAprep Spin Kit (QIAGEN) according to the manufacturer’s recommendations. PCR reactions contained 100 ng of chromosomal DNA or 1 ng of plasmid DNA, each primer at 200 nM, 200 μM each of dNTP, Expand polymerase buffer (including MgCl2 to a final concentration of 1.5 mM), and 0.6 U of Expand polymerase in a total volume of 25 μl (Roche). After initial incubation at 95°C for 3 minutes, reactions comprised 30 cycles of incubation at 50–55°C for 1 minute, 65–68°C for 1–6 minutes, and 94°C for 1 minute, with final incubations at 50–55°C for 1 minute and 65–68°C for 10 minutes.
Mutagenesis of N. meningitidis.
The porA gene (NMB1429) and flanking DNA was amplified from the chromosomal DNA of strain MC58 using primers PorA-M1 (5′-ATCAGAAACCTAAAATCCCGTCAT-3′) and PorA-M2 (5′-TCCTCTGTTTTGAAACCCTGAC-3′). The amplicon was cloned into pGEM-T Easy (Promega) to form plasmid pPorA-4d and subjected to inverse PCR mutagenesis with the primers PorA-M3 (5′-GCGGGATCCTCGGGCAAACACCCGATAC-3′) and PorA-M4 (5′-GCGGGATCCATCGGGGCGGTGAAGC-3′). This procedure resulted in removal of the porA coding sequence and introduction of a unique BamHI site, which was used to introduce an Ω cassette (encoding resistance to spectinomycin and streptomycin) (37). The resulting plasmid was used to mutate MC58 by natural transformation and allelic exchange as described previously (38). The deletion in the resulting mutant (MC58porA) was confirmed by PCR analysis. The pilQ gene (NMB1218) and its flanking sequence were amplified using the primers PilQF1 (5′-GCCGTCTGAAACAGCTGCCGACAGATGC-3′) and PilQR1 (5′-AAACCAGTACGGCGTTGCCTCGC-3′). The amplicon was cloned into pGEM-T Easy and subjected to inverse PCR mutagenesis with the primers PilQF2 (5′-CGCGGATCCCTTTCACCGTAACCTCAATCGC-3′) and PilQR2 (5′-CGCGGATCCCTGTAATGTTTCCTGCCGATGC-3′). This procedure resulted in deletion of the ORF and introduction of a unique BamHI site that was used to introduce the kanamycin resistance cassette digested with BamHI from plasmid pJMK30 (39). The resulting plasmid was used to mutate both the wild-type MC58 and the porA mutant, yielding strains MC58pilQ and MC58pilQ, porA, respectively. Successful mutagenesis of the pilQ gene in both mutants was confirmed by PCR and immunoblot analysis. The native porA gene of strain Z4682 was replaced with the MC58 porA gene. Using plasmid pPorA-4d as template, we amplified a 6.2-kDa fragment by PCR using the primers M-Kan-F (5′-AGATCTGCTTTGTTTTTGACGGCTCGC-3′) and M-Kan-R (5′-AGATCTATCCGCTTCACCGCCCCG-3′). The product was gel purified, digested with BglII, self ligated, and used to transform cells of JM109 (Promega). The resulting plasmid, pNAB10, was digested with BglII and ligated to the Ω cassette digested with BamHI to form plasmid pNAM11. pNAM11 was used to transform strain MC58 and Z4682; successful transformation of strain Z4682 was confirmed by amplification of the region containing porA and 1 kb upstream with the primers PorA-M1 (5′-ATCAGAAACCTAAAATCCCGTCAT-3′) and PorA-ER2 (5′-CCGATATTTAGAATTTGTGGCGC-3′). The sequence of the amplified product was shown to be identical to the MC58 porA sequence.
Purification of rLR.
