Abstract
Hairpin pyrrole-imidazole polyamides are a class of cell-permeable DNA-binding small molecules that can disrupt transcription factor-DNA binding and regulate endogenous gene expression. The covalent linkage of antiparallel Py-Im ring pairs with an γ-amino acid turn unit affords the classical hairpin Py-Im polyamide structure. Closing the hairpin with a second turn unit yields a cyclic polyamide, a lesser-studied architecture mainly attributable to synthetic inaccessibility. We have applied our methodology for solution-phase polyamide synthesis to cyclic polyamides with an improved high yield cyclization step. Cyclic 8-ring Py-Im polyamides 1-3 targets the DNA sequence 5′-WGWWCW-3′ which corresponds to the androgen response element (ARE) bound by the androgen receptor transcription factor to activate gene expression. We find that cyclic Py-Im polyamides 1-3 bind DNA with exceptional high affinities and regulate the expression of AR target genes in cell culture studies from which we infer the cycle is cell permeable.
Introduction
Modulating the expression of eukaryotic gene networks by small molecules is a challenge at the frontier of chemical biology. Pyrrole-imidazole polyamides are a class of cell-permeable small molecules that bind to the minor groove of DNA in a sequence specific manner.1,2 Side-by-side stacking of N-methylpyrrole (Py) and N-methylimidazole (Im) carboxamides (Im/Py pairs) distinguish G•C from C•G base pairs, whereas Py/Py pairs specify for both T•A and A•T.3 Py-Im hairpin polyamides have been programmed for a broad repertoire of DNA sequences with affinities similar to endogenous transcription factors.4 They are cell permeable and influence gene transcription by disrupting protein-DNA interfaces.2,5,6 Hairpin polyamide interference of DNA binding by transcription factors such as HIF-1α,7 androgen receptor (AR),8 and AP-19 has been described in recent years, yielding a new approach towards gene control by small molecules.
In parallel with our gene regulation studies, a significant effort has been devoted to maximizing the biological potency of hairpin Py-Im polyamides through structural modifications. In particular, we have recently demonstrated that hairpin polyamides bearing the (R)-β-amino-γ-turn, such as polyamide 4, possess favorable binding affinities to DNA and are useful in gene regulation studies (Figure 1).5g A significant effort exists in our laboratory to regulate aberrant AR-activated gene expression in prostate cancer.8 To further optimize lead oligomer 4 it would seem reasonable that closing the hairpin with an identical linker, yielding a cyclic structure 1, may further enhance DNA affinity (Figure 1). Previous syntheses of cyclic polyamides using solid-phase protocols are characterized by low reaction yields due to inefficient macrocyclization.10 We report here the solution-phase synthesis of cyclic polyamides 1-3 with an improved high yield cyclization step. In addition we examined the DNA binding properties of these compounds by thermal duplex DNA melting as well as preliminary studies of their in vitro ADMET properties. Cyclic Py-Im polyamides 1-3 were shown to regulate endogenous gene expression in cell culture experiments.
Figure 1.
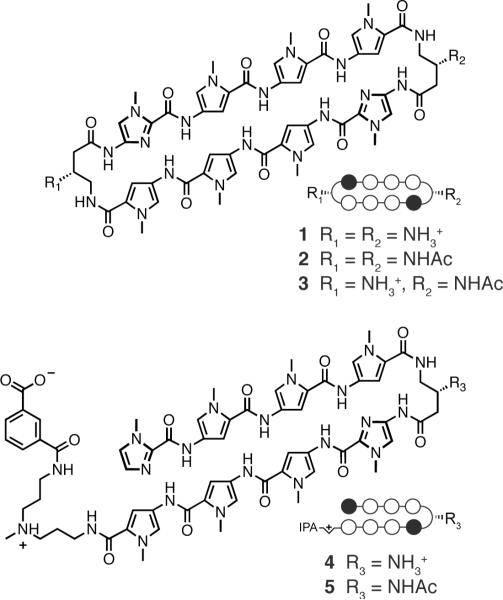
Chemical structures for cyclic and hairpin polyamides 1-5 targeted to the DNA-sequence 5′-WGWWCW-3′ are shown with ball-and-stick models. Ball-and-stick representation legend: black and white circles represent N-methylimidazole and N-methylpyrrole units, respectively, IPA denotes the terminal isophthalic acid substituent, and white half-diamond with + sign represents the triamine linker unit.
