Abstract
The forkhead box proteins A1 and A2 (Foxa1 and Foxa2) are transcription factors with critical roles in establishing the developmental competence of the foregut endoderm and in initiating liver specification. Using conditional gene ablation during a later phase of liver development, we show here that deletion of both Foxa1 and Foxa2 (Foxa1/2) in the embryonic liver caused hyperplasia of the biliary tree. Abnormal bile duct formation in Foxa1/2-deficient liver was due, at least in part, to activation of IL-6 expression, a proliferative signal for cholangiocytes. The glucocorticoid receptor is a negative regulator of IL-6 transcription; in the absence of Foxa1/2, the glucocorticoid receptor failed to bind to the IL-6 promoter, causing enhanced IL-6 expression. Thus, after liver specification, Foxa1/2 are required for normal bile duct development through prevention of excess cholangiocyte proliferation. Our data suggest that Foxa1/2 function as terminators of bile duct expansion in the adult liver through inhibition of IL-6 expression.
Introduction
Liver development is a complex process and requires the specification of hepatocytes, cholangiocytes (bile duct epithelial cells), stellate cells, Kupffer cells, and myofibroblasts in addition to the development of the circulatory and nervous systems (1–5). The liver is derived from ventral foregut endoderm, where progenitor cells differentiate into hepatoblasts beginning on E8.5 in the mouse. Liver specification is marked by initial expression of albumin (Alb), α-fetoprotein (Afp), and transthyretin (Ttr). Both hepatocytes and cholangiocytes are differentiated from bipotential hepatoblasts. Hepatoblasts start to differentiate into hepatocytes on E13.5 and into cholangiocytes on E14.5 (3–5). Hepatocyte development is completed after birth, followed by the establishment of metabolic functions such as glucose and lipid metabolism (2, 3). Bile duct development is initiated with the formation of ductal plates surrounding the portal tracts in the late gestation, which subsequently reorganize to form ducts (3–5). Complete formation of the biliary tree is followed by liver growth into adulthood (3–5).
The forkhead box protein A (Foxa) family of transcription factors controls embryonic development and organogenesis of liver, pancreas, brain, lung, thyroid, and prostate (2, 6–11). Foxa includes 3 family members, Foxa1, Foxa2, and Foxa3 (2, 6, 7, 11). Global or liver-specific ablation of Foxa1, Foxa2, or Foxa3 alone did not affect liver morphology (12–14). However, when both Foxa1 and Foxa2 (Foxa1/2) were deleted in mouse embryos at E8.5 days, progenitor cells in the foregut endoderm failed to differentiate into hepatoblasts and no liver formed (9). Thus, Foxa1 and Foxa2, though redundant, are critical for the establishment of developmental competence in foregut endoderm and the initiation of liver specification. Foxa1/2 expression in the liver persists into adulthood (7, 11, 15). Previous studies have shown that Foxa1/2 are expressed in hepatocytes and cholangiocytes (7, 11, 15). However, neither hepatocyte development nor bile duct development was affected by single ablation of either Foxa1 or Foxa2, although the ability of hepatocytes to export bile acids is dependent on Foxa2 (12–14). Here we use the Cre-loxP technology to establish a mouse model with liver-specific ablation of Foxa1/2 during late gestation in order to investigate their role in liver development after its initial specification. Our loss-of-function analysis reveals that Foxa1/2 are required for the regulation of cholangiocyte proliferation, which is mediated at least in part by inhibition of IL-6 expression.
Results
A mouse model of Foxa1/2 deficiency in the fetal liver.
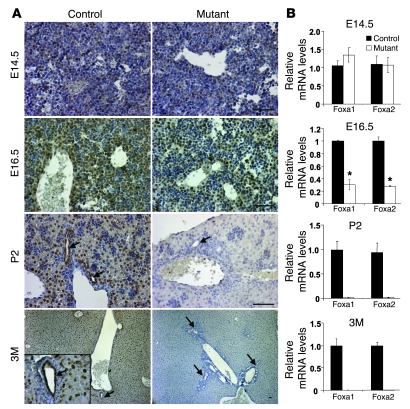
To address whether Foxa1/2 are essential for liver development after its initial specification, we derived a new mouse model, Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre (mutant) mice, in which both genes are ablated in the liver during late gestation. Foxa1loxP/loxPFoxa2loxP/loxP mice without AlfpCre were used as controls. The expression of Cre recombinase in AlfpCre mice is driven by the Alb promoter and both Alb and Afp enhancers (16, 17). Both Afp and Alb are initially expressed in the hepatoblast at E9.0. Crossing AlfpCre mice with Rosa26 mice (which allows for detection of Cre activity by lacZ staining) (17) confirmed that the AlfpCre transgene was active in the liver primordium by E10.5 (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI38201DS1). To trace the deletion of Foxa1/2 in this model, we examined protein and mRNA levels in livers by immunohistochemical staining and real-time quantitative RT-PCR (qRT-PCR), respectively. Nuclear staining of Foxa1/2 was evident in control livers as expected (Figure 1A). However, surprisingly, Foxa1/2 were not deleted in the liver of Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre embryos at E14.5, as indicated by both immunostaining and qRT-PCR (Figure 1, A and B). Ablation of Foxa1/2 was first apparent in the liver of Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre embryos at E16.5 (Figure 1, A and B), complete by P2, and maintained into adulthood (Figure 1, A and B). The late deletion observed in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice compared with the Rosa26 reporter was likely due to differential accessibility of different loxP-flanked loci, which has been reported previously (18–20).
Figure 1. Foxa1/2 are deleted in the liver of Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice after E16.5.
(A) Immunohistochemical staining of paraffin-embedded liver sections from E14.5, E16.5, P2, and 3-month-old (3M) Foxa1loxP/loxPFoxa2loxP/loxP (control) and Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre (mutant) mice with an anti-Foxa1/2 antibody. Inset shows a liver section of control 3-month-old mice (original magnification, ×400). Arrows point to bile duct epithelial cells. Scale bars: 10 μm. (B) mRNA levels of Foxa1/2 after normalization to those of Gapdh in E14.5, E16.5, P2, and 3-month-old control and mutant livers by real-time qRT-PCR. *P < 0.05 compared with control mice.
Hepatocyte differentiation after liver specification is largely independent of Foxa1/2.