E. coli BL21(DE3) containing plasmid pET28LR was cultured in LB containing kanamycin overnight at 37°C with agitation. Cells were harvested by centrifugation; the pellet was washed in PBS containing 0.05% v/v Tween-20 (PBS/T) and centrifuged at 6,500 g for 5 minutes. The supernatant was replaced with fresh PBS/T. This was repeated 3 times before cells were sonicated in an ice bath for 15 cycles of 10 seconds with 15 seconds of cooling. Samples were solubilized in 1% SDS sample buffer, and the suspension was incubated at 37°C for 30 minutes. The lysate was separated by SDS-PAGE and stained with SimplyBlue SafeStain (Invitrogen) for 30 minutes. A vertical section of the gel was removed for immunoblot analysis, and the band containing rLR was excised from the remaining gel, cut into small cubes, and placed in Dialyzer Midi D-Tubes (Calbiochem, EMD) in 1 ml PBS/T. The protein was eluted in SDS-PAGE running buffer at 100 V for 2 hours and dialyzed against PBS/T at 4°C for at least 24 hours.
Production of recombinant CbpA constructs.
All recombinant CbpA protein constructs were expressed and purified as previously described (24). Constructs included the full-length N terminus (CbpA-NR12; E39-P443), domain R1 linked to R2 (CbpA-R12; E175-P443), and the full-length N-terminal domain with a point mutation ablating adhesion to the pIgR receptor (CbpA-R12/Y205G/Y358G). To create CbpA mutants, we used R12 expressed in pET15b as a template for QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene). Clones were sequenced to confirm the presence of the desired mutation and subsequently used to transform BL21(DE3) cells for expression. All proteins were induced and purified as previously described (24).
To create pneumococci expressing CbpA with point mutations ablating binding to LR, we cloned full-length cbpA into pNE1 expression vector (24). This construct was used as a template for the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Primer MUT1 (5′-GCTAAGGAAGGGGGGAACGAGGAAAAAG-3′) was used to make the P392G and R393G substitutions and cloned as previously described (24). Clones were sequenced to confirm the presence of desired substitutions, and plasmid preparations were used to transform S. pneumoniae T4X ΔcbpA by standard methods as follows. T4X ΔcbpA was constructed by the SOEing method (40). Briefly, an erythromycin resistance cassette (1,096 bp) was amplified from the ermB gene using primers ermF (5′-GGAAATAAGACTTAGAAGCAAAC-3′) and ermR (5′-CCAAATTTACAAAAGCGACTC-3′). Approximately 1-kb regions flanking the cbpA gene were amplified with 2 primer pairs: cbp1 (5′-GGCGGGAAAGAATTTGGAG-3′), CbpASOEup (5′-GTTTGCTTCTAAGTCTTATTTCCGTTTATTTCCTTCTATATTTTTTCTTTAACC-3′) and cbpA4 (5′-CGCCAACGGTGATATCCGTAC-3′), CbpASOEdn (5′-GAGTCGCTTTTGTAAATTTGGACCTAATATAACTAGTTAATACTGACTTCC-3′). Primers CbpAErmSOEup and CbpAErmSOEdn served as fusion primers and consisted of primers ermF and ermR, respectively, and the target gene–specific sequence. A mixture of gel-purified preparations of the 2 flanking products and the erm cassette was used as a template for the SOEing PCR, using the outer CbpA1 and CbpA2 primers. The full-length PCR product consisting of the erm cassette flanked by the CbpA-flanking sequences served as the knockout construct. The PCR fusion product was introduced into the chromosome of S. pneumoniae T4X by transformation and homologous recombination based on standard methods. Loss of CbpA in transformants was confirmed by PCR, sequencing, and immunoblot analysis.
Affinity purification with a CbpA column.