Results and Discussion
Solution-Phase Synthesis of Cyclic Polyamides
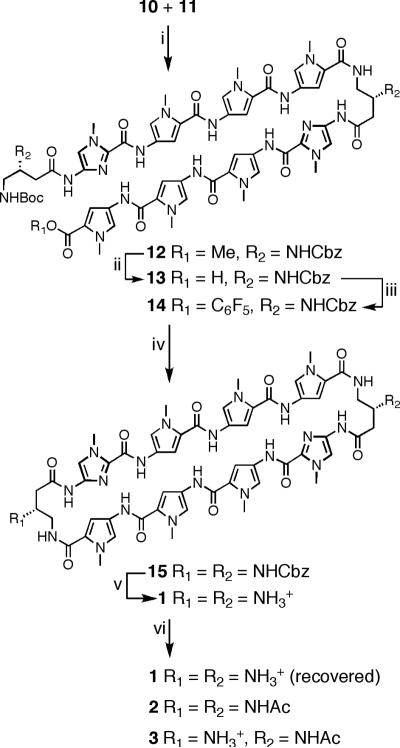
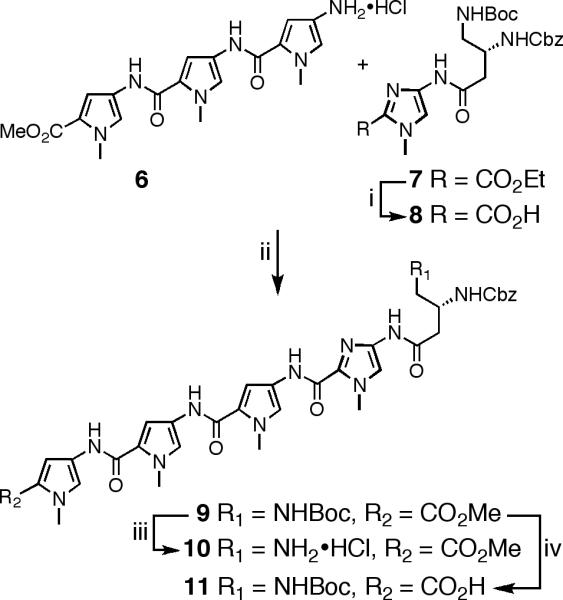
Due to the symmetrical nature of cyclic polyamides 1-3 and their sequence similarity to previously described hairpin polyamide 4,11 PyPyPy trimer 6 and Im-turn dimer 7 provide all the necessary atoms to synthesize 1-3. The preparation of advanced intermediates 6 and 7 have been detailed in the preceding paper11 from readily available building blocks.12 The cornerstone of our synthesis strategy capitalizes on the disparate physical properties of starting materials versus products, which permit purification of most intermediates to be achieved by combinations of precipitation, trituration, and crystallization. In addition, in situ deprotection of advanced pentafluorophenyl ester polyamide 14 at high dilution leads to macrocyclization in high yield affording cyclic polyamide 15.
The synthesis of tetramer-turn 9 begins with Im-turn dimer 7 (Scheme 1). Saponification of 7 with aqueous KOH in methanol at 37 °C, followed by neutralization, precipitation, and Et2O trituration, yields Im-turn acid 8 in 95% yield. Amide coupling of 8 with pyrrole trimer 6 provides pentamer 9 in 96% yield. The utilization of a small excess of 6 relative to 8 drives the reaction to completion, and residual 6 is readily separated from 9 following precipitation in water and aqueous washing of residual solid 9. With all atoms in place for the target cyclic polyamide 1, compound 9 was elaborated to amine salt 10 (99% yield) by reaction with HCl in 1,4-dioxane. Carboxylic acid 11 was generated by saponification of 10 with NaOH in 1,4-dioxane in 95 % yield.