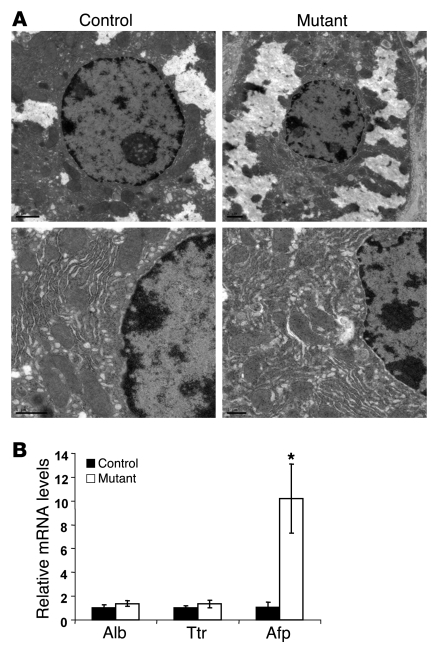
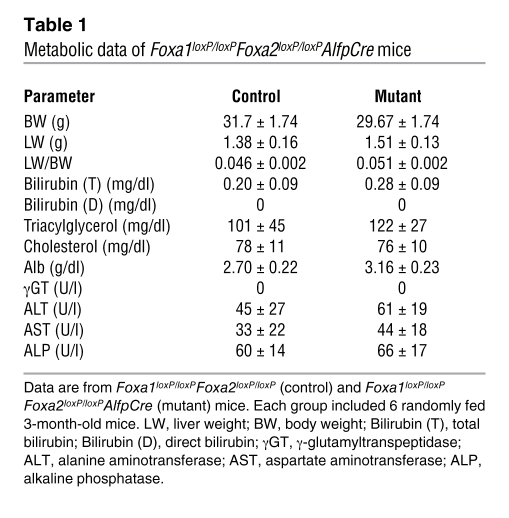
Foxa1/2 are highly expressed in hepatocytes (Figure 1A) (7, 15). We therefore investigated whether hepatocyte morphology was altered in Foxa1/2-deleted livers using immunohistochemical staining (Figure 1A) and transmission EM (Figure 2A). Hepatocytes from Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice were indistinguishable from those in controls with regard to both overall morphology and ultrastructure (Figure 1A and Figure 2A). In addition, adult Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice showed normal body weight, liver weight, and ratios of liver to body weight in both genders as compared with controls (Table 1). Mutant mice did not show significant changes in blood biochemistry, including levels of Alb, bilirubin, triacylglycerol, cholesterol, alanine aminotransferase, aspartate aminotransferase, γ-glutamyltranspeptidase, and alkaline phosphatase, compared with controls (Table 1). Expression of the hepatoblast/hepatocyte markers Alb, Afp, and Ttr was assayed by qRT-PCR. Expression of Alb and Ttr was not altered by Foxa1/2 deficiency (Figure 2B). However, Afp mRNA levels were increased about 10-fold in mutant livers (Figure 2B). Therefore, deletion of Foxa1/2 in late gestation did not affect hepatocyte morphology, and hepatocyte differentiation after liver specification was largely independent of Foxa1/2.
Figure 2. Hepatocyte differentiation is not affected by Foxa1/2 deficiency.
(A) Ultrastructure of hepatocytes in adult Foxa1loxP/loxPFoxa2loxP/loxP and Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice as determined by transmission EM analysis. Scale bars: 2 μm. (B) Alb, Afp, and Ttr mRNA levels relative to Gapdh in control and mutant adult livers, as assessed by real-time qRT-PCR. *P < 0.05 compared with control mice.
Table 1 .
Metabolic data of Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice
Liver-specific ablation of Foxa1/2 results in bile duct hyperplasia and fibrosis.
Differentiation of hepatoblasts into cholangiocytes initiates during mid-gestation (E14.5) in the mouse (3–5). We used cytokeratin 19 (CK19), a well-characterized marker for biliary epithelial cells (21), to analyze whether Foxa1/2 deletion was associated with abnormal bile duct development. CK19-positive cholangiocytes were found in portal tracts of both Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre and Foxa1loxP/loxPFoxa2loxP/loxP mice (Figure 3, A and B). Immunostaining showed that Foxa1/2 were highly expressed in both hepatocytes and cholangiocytes (Figure 1A) in control mice, consistent with previous studies (15). Both proteins were absent from hepatocytes and cholangiocytes in adult Foxa1/2 mutant mice (Figure 1A).
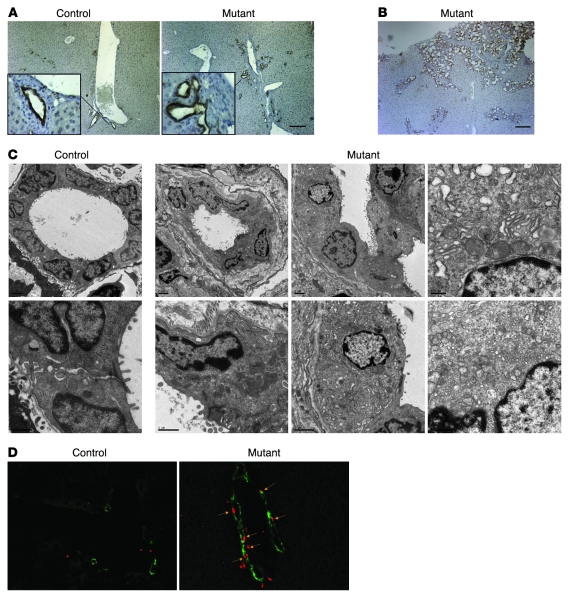
Figure 3. Hepatic deficiency of Foxa1/2 results in bile duct hyperplasia.
(A) Immunohistochemical staining of paraffin-embedded liver sections from 3-month-old Foxa1loxP/loxPFoxa2loxP/loxP and Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice with anti-CK19 antibody. Scale bar: 50 μm. Original magnification of insets, ×400. (B) Immunohistochemical staining of paraffin-embedded liver sections from 3-month-old mutant adult mice with anti-CK19 antibody. Scale bar: 50 μm. (C) Ultrastructure of bile ducts in adult control and mutant mice as determined by transmission EM analysis. Scale bars: 2 μm. (D) Immunofluorescent staining of proliferating cells in OCT-embedded liver sections from 3-month-old control and mutant mice. Mice were injected with BrdU 1 hour before sacrifice to label cells in S phase. BrdU-positive nuclei are shown in red; cholangiocytes were labeled with anti-CK19 antibody (green). Arrows point to BrdU-positive cholangiocytes. Original magnification, ×200.