CbpAR12 (residues E39-P443) was dialyzed against 0.1 M MOPS (pH 7.5) and concentrated to 2.5 mg/ml using Centriplus columns (Amicon, Millipore). CbpA-NR12 (10 mg) was covalently conjugated to 1 ml of Affi-Gel 15 (Bio-Rad) in 0.1 M MOPS (pH 7.5) at 4°C as instructed by the manufacturer. The column was sequentially washed with 20 ml of 100 mM glycine (pH 2.5) and 20 ml of PBS containing 1 mM MgCl2, 0.5 mM CaCl2, and 0.1% Triton X-100 (PBS-Ca2+/Mg2+/T), respectively. rBCEC6 cells (41) were grown to complete confluence in 150 × 25 mm tissue culture dishes and preincubated with TNF-α at 10 ng/ml for 2 hours. Cell lysates were prepared in 1% Triton X-100 lysis buffer containing EDTA-free protease inhibitors (Roche). Cellular debris was removed by centrifugation at 135,000 g for 1 hour at 4°C. The supernatant was recycled through the CbpA-NR12 affinity column for a period of 4 hours at 4°C. After the column was washed with 20 ml of PBS/Ca2+/Mg2+/T, bound proteins were eluted in 300-μl fractions with 100 mM glycine-HCl (pH 2.5). Eluted proteins were visualized by SDS-PAGE and silver staining. Bands corresponding to eluted proteins were excised, and proteins were identified by MALDI-TOF analysis in conjunction with a database search performed with Applied Biosystems GPS Explorer software by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children’s Research Hospital. Protein assignment was based on both mass spectrometry (MS) and MS/MS spectra and the NCBI nonredundant database.
Coimmunoprecipitation of LR with rCbpA.
rBCEC6 cells were activated with TNF-α (10 ng/ml) for 1 hour, lysed with 3 ml NP-40 buffer (0.5% NP-40, 500 mM Tris, pH 7.4, 150 mM NaCl), and briefly sonicated. The lysate was combined with 4 μg CbpA-AR12 and incubated overnight with 35 μg of either anti-LR (ab711; Abcam; 1:1000) or anti-CbpA (1:2,000). Rabbit antibody against the N terminus (amino acids 39–443) of CbpA was made by Covance. Protein A beads (Invitrogen) prepared as recommended by the manufacturer were added, and the mix was incubated for 1 hour. Samples were washed 7 times with 1 ml NP-40 buffer, resuspended in 40 μl 2× sample buffer, and boiled for 10 minutes. Samples were analyzed by SDS-PAGE and immunoblotting in which samples pulled down with anti-LR were probed with anti-CbpA, and those pulled down with CbpA were probed with anti-LR.
Adhesion assay.
Adhesion assays were performed in several cell culture systems. Given the technical difficulty of obtaining primary mouse brain endothelial cells, the rat brain capillary endothelial cell line rBCEC6 (41) was used in most in vitro tests. Human brain microvascular endothelial cells (ScienCell Research Laboratories) were also tested to confirm the relevance of all binding events studied in the rBCEC6 cells. A549 human lung epithelial cells were used to study CbpA-mediated adherence independent of the domain required for endothelial adherence. Cells were seeded to 80% confluence in a 24-well plate (Costar, Corning) and activated for 1 or 2 hours with TNF-α (10 ng/ml). Cells were washed to remove inhibitors and incubated with 1 × 107 CFU/ml bacteria. For adhesion assays, cells were washed 3 times with PBS after 30 minutes, trypsinized, and diluted 1:10 in PBS; 10 μl was plated on blood agar and incubated overnight. All peptides used in inhibition experiments (LR amino acids 161–180, 263–282, a scrambled version of 263–282, LMWWML, YIGSR, LGTIPG, and VGVAPG) were manufactured by Invitrogen. To block bacterial adhesion to rBCEC6 endothelia, 107 CFU bacteria were incubated for 1 hour at room temperature with 25 μg peptide. Bacteria were pelleted by centrifugation and washed twice with Dulbecco’s PBS (DPBS). Alternatively, monolayers were treated with antibody anti-LR 711 (Abcam) recognizing amino acids 263–282 of LR at a dilution of 1:500 or laminin from human placenta (Sigma-Aldrich; 5 μg/ml) for 1 hour and the assay continued as above.
Adhesion assays using microspheres were performed as previously described (42). Inhibition assays were performed in a similar manner with a 1-hour preincubation with purified laminin, anti-LR 711, naive rabbit serum, LR peptides, or complex sugars dissolved in PBS prior to addition of the microspheres. For all experiments, 4 wells per sample were used, and experiments were performed in triplicate.