Scheme 1.
Preparation of 10 and 11a
aReagents: (i) 1.0 M KOH (aq), MeOH, 37°C, 2 h, 95%; (ii) 8, PyBOP, DMF, DIEA, 6, 23°C, 4 h, 96%; (iii) 4.0 M HCl in 1,4-dioxane, 23°C, 2 h, 99% (iv) 1.0 M NaOH (aq), 1,4-dioxane, 42°C, 3 h, 95%.
Assembly of the acyclic advanced intermediate 12 was achieved by PyBOP-mediated coupling of intermediates 10 and 11 in 94 % yield (Scheme 2). A small excess of amine salt 10 was utilized to drive the reaction to completion. Saponification of ester 12 proceeded smoothly with aqueous NaOH in 1,4-dioxane, yielding 13 in 93% yield. Activation of acid 13 as the pentafluorophenol ester 14 provided the necessary functionality to afford macrocyclization following removal of the terminal t-butyl carbamate (Boc) protecting group. In our hands, we found the pentafluorophenol ester sufficiently activated the terminal acid for amide coupling while avoiding undesired oligomerization and/or decomposition processes that are conceivable with more reactive functionalities, such as acid chlorides. Premature initiation of the macrocyclization reaction was tempered by keeping the terminal amine protonated until transferred into a dilute solution of acetonitrile. Addition of an amine base (DIEA) then generated the free terminal amine, which could then undergo macrocyclization in dilute solvent conditions to deliver 15, which was directly deprotected. The benzyl carbamate protecting groups were cleaved by treatment with superacid conditions (trifluoromethylsulfonic acid-trifluoroacetic acid) to provide 1 in 68% yield over 3 steps. Controlled acetylation of 1 by reaction with sub-stoichiometric quantities of Ac2O in NMP/DIEA provided a statistical population of 1 (18%), 2 (40%), and 3 (22%) that were easily separable by preparative HPLC. Acetylated hairpin 5 was prepared using excess Ac2O/pyridine in 95% yield from previously reported amine hairpin 4.11
Scheme 2.
Preparation of 1, 2, and 3a
aReagents: (i) PyBOP, DMF, DIEA, 23°C, 2 h, 94%; (ii) 1.0 M NaOH (aq), 1,4-dioxane, 40°C, 4 h, 93%; (iii) DCM, DCC, pentafluorophenol, DMAP, 23°C, 12 h, 80%; (iv) a) TFA, DCM, 23°C, concentrate; b) DMF, acetonitrile, DIEA, 0-23°C, 3 days; (v) CF3SO3H, CF3CO2H, 23°C, 5 min, 68% over 3 steps; (vi) NMP, Ac2O, 23°C, 18% of 1 (recovered), 40% of 2, 22% of 3.
Thermal Stabilization of DNA-duplexes by Polyamides
Quantitative DNase I footprint titrations have historically been utilized to measure polyamide-DNA binding affinities and specificities.13 However, this method is limited to measuring Ka values ≤ 2 × 1010 M-1, which invalidates this technique for quantifying the exceptionally high DNA-binding affinities of cycles 1-3.14 The magnitude of DNA thermal stabilization (ΔTm) of DNA-polyamide complexes has been utilized to rank order polyamides with high DNA binding affinities.5g,15 Accordingly, we have employed melting temperature analysis (ΔTm) for dissecting differences in DNA-binding affinities of hairpin versus cyclic polyamides. Spectroscopic analyses were performed on a 14-mer duplex DNA mimicking the androgen response element (ARE) DNA sequence, 5′-TTGCTGTTCTGCAA-3′ DNA duplex, which contains one polyamide binding site. As shown in Table 1 polyamides 1-5 provided an increase in the duplex DNA melting temperature relative to the individual DNA duplex, thereby confirming polyamide-DNA binding. Chiral hairpin 4 led to an increased melting temperature ΔTm = 18.4 °C whereas cyclic polyamide 1 yielded a higher ΔTm-value of 23.6 °C. Cyclic polyamides 1-3 reveal stronger stabilizations than parent hairpin analogs 4 and 5. Acylation of the β-amino turns was shown to decrease the thermal stabilization values in both hairpin and cyclic motifs, presumably due to the loss of the electrostatic benefit of the cation protonated amine on the turn unit.