Biliary development was dramatically affected by the absence of Foxa1/2 (Figure 3, A–C). CK19 immunostaining showed dilated, disorganized, and expanded bile ducts in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice as compared with controls (Figure 3, A and B). Ultrastructural analysis demonstrated that mutant bile ducts had irregular lumens and increased extracellular matrix deposition (Figure 3C). In addition, multiple branched bile ducts, sharing a single layer of cholangiocytes, were present (Figure 3C). Moreover, compared with littermate controls, mutant cholangiocytes contained disorganized and small mitochondria and less endoplasmic reticulum but normal microvilli (Figure 3C). Furthermore, mutant bile ducts appeared to be surrounded by an increased deposition of extracellular matrix (see below). BrdU incorporation, phosphorylated histone 3 (p–histone 3), and Ki67 immunostaining were used to identify cells in S phase, M phase, and G1 to M phase of the cell cycle, respectively (Figure 3D and Supplemental Figure 2). In addition, we quantified Ki67 mRNA levels (Supplemental Figure 2). All proliferation markers showed that while the proliferation rate of hepatocytes was unaffected by Foxa1/2 deficiency, that of cholangiocytes was dramatically increased (Figure 3D and Supplemental Figure 2). Thus, bile duct hyperplasia in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice resulted from increased cycling of cholangiocytes.
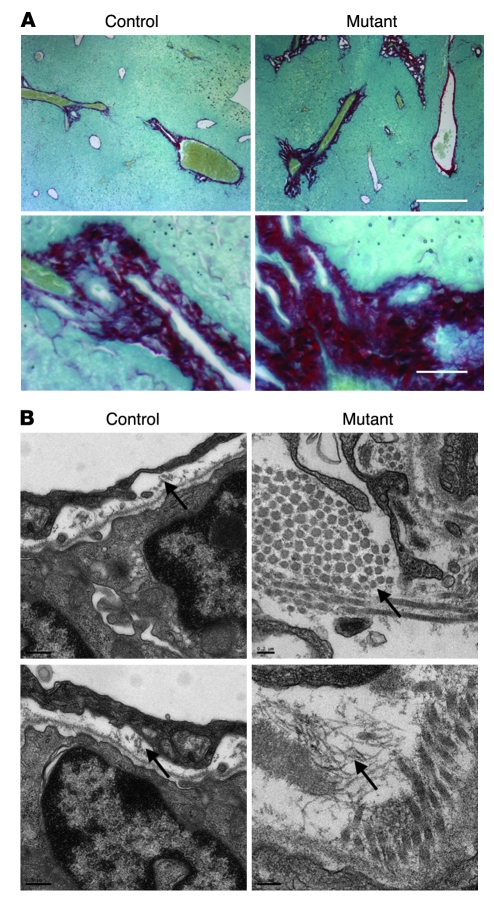
Next, we investigated the nature of the extracellular matrix deposits surrounding the bile ducts of Foxa1/2-deficient livers. Strong Sirius red staining, which labels collagen bundles, was found surrounding bile ducts in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice (Figure 4A). We confirmed this finding by ultrastructural analysis, which showed increased collagen fibers in portal tracts of Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice as compared with controls (Figure 4B). In addition, hepatic mRNA expression of multiple members of the collagen gene family was increased 2-fold or more in mutant mice compared with controls, including that of Col1a1, Col2a1, Col3a1, Col4a3, Col4a6, and Col6a3 (data not shown). Immunostaining of TGF-β did not show a significant difference between control and mutant mice (Supplemental Figure 3). Likewise, Tgfb mRNA levels were unchanged (Figure 5A). In addition, α–smooth muscle actin levels were unchanged between control and mutant mice (Supplemental Figure 3).
Figure 4. Hepatic deficiency of Foxa1/2 results in fibrosis.
(A) Sirius red staining of paraffin-embedded liver sections from 3-month-old control and mutant mice. Fast green FCF was used to counterstain. Scale bars: 10 μm. (B) Ultrastructure of collagen fibers (arrows) in adult control and mutant livers as determined by transmission EM analysis. Scale bars: 0.2 μm.
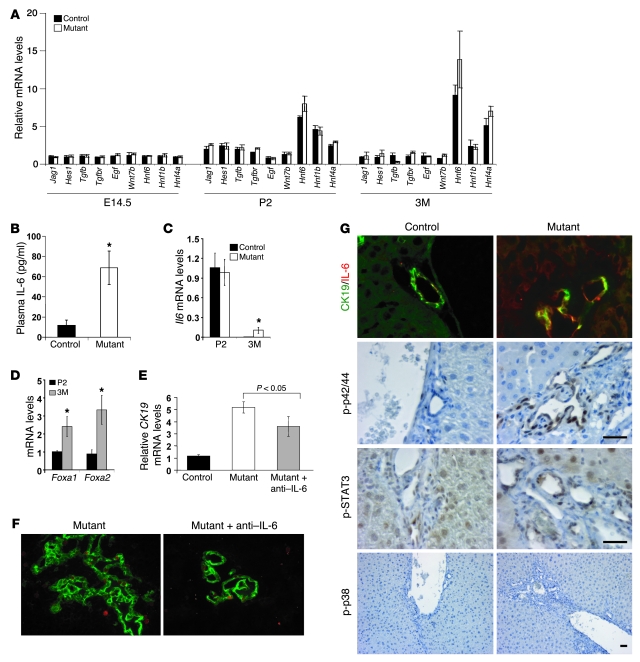
Figure 5. Bile duct proliferation in Foxa1/2-deficient mice correlates with IL-6 signaling.