LR siRNA (LamR1, 5′-TTGGTTCCCATCGTAACTTAA-3′; QIAGEN) was prepared according to the manufacturer’s instructions. rBCEC6 cells (105 per well) were seeded into 24-well plates and grown to 50% confluence. For each well, 10 nM siRNA was diluted in 100 μl serum-free medium. HiPerFect Transfection Reagent (3 μl; QIAGEN) was incubated with the siRNA for 10 minutes, and the mixture was added dropwise to cells and incubated at 37°C in 5% CO2 for 48 hours. To document decreased expression of LR, 1 well per sample was lysed with NP-40 buffer and sonicated, and equal amounts of protein were loaded for SDS-PAGE and Western blot analysis with polyclonal antibody against LR (Abcam) (Supplemental Figure 3). The remaining transfected cells were used for adhesion assays with bacteria as described above.
Intravital fluorescence microscopy and mouse challenge studies.
All experiments involving animals were performed with the prior approval of and in accordance with guidelines of the St. Jude Institutional Animal Care and Use Committee. Female BALB/c (The Jackson Laboratory), C57BL/6 Pafr–/– (provided by P. Murray, St. Jude Children’s Research Hospital), and B6N10 mice and their respective wild-type littermates were maintained in Biosafety Level 1 and 2 facilities in the St. Jude Children’s Research Hospital Animal Facility. Mice were anesthetized for all experimental procedures by inhalation of 2.5% isoflurane (Baxter). Cranial windows overlying the parietal cortex on one side of the midsagittal suture and extending from the bregma to the lamboid sutures were created in 8- to 10-week-old BALB/c mice as described previously (43). After surgery, mice were allowed to recover for 10 days before data collection was initiated. Intravital microscopy was used to visualize attachment of fluorescent latex microspheres (Polysciences Inc.) to the microvasculature of the brain. Mice with cranial windows were anesthetized, immobilized on a stereotactic frame, and placed under an industrial-scale microscope (model MM-11; Nikon) with a camera assembly and bright-field and fluorescent light sources. For some experiments, mice were injected with recombinant mouse TNF-α (12 μg/kg i.v.) 1 hour before injection of microspheres. Yellow-green fluorescent 2-μm spheres were coated with the recombinant CbpA derivatives or BSA according to the supplier’s instructions. Mice were injected with 2.5 μl/g body weight of a 108 sphere/ml suspension. Video images were followed for up to 3 hours, and still pictures were captured during injection and 3 and 10 minutes after injection at an excitation wavelength of 490 nm by using MetaMorph software (Universal Imaging, Molecular Devices). Beads attached to the microvasculature were counted, and counts were confirmed by comparison of the cerebral microvasculature with baseline pictures taken prior to injection. For some experiments, mice were pretreated with 200 μl of either undiluted antibody anti–LR 711 (Abcam) or control rabbit serum i.v. 5 minutes prior to challenge with CbpA beads.
For immunohistochemical staining for LR with and without infection, mice were challenged with PBS or 107 T4 pneumococci intranasally, and pneumonia was allowed to develop over 48 hours. Lungs were harvested, and frozen sections were stained with mouse anti-LR antibody MLuC5 (ab3099; Abcam) for 1 hour, followed by detection of binding with HRP-conjugated goat anti-mouse IgM (ab5930; Abcam) and the VIP Substrate Kit (Vector Laboratories). Sections were counterstained with methyl green.
Identification of H. influenzae and N. meningitidis LR ligands by retagging.
Bacterial LR-binding proteins were purified as previously described for the H. pylori SabA adhesion protein (44), with some modifications. N. meningitidis, H. influenzae, and S. pneumoniae were incubated with purified rLR to which the Sulfo-SBED cross-linker (Pierce, Thermo Scientific) had been conjugated according to the manufacturer’s recommendations. The photoreactive cross-linker group was activated by 2 minutes of UV irradiation, and the biotin-(re)tagged proteins were purified with streptavidin-coated magnetic beads as described previously (44). Extracted biotin-tagged proteins were separated by SDS-PAGE, and bands were digested with sequencing grade trypsin (Promega) and analyzed using a Micromass TofSpec E. The nanoflow liquid chromatography–MS/MS was done on a 7-Tesla linear trap quadrupole Fourier transform (LTQ-FT) mass spectrometer (Thermo Scientific) equipped with a nanospray source modified in-house. The spectrometer was operated in data-dependent mode, automatically switching to MS/MS mode. MS spectra were acquired in the FT ion cyclotron resonance analyzer (FTICR), while MS/MS-spectra were acquired in the LTQ-trap. For each scan of FTICR, the 3 most intense, doubly or triply charged, ions were sequentially fragmented in the linear trap by collision-induced dissociation. All the tandem mass spectra were searched by Mascot (Matrix Science) against the databases for the N. meningitidis strain MC58, H. influenzae strain Rd KW20, and S. pneumoniae strain T4, as appropriate.