Table 1.
Tm values for polyamides for 1 - 5a
| Polyamides | Tm/°C | ΔTm/°C |
|---|---|---|
| - | 60.0 (±0.3) | - |
| 83.5 (±0.5) | 23.6 (±0.6) | |
| 81.2 (±0.2) | 21.3 (±0.4) | |
| 82.0 (±0.0) | 22.1 (±0.3) | |
| 78.4 (±0.5) | 18.4 (±0.6) | |
| 76.0 (±0.5) | 16.1 (±0.6) | |
All values reported are derived from at least three melting temperature experiments with standard deviations indicated in parentheses. ΔTm values are given as Tm(DNA/polyamide) - Tm(DNA). The propagated error in ΔTm measurements is the square root of the sum of the square of the standard deviations for the Tm values.
Biological Assay for Cell Permeability
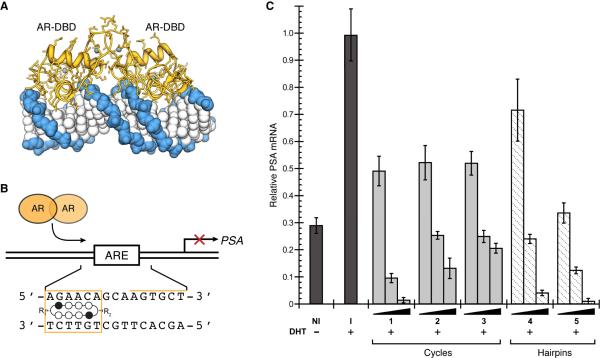
Hairpin polyamides have been shown to modulate endogenous gene expression in living cells by disrupting transcription factor-DNA binding in gene promoters.2,7-9 Recently, hairpin polyamide 4 was shown to inhibit androgen receptor-mediated expression of prostate specific antigen (PSA) in LNCaP cells by targeting the DNA sequence 5′-AGAACA-3′ found in the ARE.8 We utilized this cell culture transcription assay to investigate the biological activity of cyclic polyamides 1-3 in comparison to hairpin polyamides 4 and 5. Since small structural changes to polyamides have been shown to correlate with differences in cellular uptake properties,5 it was not obvious whether cyclic polyamides 1-3 would permeate cell membranes and exhibit biological activity comparable to hairpin polyamides 4 and 5. Quantitative real-time RT-PCR analysis of DHT-induced PSA expression revealed cyclic polyamides 1-3 all decreased PSA mRNA levels in LNCaP cells, with cycle 1 exhibiting comparable activity to acetylated hairpin polyamide 5 (Figure 2). Based on these results we can infer that this class of cyclic Py-Im polyamides are cell permeable and can regulate endogenous gene expression in cell culture.
Figure 2.
Targeting the ARE with DNA-binding polyamides. (a) X-ray crystal structure of androgen receptor homodimer DNA-binding domain bound to the sequence 5′-CTGTTCTTGATGTTCTGG-3′ (PDB 1r4i).16 (b) Map of the PSA-ARE site (top) and schematic representation of a cyclic polyamide targeting the PSA-ARE site 5′-AGAACA-3′. (c) Inhibition of induced PSA mRNA expression in LNCaP cells by cyclic Py-Im polyamides 1-3 and hairpin polyamides 4 and 5 (dosed at 0.3, 3, and 30 μM) by real-time quantitative PCR. The results were normalized to a DHT-induced, untreated control (control=1), and the error bars represent the standard error of the mean of a single experiment performed in biological triplicate. The entire experiment was reproduced four times, with similar results. NI = non-induced, I = induced, DHT = dihydrotestosterone.