(A) Hepatic mRNA levels of Jag1, Hes1, Tgfb, Tgfbr, Egf, Wnt7b, Hnf6, Hnf1b, and Hnf4a from E14.5, P2, and 3-month old Foxa1loxP/loxPFoxa2loxP/loxP and Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice were measured by real-time qRT-PCR. (B) Plasma IL-6 levels were assayed by sandwich ELISA. *P < 0.05 from comparison between mutant and control mice. (C) Hepatic Il6 mRNA levels in P2 and 3-month-old control and mutant mice. *P < 0.05 compared with control mice. (D) Hepatic Foxa1 and Foxa2 mRNA levels in P2 and 3-month-old control mice. *P < 0.05, 3M versus P2 mice. (E) Hepatic Ck19 mRNA levels in 3-month-old control and mutant mice and mutant mice after weekly treatment with a neutralizing anti–IL-6 antibody. (F) Reduced cholangiocyte proliferation after the treatment of IL-6 antagonist. Immunofluorescence staining for anti-CK19 (green) and anti-Ki67 (red) in OCT-embedded liver sections from 3-month-old mutant mice and mutant mice after treatment with anti–IL-6 antibody. Original magnification, ×100. (G) Immunofluorescence staining of OCT-embedded liver sections from control and mutant adult mice with anti–IL-6 (red) and anti-CK19 (green) antibodies. Immunohistochemical staining of paraffin-embedded liver sections from control and mutant adult mice with anti–p-p42/44 MAPK, anti–p-STAT3, and anti–p-p38 MAPK antibodies. Original magnification of top panel, ×400. Scale bars: 5 μm.
Bile duct proliferation is due to increased expression of IL-6.
Bile duct growth and proliferation are regulated by bile acids, hormones, and cytokines including TGF, epithelial growth factor (EGF), and IL-6 (22–27). In addition, abnormal bile duct proliferation can result from bile duct obstruction, inflammation, or developmental defects (25–27). To explore the mechanism of bile duct proliferation in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice, livers from E14.5, P2, and 3-month-old animals were used to screen the gene expression profiles of important regulatory pathways of bile duct development, including the Wnt, Notch, TGF, and EGF signaling pathways. Surprisingly, none of these pathways seem to be altered in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice (Figure 5A).
Next, we investigated early postnatal (P2) mice, in which biliary development had just completed, to test whether the ablation of Foxa1/2 causes a developmental defect. By CK19 immunostaining, bile duct formation was observed at P2 in both mutant and control mice (Supplemental Figure 4). In addition, mutant mice did not show abnormal formation of ductal plates and early bile ducts (Supplemental Figure 4), even though Foxa1/2 were completely ablated by P2 (Figure 1, A and B).
IL-6 is a key regulator of normal bile duct growth and bile duct proliferation by both in vitro and in vivo models (22, 23, 25–30). We found that plasma IL-6 levels were dramatically elevated in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice (Figure 5B), which correlated with increased hepatic mRNA levels of IL-6 in adult but not P2 mice (Figure 5C). We also found that IL-6 was expressed in P2 but absent in adult control livers (Figure 5C) and inversely correlated with Foxa1/2 expression, which increased from P2 to adult in the liver of control mice (Figure 5D). IL-6 immunostaining showed that increased IL-6 protein expression was found mostly in cholangiocytes rather than hepatocytes in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice (Figure 5G and Supplemental Figure 6). To investigate whether increased IL-6 was causally linked to bile duct proliferation in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice, we treated mice with a carrier- and endotoxin-free anti–IL-6 antibody to inhibit IL-6 activity. This IL-6 antagonist significantly reduced bile duct proliferation in Foxa1/2 mutants as shown by Ck19 and Ki67 mRNA levels and CK19/Ki67 double staining (Figure 5, E and F, and Supplemental Figure 2B). Ck19 mRNA levels were increased 5-fold in mutant mice as compared with controls, but decreased 27% after the treatment with IL-6 antagonist (Figure 5E). Consistent with the expression change seen for CK19, 2 other ductal cell markers, CK7 and CK20, were also significantly increased in mutant mice and decreased after IL-6 antagonist treatment (Supplemental Figure 5). Ki67 mRNA levels decreased about 50% (Supplemental Figure 2B), and Ki67-positive cholangiocytes decreased by about half in mutants after treatment with the IL-6 antagonist (Figure 5F). Thus, IL-6 antagonist administration blocked cholangiocyte growth by inhibiting proliferation. The fact that expression of the cytokeratin genes was not normalized completely by the anti–IL-6 antibody is likely due to incomplete blockage of the elevated hepatic IL-6 levels.
Both hepatocytes and cholangiocytes synthesize and secrete IL-6 (22, 31). IL-6 acts in both autocrine and paracrine fashions through the IL-6 receptor, which is composed by a heterodimer of gp80 and gp130 proteins (22, 32, 33). Levels of gp80 and gp130 mRNA and proteins were not significantly changed in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice as compared with controls (Figure 6A and Supplemental Figure 7). Previous in vitro studies had shown that IL-6–dependent cholangiocyte proliferation could be mediated via the STAT3, p42/44 MAPK, or p38 MAPK pathways (29, 32–37). Immunostaining for active or phosphorylated STAT3, p42/44 MAPK, or p38 MAPK showed that both the STAT3 and p42/44 MAPK pathways, but not the p38 MAPK pathway, were activated in cholangiocytes of Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice (Figure 5G). Interestingly, many hepatocytes were also positive for p-STAT3 (Figure 5G). This suggests that STAT3 signaling might be blocked in hepatocytes, as we did not observe an increase in hepatocyte proliferation in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice (Figure 3D). Increased expression of p21, cytokine-inducible SH2-containing protein (Cish), and retinoblastoma might counteract increased STAT3 signaling to maintain normal proliferation in hepatocytes of mutant mice (Supplemental Figure 8). However, IL-6 signaling in cholangiocytes promoted cell proliferation, which was also evidenced by the upregulation of IL-6 target genes such as fibrinogen-like 1 (Fgl1) (Supplemental Figure 2B) (38). Once IL-6 signaling was blocked by antagonist treatment, Fgl1 expression was also reduced significantly (Supplemental Figure 2B)
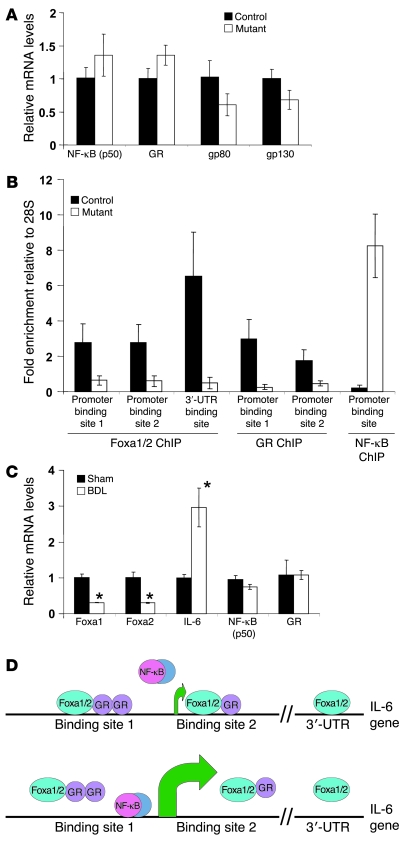
Figure 6. Foxa1/2 and GR corporately regulate IL-6 transcription.