ELISA of rLR.
Purified rLR or BSA (2–50 μg/ml) diluted in carbonate buffer (150 mM; 142 mM NaHCO3, 8 mM Na2SO3, pH 9.0) was used to coat amino-reactive 96-well microtiter plates (Immoblizer Amino; Nunc) for 2 hours at room temperature. Bacterial strains were grown in liquid culture and washed 3 times in PBS/T before being resuspended in carbonate buffer to an OD of 0.1 at 600 nm. Bacteria were added to ELISA plates unlabeled (N. meningitidis) or labeled by adding 10 μg of digoxigenin (Roche) per 1 ml bacterial suspensions for 2-4 hours at room temperature (H. influenzae and S. pneumoniae). Liquid was removed from the plates, plates were washed 3 times with PBS/T, 100 μl of bacterial suspensions were added to each well, and plates were incubated for 2–4 hours at room temperature. Plates were washed several times with PBS/T and incubated with polyclonal anti-digoxigenin Fab fragment–conjugated antibody (1:5,000; Roche) in 1% BSA in PBS/T (blocking buffer) at 100 μl per well (digoxigenin-labeled cells) or with rabbit anti–whole N. meningitidis followed by goat anti-rabbit HRP. Plates were incubated at room temperature for an additional 1 hour and washed several times as described above. One hundred microliters of ABTS tablets (5 mg/ml; Roche) were added to each well, and the absorbance was measured at 405 nm after 30 minutes using an ELISA plate reader. Inhibition assays were performed as described above, except that bacteria were preincubated with 20 μg/ml rLR for 2 hours at room temperature and washed twice before being added to the ELISA plates. In the reverse inhibition assay, wells were coated with either N. meningitidis or H. influenzae (OD600, 0.1), and then incubated with digoxigenin-labeled rLR. In this experiment, rLP was preincubated with rPorA, rPilQ, rOmpP2, or nothing. Wells coated with BSA or ethanolamine were used as negative controls.
For studies on Factor H binding to CbpAs, plates were coated with 106 bacteria expressing CbpAWT or CbpAP392G-R393G and blocked with 10% bovine serum. Wells were exposed to 1 μg Factor H (Sigma-Aldrich), washed, and treated with 1:100 or 1:1,000 anti–Factor H antibody (Sigma-Aldrich). Binding was quantitated as the OD405 developed by anti-goat IgG alkaline phosphatase (AP) and yellow AP substrate (Sigma-Aldrich).
Confocal microscopy.
Bacterial suspensions were adjusted to an OD of 0.1 at 600 nm, and 1 ml of suspension was labeled with 10 μg CFSE (Molecular Probes, Invitrogen) for 10 minutes at room temperature, after which the cells were harvested by centrifugation and washed in 1 ml PBS/T. Purified rLR was labeled with Cy5 (Amersham, GE Healthcare) according to the manufacturer’s recommendations. CFSE-labeled bacterial cells were incubated with Cy5-labeled rLR for 2–4 hours. Cells were washed 2 times in PBS/T and transferred to the confocal plates. Images were captured using a Zeiss LSM 510uv META combi confocal system on a Zeiss Axiovert 100 microscope with a Zeiss C-Apochromat 63×/1.2 W objective lens. Sequential scanning with an argon 488-nm laser was used to excite the CFSE with a 505- to 550-nm band pass filter and an HeNe 633-nm laser with a 650-nm-long pass filter for the Cy5. The image size was 1,024 × 1,024 pixels, with 4 averages taken per line and a zoom of 3. All images were captured with the same laser, gains, and zoom settings. Colocalization analysis was performed using Zeiss LSM software and its Pixel Spreadgram function.