(A) Hepatic mRNA levels of NF-κB (p50), GR, and IL-6 receptor gp80 and gp130 subunits from 3-month-old Foxa1loxP/loxPFoxa2loxP/loxP and Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice by real-time qRT-PCR. (B) ChIP assays for the occupancy of Foxa1/2, GR, and NF-κB on the mouse IL-6 promoter. (C) Hepatic mRNA levels from Foxa1, Foxa2, Il6, Nfkb (p50), and GR from wild-type mice after sham or BDL for 5 days, as assessed by real-time qRT-PCR. *P < 0.05 compared with sham-treated mice. (D) Model for the control of IL-6 transcription by Foxa1/2, GR and NF-κB. Green arrows indicate translational start sites. Top panel shows that GR-mediated IL-6 inhibition is dependent on Foxa1/2 occupancy. Bottom panel shows that once Foxa1/2 is deleted, NF-κB replaces the GR to bind to the IL-6 promoter and initiate IL-6 expression.
Glucocorticoid receptor–mediated IL-6 suppression is dependent of Foxa1/2.
Next, we investigated the molecular mechanism of the hyper-induction of IL-6 in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice. We analyzed the transcription factors known to regulate IL-6 expression by qRT-PCR and ChIP assays. The glucocorticoid receptor (GR) and NF-κB have been shown to competitively regulate the expression of many genes, including IL-6 (39–41). Under normal circumstances, GR binds to and suppress the IL-6 promoter. Upon stimulation, NF-κB replaces GR and activates IL-6 expression. We found no significant changes in GR and NF-κB (p50) mRNA levels between mutants and controls (Figure 6A). The mouse IL-6 promoter contained closely spaced binding sites for GR, Foxa1/2, and NF-κB (Figure 6D). We investigated the occupancy of all these factors on the IL-6 promoter in control and Foxa1/2 mutant livers by ChIP. Foxa1/2 were bound to IL-6 cis-regulatory elements at multiple sites, 2 in the promoter region and 1 in the 3′-UTR, but were absent from these sites in mutant liver as expected (Figure 6B). GR and Foxa1/2 have been shown to cooperatively regulate the expression of many genes including phosphoenolpyruvate carboxykinase and carbamoylphosphate synthetase-I (16, 42). Strikingly, NF-κB binding to the IL-6 promoter was greatly enhanced, whereas GR binding to IL-6 promoter was significantly decreased in Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice (Figure 6B). Thus, these data demonstrate that GR-mediated IL-6 suppression requires Foxa1/2.
To investigate whether Foxa1/2 regulate IL-6 expression in a pathophysiologic condition, we analyzed another paradigm of IL-6–mediated bile duct proliferation, the bile duct ligation (BDL) model. Consistent with previous findings, IL-6 expression was greatly induced after 5 days of BDL (Figure 6C). At the same time, Foxa1/2 expression was significantly downregulated in mice following BDL, whereas GR and NF-κB (p50) expression was not altered (Figure 6C). This suggests that the induction of IL-6 levels in bile duct obstruction is related to the suppression of Foxa1/2 expression. Our model of Foxa1/2-regulated IL-6 transcription is summarized in Figure 6D.
Discussion
Foxa1/2: terminators of bile duct expansion.
Using conditional gene ablation of Foxa1/2 in the late gestation liver, we have uncovered what we believe is a novel and unexpected role for these transcription factors in the control of bile duct proliferation. Foxa1/2-deficient mice exhibit dramatic bile duct hyperplasia and fibrosis. This phenotype is due, at least in part, to inappropriate induction of IL-6. Absence of Foxa1/2 causes loss of inhibitory regulation by the GR on IL-6 expression. However, hepatocyte differentiation was not dependent on Foxa1/2 deficiency after liver specification, in contrast to the situation in which these genes are ablated in foregut endoderm (9). The fact that reduced IL-6 expression in adult livers compared with neonatal livers correlates inversely with Foxa1/2 levels (Figure 5, C and D) suggests that increased Foxa1/2 expression may act as a terminating signal ending bile duct expansion via inhibition of IL-6 expression in the maturing liver.
Mutant mice at P2 showed normal IL-6 expression (Figure 5C) despite the fact that ablation of Foxa1/2 was already complete (Figure 1B). Therefore, at this stage, Foxa1/2 was not required for the control of IL-6 expression. In contrast, by 3 months of age, the absence of Foxa1/2 led to IL-6 overexpression. In addition, in control mice the expression of Foxa1/2 increased 2- to 3.5-fold between the postnatal period and adulthood (Figure 5D), and we suggest that this increased expression in the mature liver is one of the mechanisms that limits bile duct expansion by restricting IL-6 expression via increased GR occupancy. Dramatic phenotypic consequences of small changes in transcription factor expression are quite common, as evidenced by human autosomal dominant diseases such as maturity onset diabetes of the young (MODY), in which haploinsufficiency of at least 5 different transcription factors has been shown to cause disease.
IL-6 regulates hepatocyte and cholangiocyte proliferation differentially.
Both hepatocytes and cholangiocytes express Foxa1/2, thus, it is not surprising that we observed IL-6 induction in both cell types in Foxa1/2-deficient mice (Figure 5G and Supplemental Figure 6). However, Foxa1/a2 ablation only stimulated proliferation of cholangiocytes but not of hepatocytes (Figure 3D). This is consistent with previous studies, which showed that IL-6 addition to primary hepatocytes in culture stimulated expression of p21, thereby inhibiting hepatocyte proliferation (43). Thus, upregulation of p21 as well as Cish and retinoblastoma (Supplemental Figure 8) might prevent the proliferation of hepatocytes in mutant livers. In contrast, IL-6 administration triggers multiple proliferation pathways in cholangiocytes (29, 32–37). Although IL-6–null mice do not exhibit defect of bile duct development, BDL IL-6–null mice show attenuated bile duct proliferation (23, 44). In addition, IL-6 levels have been related to the progression of hepatocellular carcinoma (45), suggesting that the differential regulation of hepatocyte and cholangiocyte proliferation by IL-6 may be altered in cancer.