Statistics.
In instances where values from 2 experimental conditions were compared with each other, we used a 2-tailed Student’s t test to test for statistical significance. In instances where multiple experimental conditions were compared with a single control group, we tested for statistical significance using 1-way ANOVA. In regard to the latter, a Holm-Sidak ANOVA test was used for experiments with values passing normality and equal variance tests; otherwise, a Kruskal-Wallis ANOVA by ranks test was used. A P value less than 0.01 was considered significant.
Supplementary Material
Acknowledgments
We thank G. Gao and Y. Hu for technical assistance and M. Johnson and C. Calabrese for help with animal experiments. We thank J. Reidl and J. McCullers for providing the H. influenzae ompP2– mutant and S. aureus NRS193, respectively; and P. Murray for Pafr–/– mice. We thank S. Naron for editorial assistance. This work was supported by NIH grants R01 AI27913 (to E.I. Tuomanen) and CA21765; by the American Lebanese Syrian Associated Charities (ALSAC); and by the James Tudor Foundation.
Footnotes
Authorship note: Dlawer A.A. Ala’Aldeen and Elaine I. Tuomanen are co–senior authors. Carlos J. Orihuela, Jafar Mahdavi, Justin Thornton, and Beth Mann contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: BBB, blood brain barrier; CbpA, choline-binding protein A; LR, laminin receptor; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; PAFr, platelet-activating factor receptor; PBS/T, 0.05% Tween-20 in PBS; pIgR, polymeric immunoglobulin receptor; rBCEC6, rat brain endothelial cell line; rLR, recombinant LR.
Citation for this article: J. Clin. Invest. 119:1638–1646 (2009). doi:10.1172/JCI36759
References
- 1.Jaseja M., et al. Structure-function studies of the functional and binding epitope of the human 37 kDa laminin receptor precursor protein. J. Pept. Res. 2005;66:9–18. doi: 10.1111/j.1399-3011.2005.00267.x. [DOI] [PubMed] [Google Scholar]
- 2.Mecham R. Receptors for laminin on mammalian cells. FASEB J. 1991;5:2538–2546. doi: 10.1096/fasebj.5.11.1651264. [DOI] [PubMed] [Google Scholar]
- 3.Hilario E., et al. Presence of laminin and 67kDa laminin-receptor on endothelial surface of lung capillaries. Histol. Histopathol. 1996;11:915–918. [PubMed] [Google Scholar]
- 4.Nelson J., et al. The 67 kDa laminin receptor: structure, function and role in disease. Biosci. Rep. 2008;28:33–48. doi: 10.1042/BSR20070004. [DOI] [PubMed] [Google Scholar]
- 5.Jamieson K., Wu J., Hubbard S., Meruelo D. Crystal structure of human laminin receptor precursor. J. Biol. Chem. 2008;283:3002–3005. doi: 10.1074/jbc.C700206200. [DOI] [PubMed] [Google Scholar]
- 6.Menard S., Castronovo V., Tagliabue E., Sobel M. New insights into the metastasis-associated 67kD laminin receptor. J. Cell. Biochem. 1997;67:155–165. doi: 10.1002/(SICI)1097-4644(19971101)67:2<155::AID-JCB1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Wang K., Kuhn R., Strauss E., Ou S., Strauss J. High affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J. Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thepparit C., Smith D. Serotype-specific entry of dengue virus into liver cells: identification of the 37 kDa/67 kDa high affinity laminin receptor as a dengue virus serotype 1 receptor. J. Virol. 2004;78:12647–12656. doi: 10.1128/JVI.78.22.12647-12656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akache B., Grimm D., Pandey K., Yant S., Zu H., Kay M. The 37/67 kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8,2,3 and 9. J. Virol. 2006;80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig G., Kondig J., Smith J. A putative receptor for Venezuelan equine encephalitis virus from mosquito cells. J. Virol. 