Fibrosis associated with bile duct proliferation has been found in many models including BDL. Cytokine-induced bile duct proliferation has been suggested to be a cause of fibrosis (46, 47). Therefore, overexpression of IL-6 might trigger fibrosis in our mutant mice, which may be independent of TGF-β signaling, since we did not observe a significant change in TGF-β levels between control and mutant mice (Supplemental Figure 3). Interestingly, the number of α–smooth muscle actin–positive cells was not altered between control and mutant mice (Supplemental Figure 3). This suggests that increased collagen deposition in Foxa1/2 mutant mice is not caused by an expansion of activated myofibroblasts; however, we cannot exclude the possibility of an increased per-cell production of extracellular matrix by myofibroblasts in our mutant mice. Ultrastructural analysis by EM showed that increased collagen fibers were adjacent to bile duct epithelia in Foxa1/2 mutant mice (Figure 4B). It has been suggested that cholangiocytes themselves might be involved in bile duct fibrogenesis (48, 49), thus it is conceivable that bile duct epithelia contribute directly to fibrogenesis in our mutant mice.
The role of Foxa1/2 in biliary disorders.
Abnormal cholangiocyte proliferation and growth is associated with many biliary disorders (25, 27, 28, 50). In addition, cholangiocyte proliferation occurs in most pathologic conditions of liver injury (27, 28). We recently showed that hepatic deletion of Foxa2 causes mild cholestasis in mice fed a cholic acid–enriched diet, and that FOXA2 is downregulated in human subjects with primary sclerosing cholangitis and biliary atresia (51). We show here that Foxa1/2 are downregulated in a mouse model of obstructive intrahepatic cholestasis, the BDL model (Figure 6C). Thus, it seems likely that FOXA1/2 play a role in patients with cholestatic disorders.
In summary, we have shown that lack of Foxa1/2 leads to abnormal bile duct development, a defect not observed in either Foxa1 or Foxa2 deficiency alone (12–14). We delineate the molecular defect to abnormal regulation of the Il6 gene by the GR, which requires Foxa1/2 for target occupancy. Thus, Foxa1/2 coordinately regulate the differentiation of the biliary tree and provide what we believe is a novel mechanism to limit the proliferation of cholangiocytes.
Methods
Animals.
Foxa1loxP/loxP mice (C57BL/6;129J) (52) were bred with Foxa2loxP/loxPAlfpCre (C57BL/6;129J) (16) mice to obtain Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mutant mice. Foxa1loxP/loxPFoxa2loxP/loxP mice without AlfpCre were used as controls. For the IL-6 antagonist study, 4 Foxa1loxP/loxPFoxa2loxP/loxPAlfpCre mice at 2 weeks of age were injected i.p. with 50 μg anti–IL-6 antibody (R&D Systems) weekly for 8 weeks. Mice were fed standard rodent chow diet and water ad libitum under standard 12-h light/12-h dark cycles. All mice were sacrificed at around 2 pm. Immediately after sacrifice, the liver was dissected, snap-frozen in liquid N2, and stored at –70°C for further analysis. Blood was collected from portal veins and plasma separated by immediate centrifugation and stored at –70°C. Blood biochemistry was assayed by AnaLytics Inc. Unless otherwise noted, 6 mice were used in each group. All procedures involving mice were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania and were in accordance with NIH guidelines.
Morphological analysis.
Immediately after mice were sacrificed, livers were fixed in 4% paraformaldehyde or OCT. Paraffin-embedded liver sections were analyzed by H&E staining. Liver sections were also analyzed by immunohistochemical or immunofluorescent staining with anti-Foxa1/2 (1:200; Santa Cruz Biotechnology Inc.), anti-CK19 (Troma-III, 1:20; Developmental Studies Hybridoma Bank), anti-BrdU (1:1,000; Zymed), anti-Ki67 (1:1,500; Vector Laboratories), anti–p–histone 3 (1:5,000; Vector Laboratories), anti–IL-6 (1;200; R&D Systems), anti–p-p42/44 (1:100; Cell Signaling Technology), anti–p-STAT3 (1:200; Cell Signaling Technology), and anti–p-p38 (1:200; Cell Signaling Technology) antibodies and 3,3′-diaminobenzidine (DAB) color staining, followed by counterstaining with hematoxylin. In addition, liver tissues were also fixed in 2.5% paraformaldehyde and 0.5% glutaraldehyde for transmission EM analysis. Sirius red staining was performed with the standard protocol (53).
Gene expression analysis by real-time qRT-PCR.
Total RNA was extracted from liver tissues with TRIzol (Invitrogen) and RNeasy Mini Kit (Qiagen). Standard real-time qRT-PCR was performed to quantify mRNA levels (54). All reactions were performed using 4 biological replicates and 3 technical replicates. A median Ct value was used for analysis, and all Ct values were normalized to expression of the housekeeping gene Gapdh. Primer sets used for qRT-PCR are listed in the Supplemental Methods.
ChIP assays.
ChIP assays were performed as described before (55). Input and precipitated DNA fragments were subjected to real-time qPCR with primers specific for putative binding sites of Foxa1/2, GR, and NF-κB in the mouse IL-6 promoter. Three putative Foxa1/2 binding sites at –660 bp (site 1), 119 bp (site 2), and 11,731 bp (3′-UTR site); 3 putative GR binding sites at –627/–590 bp (site 1) and 366 bp (site 2); and 1 putative NF-κB binding site at –90 bp from the transcriptional starting site were analyzed. Enrichment of the targets was calculated as follows: fold enrichment = 2 × ([28SChIP – interested geneChIP] – [28Sinput – interested geneinput]). Primer sets used for qPCR are listed in the Supplemental Methods.
Statistics.
Values are presented as mean ± SD. P values were determined using the 2-tailed Student’s t test with unequal variance. P < 0.05 was accepted as statistically significant.