1996;70:5592–5599. doi: 10.1128/jvi.70.8.5592-5599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leucht C., et al. The 37kDa/67 kDa laminin receptor is required for PrpSc propagation in scrapie-infected neuronal cells. EMBO Rep. 2003;4:290–295. doi: 10.1038/sj.embor.embor768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gauczynski S., et al. The 37 kDa/67 kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. EMBO J. 2001;20:5863–5875. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNichol B., Rasmussen S., Carvalho H., Myesick K., O’Brien A. Two domains of cytotoxic necrotizing factor 1 bind the cellular receptor, laminin receptor precursor protein. Infect. Immun. 2007;75:5095–5104. doi: 10.1128/IAI.00075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K., Chung J., Kim K. 67 kDa laminin receptor promotes internalization of cytotoxic necrotizing factor-1expressing Escherichia coli K1 into human brain microvascular endothelial cells. . J. Biol. Chem. 2005;280:1360–1368. doi: 10.1074/jbc.M410176200. [DOI] [PubMed] [Google Scholar]
- 15.Cundell D., Gerard N., Gerard C., Idanpaan-Heikkila I., Tuomanen E. Streptococcus pneumoniae anchors to activated eukaryotic cells by the receptor for platelet activating factor. . Nature. 1995;377:435–438. doi: 10.1038/377435a0. [DOI] [PubMed] [Google Scholar]
- 16.Swords W., et al. Binding of the non-typeable Haemophilus influenzae lipooligosaccharide to the PAF receptor initiates host cell signalling. . Cell. Microbiol. 2001;3:525–536. doi: 10.1046/j.1462-5822.2001.00132.x. [DOI] [PubMed] [Google Scholar]
- 17.Gould J., Weiser J. The inhibitory effect of C-reactive protein on bacterial phosphorylcholine platelet-activating factor receptor-mediated adherence is blocked by surfactant. J. Infect. Dis. 2002;186:361–371. doi: 10.1086/341658. [DOI] [PubMed] [Google Scholar]
- 18.Radin J., et al. β-arrestin 1 determines the traffic pattern of PAFr-mediated endocytosis of Streptococcus pneumoniae. . Infect. Immun. 2005;73:7827–7835. doi: 10.1128/IAI.73.12.7827-7835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raghunath P., et al. Interferons upregulate the expression of laminin and its receptor LBP-32 in cultured cells. J. Biol. Regul. Homeost. Agents. 1993;7:22–30. [PubMed] [Google Scholar]
- 20.Castronovo V., Taraboletti G., Sobel M. Functional domains of the 67 kD laminin receptor precursor. J. Biol. Chem. 1991;266:20440–20446. [PubMed] [Google Scholar]
- 21.Ring A., Weiser J., Tuomanen E. Pneumococcal penetration of the blood brain barrier: molecular analysis of a novel re-entry path. J. Clin. Invest. 1998;102:347–360. doi: 10.1172/JCI2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orihuela C., et al. Organ specific models of Streptococcus pneumoniae disease. . Scand. J. Infect. Dis. 2003;35:647–652. doi: 10.1080/00365540310015854. [DOI] [PubMed] [Google Scholar]
- 23.Ilver D., et al. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. . Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 24.Luo R., et al. Solution structure of choline binding protein A of Streptococcus pneumoniae reveals a novel mode of interaction with its human receptor, plgR. . EMBO J. 2005;24:34–43. doi: 10.1038/sj.emboj.7600490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J.-R., et al. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell. 2000;102:827–837. doi: 10.1016/S0092-8674(00)00071-4. [DOI] [PubMed] [Google Scholar]
- 26.Hammerschmidt S., Tillig M., Wolff S., Vaerman J., Chhatwal G. Species-specific binding of human secretory component to SpsA protein of Streptococcus pneumoniae via a hexapeptide motif. . Mol. Microbiol. 2000;36:726–736. doi: 10.1046/j.1365-2958.2000.01897.x. [DOI] [PubMed] [Google Scholar]
- 27.Cope L., Pelzel S., Latimer J., Hansen E. Characterization of a mutant of Haemophilus influenzae type b lacking the P2 major outer membrane protein. . Infect. Immun. 1990;58:3312–3318. doi: 10.1128/iai.58.10.3312-3318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenow C., et al. Contribution of a novel choline binding protein to adherence, colonization, and immunogenicity of Streptococcus pneumoniae. . Mol. Microbiol. 1997;25:819–829. doi: 10.1111/j.1365-2958.1997.mmi494.x. [DOI] [PubMed] [Google Scholar]
- 29.Andersen C., et al. Porin OmpP2 of Haemophilus influenzae shows specificity for nicotinamide-derived nucleotide substrates. . J. Biol. Chem. 2003;278:24269–24276. doi: 10.1074/jbc.M213087200. [DOI] [PubMed] [Google Scholar]
- 30.van der Ende A., Hopman C., Dankert J. Multiple mechanisms of phase variation of PorA in Neisseria meningitidis. . Infect. Immun. 2000;68:6685–6690. doi: 10.1128/IAI.68.12.6685-6690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Virji M., et al. Pilus-facilitated adherence of Neisseria meningitidis to human epithelial and endothelial cells: modulation of adherence phenotype occurs concurrently with changes in primary amino acid sequence and the glycosylation status of pilin. . Mol. Microbiol. 1993;10:1013–1028. doi: 10.1111/j.1365-2958.1993.tb00972.x. [DOI] [PubMed] [Google Scholar]
- 32.Rowe H., Griffiths N., Hill D., Virji M. Coordinate action of bacterial adhesins and human carcinoembryonic antigen receptors in endhanced cellular invasion by capsulate serum resistant Neisseria meningitidis. . Cell. Microbiol. 2007;9:154–168. doi: 10.1111/j.1462-5822.2006.00775.x. [DOI] [PubMed] [Google Scholar]
- 33.Fujimura Y., Yamada K., Tachibana H. A lipid raft-associated 67kDa laminin receptor mediates suppressive effect of epigallocatechin-3-O-gallate on FcepsilonRI expression. Biochem. Biophys. Res. Commun. 2005;336:674–681. doi: 10.1016/j.bbrc.2005.08.146. [DOI] [PubMed] [Google Scholar]
- 34.Zaas D., Duncan M., Rae Wright J., Abraham S. The role of lipid rafts in the pathogenesis of bacterial infections. Biochim. Biophys. Acta. 2005;1746:305–313. doi: 10.1016/j.bbamcr.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Tettelin H., et al. Complete genome sequence of virulent isolates of Streptococcus pneumoniae. . Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 36.Tomasz A. Choline in the cell wall of a bacterium: novel type of polymer-linked choline in pneumococcus. Science. 1967;157:694–697. doi: 10.1126/science.157.3789.694. [DOI] [PubMed] [Google Scholar]
- 37.Prentki P., Krisch H. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 38.Hadi H., Wooldridge K., Robinson K., Ala’Aldeen D. Identification and characterization of App: an immunogenic autotransporter protein of Neisseria meningitidis. . Mol. Microbiol. 2001;41:611–623. doi: 10.1046/j.1365-2958.2001.02516.x. [DOI] [PubMed] [Google Scholar]
- 39.van Vliet. A., Wooldridge K., Ketley J.1998Iron-responsive gene regulation in a Campylobacter jejuni fur mutan t . J. Bacteriol. 1805291–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho S., Hunt H., Horton R., Pullen J., Pease L. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 41.Blasig I., et al. *NO and oxyradical metabolism in new cell lines of rat brain capillary endothelial cells forming the blood brain barrier. Microvasc. Res. 2001;62:114–127. doi: 10.1006/mvre.2001.2318. [DOI] [PubMed] [Google Scholar]
- 42.Orihuela C., et al. Cell wall-mediated neuronal damage in early sepsis. Infect. Immun. 2006;74:3783–3789. doi: 10.1128/IAI.00022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fillon S., et al. Platelet activating factor and innate immunity: uptake of Gram positive bacterial cell wall into host cells and cell-specific pathophysiology. J. Immunol. 2006;177:6182–6191. doi: 10.4049/jimmunol.177.9.6182. [DOI] [PubMed] [Google Scholar]
- 44.Mahdavi J., et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. . Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.