Supplementary Material
Acknowledgments
We are grateful to Linda Greenbaum and Ben Stanger for critical reading of the manuscript. We thank Beth Helmbrecht, Karrie Brondell, Alan Fox, and Olga Smirnova for their excellent technical support and Gary Swain and the Morphology Core at the University of Pennsylvania School of Medicine for their advice on immunohistochemical staining. We thank Qian-Chun Yu and the Electron Microscopy Core for their help with the EM analysis. This study was supported by NIDDK grant P01-049210 (to K.H. Kaestner). Z. Li was supported by a Natural Sciences and Engineering Research Council of Canada postdoctoral fellowship.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Afp, α-fetoprotein; Alb, albumin; BDL, bile duct ligation; CK, cytokeratin; Foxa1, forkhead box protein A1; GR, glucocorticoid receptor; qRT-PCR, quantitative RT-PCR; Ttr, transthyretin.
Citation for this article: J. Clin. Invest. 119:1537–1545 (2009). doi:10.1172/JCI38201
References
- 1.Kaestner K.H. The making of the liver: developmental competence in foregut endoderm and induction of the hepatogenic program. Cell Cycle. 2005;4:1146–1148. doi: 10.4161/cc.4.9.2033. [DOI] [PubMed] [Google Scholar]
- 2.Friedman J.R., Kaestner K.H. The Foxa family of transcription factors in development and metabolism. Cell. Mol. Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaret K.S. Regulatory phases of early liver development: paradigms of organogenesis. Nat. Rev. Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 4.Duncan S.A. Mechanisms controlling early development of the liver. Mech. Dev. 2003;120:19–33. doi: 10.1016/S0925-4773(02)00328-3. [DOI] [PubMed] [Google Scholar]
- 5.Lemaigre F.P. Development of the biliary tract. Mech. Dev. 2003;120:81–87. doi: 10.1016/S0925-4773(02)00334-9. [DOI] [PubMed] [Google Scholar]
- 6.Kaestner K.H., et al. Six members of the mouse forkhead gene family are developmentally regulated. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7628–7631. doi: 10.1073/pnas.90.16.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaestner K.H., Hiemisch H., Luckow B., Schutz G. The HNF-3 gene family of transcription factors in mice: gene structure, cDNA sequence, and mRNA distribution. Genomics. 1994;20:377–385. doi: 10.1006/geno.1994.1191. [DOI] [PubMed] [Google Scholar]
- 8.Wan H., et al. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J. Biol. Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- 9.Lee C.S., Friedman J.R., Fulmer J.T., Kaestner K.H. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 10.Ferri A.L., et al. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134:2761–2769. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- 11.Lai E., et al. Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991;5:416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- 12.Kaestner K.H., Hiemisch H., Schutz G. Targeted disruption of the gene encoding hepatocyte nuclear factor 3gamma results in reduced transcription of hepatocyte-specific genes. Mol. Cell. Biol. 1998;18:4245–4251. doi: 10.1128/mcb.18.7.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaestner K.H., Katz J., Liu Y., Drucker D.J., Schutz G. Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sund N.J., et al. Hepatocyte nuclear factor 3beta (Foxa2) is dispensable for maintaining the differentiated state of the adult hepatocyte. Mol. Cell. Biol. 2000;20:5175–5183. doi: 10.1128/MCB.20.14.5175-5183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Besnard V., Wert S.E., Hull W.M., Whitsett J.A. Immunohistochemical localization of Foxa1 and Foxa2 in mouse embryos and adult tissues. Gene Expr. Patterns. 2004;5:193–208. doi: 10.1016/j.modgep.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L., Rubins N.E., Ahima R.S., Greenbaum L.E., Kaestner K.H. Foxa2 integrates the transcriptional response of the hepatocyte to fasting. Cell Metab. 2005;2:141–148. doi: 10.1016/j.cmet.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 18.Krupczak-Hollis K., et al. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev. Biol. 2004;276:74–88. doi: 10.1016/j.ydbio.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 19.Parviz F., et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 20.Loomes K.M., et al. Bile duct proliferation in liver-specific Jag1 conditional knockout mice: effects of gene dosage. Hepatology. 2007;45:323–330. doi: 10.1002/hep.21460. [DOI] [PubMed] [Google Scholar]
- 21.Fava G., Glaser S., Francis H., Alpini G. The immunophysiology of biliary epithelium. Semin. Liver Dis. 2005;25:251–264. doi: 10.1055/s-2005-916318. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z., et al. Interleukin-6, hepatocyte growth factor, and their receptors in biliary epithelial cells during a type I ductular reaction in mice: interactions between the periductal inflammatory and stromal cells and the biliary epithelium. Hepatology. 1998;28:1260–1268. doi: 10.1002/hep.510280514. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z., et al. Acute obstructive cholangiopathy in interleukin-6 deficient mice: compensation by leukemia inhibitory factor (LIF) suggests importance of gp-130 signaling in the ductular reaction. Liver. 2000;20:114–124. doi: 10.1034/j.1600-0676.2000.020002114.x. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto K., Fujii H., Michalopoulos G., Fung J.J., Demetris A.J. Human biliary epithelial cells secrete and respond to cytokines and hepatocyte growth factors in vitro: interleukin-6, hepatocyte growth factor and epidermal growth factor promote DNA synthesis in vitro. Hepatology. 1994;20:376–382. doi: 10.1002/hep.1840200217. [DOI] [PubMed] [Google Scholar]
- 25.LeSage G., Glaser S., Alpini G. Regulation of cholangiocyte proliferation. Liver. 2001;21:73–80. doi: 10.1034/j.1600-0676.2001.021002073.x. [DOI] [PubMed] [Google Scholar]
- 26.Alvaro D., Gigliozzi A., Attili A.F. Regulation and deregulation of cholangiocyte proliferation. J. Hepatol. 2000;33:333–340. doi: 10.1016/S0168-8278(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 27.Strazzabosco M., Spirli C., Okolicsanyi L. Pathophysiology of the intrahepatic biliary epithelium. J. Gastroenterol. Hepatol. 2000;15:244–253. doi: 10.1046/j.1440-1746.2000.02091.x. [DOI] [PubMed] [Google Scholar]
- 28.Alvaro D., et al. Proliferating cholangiocytes: a neuroendocrine compartment in the diseased liver. Gastroenterology. 2007;132:415–431. doi: 10.1053/j.gastro.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 29.Meng F., Yamagiwa Y., Ueno Y., Patel T. Over-expression of interleukin-6 enhances cell survival and transformed cell growth in human malignant cholangiocytes. J. Hepatol. 2006;44:1055–1065. doi: 10.1016/j.jhep.2005.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ezure T., et al. The development and compensation of biliary cirrhosis in interleukin-6-deficient mice. Am. J. Pathol. 2000;156:1627–1639. doi: 10.1016/s0002-9440(10)65034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J., Gores G.J., Patel T. Lipopolysaccharide induces cholangiocyte proliferation via an interleukin-6-mediated activation of p44/p42 mitogen-activated protein kinase. Hepatology. 1999;29:1037–1043. doi: 10.1002/hep.510290423. [DOI] [PubMed] [Google Scholar]
- 32.Kamimura D., Ishihara K., Hirano T. IL-6 signal transduction and its physiological roles: the signal orchestration model. Rev. Physiol. Biochem. Pharmacol. 2003;149:1–38. doi: 10.1007/s10254-003-0012-2. [DOI] [PubMed] [Google Scholar]
- 33.Hirano T., Nakajima K., Hibi M. Signaling mechanisms through gp130: a model of the cytokine system. Cytokine Growth Factor Rev. 1997;8:241–252. doi: 10.1016/S1359-6101(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 34.Park J., Tadlock L., Gores G.J., Patel T. Inhibition of interleukin 6-mediated mitogen-activated protein kinase activation attenuates growth of a cholangiocarcinoma cell line. Hepatology. 1999;30:1128–1133. doi: 10.1002/hep.510300522. [DOI] [PubMed] [Google Scholar]
- 35.Nozaki I., et al. Regulation and function of trefoil factor family 3 expression in the biliary tree. Am. J. Pathol. 2004;165:1907–1920. doi: 10.1016/S0002-9440(10)63243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokoyama T., et al. Human intrahepatic biliary epithelial cells function in innate immunity by producing IL-6 and IL-8 via the TLR4-NF-kappaB and -MAPK signaling pathways. Liver Int. 2006;26:467–476. doi: 10.1111/j.1478-3231.2006.01254.x. [DOI] [PubMed] [Google Scholar]
- 37.Chen L.P., et al. Activation of interleukin-6/STAT3 in Rat cholangiocyte proliferation induced by lipopolysaccharide. Dig. Dis. Sci. 2009;54:547–554. doi: 10.1007/s10620-008-0401-0. [DOI] [PubMed] [Google Scholar]
- 38.Liu Z., Ukomadu C. Fibrinogen-like protein 1, a hepatocyte derived protein is an acute phase reactant. Biochem. Biophys. Res. Commun. 2008;365:729–734. doi: 10.1016/j.bbrc.2007.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pascussi J.M., et al. Pathophysiological factors affecting CAR gene expression. Drug Metab. Rev. 2003;35:255–268. doi: 10.1081/DMR-120026394. [DOI] [PubMed] [Google Scholar]
- 40.De Bosscher K., et al. Glucocorticoids repress NF-kappaB-driven genes by disturbing the interaction of p65 with the basal transcription machinery, irrespective of coactivator levels in the cell. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3919–3924. doi: 10.1073/pnas.97.8.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhavsar P.K., Sukkar M.B., Khorasani N., Lee K.Y., Chung K.F. Glucocorticoid suppression of CX3CL1 (fractalkine) by reduced gene promoter recruitment of NF-kappaB. FASEB J. 2008;22:1807–1816. doi: 10.1096/fj.07-094235. [DOI] [PubMed] [Google Scholar]
- 42.Christoffels V.M., et al. Glucocorticoid receptor, C/EBP, HNF3, and protein kinase A coordinately activate the glucocorticoid response unit of the carbamoylphosphate synthetase I gene. Mol. Cell. Biol. 1998;18:6305–6315. doi: 10.1128/mcb.18.11.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun R., Jaruga B., Kulkarni S., Sun H., Gao B. IL-6 modulates hepatocyte proliferation via induction of HGF/p21cip1: regulation by SOCS3. Biochem. Biophys. Res. Commun. 2005;338:1943–1949. doi: 10.1016/j.bbrc.2005.10.171. [DOI] [PubMed] [Google Scholar]
- 44.Cressman D.E., et al. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 45.Naugler W.E., et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 46.Lazaridis K.N., Strazzabosco M., Larusso N.F. The cholangiopathies: disorders of biliary epithelia. Gastroenterology. 2004;127:1565–1577. doi: 10.1053/j.gastro.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Strazzabosco M., Fabris L., Spirli C. Pathophysiology of cholangiopathies. J. Clin. Gastroenterol. 2005;39:S90–S102. doi: 10.1097/01.mcg.0000155549.29643.ad. [DOI] [PubMed] [Google Scholar]
- 48.Xia J.L., Dai C., Michalopoulos G.K., Liu Y. Hepatocyte growth factor attenuates liver fibrosis induced by bile duct ligation. Am. J. Pathol. 2006;168:1500–1512. doi: 10.2353/ajpath.2006.050747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diaz R., et al. Evidence for the epithelial to mesenchymal transition in biliary atresia fibrosis. Hum. Pathol. 2008;39:102–115. doi: 10.1016/j.humpath.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 50.Alpini G., McGill J.M., Larusso N.F. The pathobiology of biliary epithelia. Hepatology. 2002;35:1256–1268. doi: 10.1053/jhep.2002.33541. [DOI] [PubMed] [Google Scholar]
- 51.Bochkis I.M., et al. Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nat. Med. 2008;14:828–836. doi: 10.1038/nm.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao N., et al. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22:3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rojkind M., Valadez G. Regulation of fibroblast proliferation by Kupffer cells and monocytes. Ciba Found. Symp. 1985;114:208–221. doi: 10.1002/9780470720950.ch14. [DOI] [PubMed] [Google Scholar]
- 54.Gupta R.K., et al. The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. J. Clin. Invest. 2005;115:1006–1015. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubins N.E., et al. Transcriptional networks in the liver: hepatocyte nuclear factor 6 function is largely independent of Foxa2. Mol. Cell. Biol. 2005;25:7069–7077. doi: 10.1128/MCB.25.16.7069-7077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.