ADMET Studies of Polyamides 1 and 5
Due to the promising cell culture results obtained with cyclic polyamide 1 and hairpin polyamide 5 against PSA gene expression, we contracted preclinical in vitro absorption, distribution, metabolism, excretion and toxicity (ADMET)17 studies for both compounds.18 Results from this study are summarized as follows and additional detail can be found in the Supporting Information. Polyamides 1 and 5 were both found to exhibit low Caco-2 permeability, suggesting neither compound may be orally available. Both 1 and 5 were found to be almost exclusively protein bound in plasma with half-lives greater than 2 h. A recent positron emission tomography (PET)-based biodistribution study of a related hairpin polyamide in mice revealed high levels of liver occupancy following tail vein dosage.19 Based on this result we investigated the liver stability of candidate polyamides 1 and 5. Microsomal intrinsic clearance studies found half-lives greater than 3 hr for 1 and 5 in both human and rat liver microsomes, and no significant inhibition was measured against any cytochrome P450 isoform examined (Cyp1A2/CEC, Cyp2C8/DBF, Cyp2C9/DBF, Cyp2C19/DBF, Cyp2D6/AMMC, Cyp3A4/BFC, Cyp3A4/DBF). Furthermore, no obvious toxicity (IC50 > 100 μM) was observed in the human hepatocellular carcinoma cell line HepG2. In addition, standard hERG FastPatch assays of cardiac toxicity found both polyamides (1 and 5) were devoid of unwanted inhibition (IC50 > 100 μM).
Conclusion
We describe a solution-phase synthesis methodology for preparing cyclic Py-Im polyamides, highlighted by an efficient macrocyclization between the alkyl linker amine and a pentafluorophenyl ester-activated amino acid. The three cyclic Py-Im polyamides possessed high DNA-binding affinities and were capable of accessing the nucleus in cell culture as judged by their ability to downregulate AR-activated PSA expression in cell culture. Preclinical ADMET analysis of cyclic polyamide 1 and hairpin polyamide 5 revealed favorable drug-like properties such as high liver stability and low toxicity. Ongoing work is focused on characterizing the precise molecular interactions between cyclic polyamides and their cognate DNA sequences by high-resolution crystallographic studies.
Experimental Section
General
Chemicals and solvents were purchased from Sigma-Aldrich and were used without further purification. (R)-3,4-Cbz-Dbu(Boc)-OH was purchased from Senn Chemicals AG (code number 44159). All DNA oligomers were purchased HPLC purified from Integrated DNA Technologies. Water (18 MΩ) was purified using a Millipore MilliQ purification system. Centrifugation was performed in a Beckman Coulter bench-top centrifuge (Allegra 21R) equipped with a Beckman swing-out rotor (model S4180). Analytical HPLC analysis was conducted on a Beckman Gold instrument equipped with a Phenomenex Gemini analytical column (250 × 4.6 mm, 5 μm), a diode array detector, and the mobile phase consisted of a gradient of acetonitrile (MeCN) in 0.1% (v/v) aqueous CF3CO2H. Preparative HPLC was performed on an Agilent 1200 system equipped with a solvent degasser, diode array detector, and a Phenomenex Gemini column (250 × 21.2 mm, 5 μm). A gradient of MeCN in 0.1% (v/v) aqueous CF3CO2H was utilized as the mobile phase. UV-Vis measurements were made on a Hewlett-Packard diode array spectrophotometer (Model 8452 A) and polyamide concentrations were measured in 0.1% (v/v) aqueous CF3CO2H using an extinction coefficient of 69200 M-1·cm-1 at λmax near 310 nm. NMR spectroscopy was performed on a Varian instrument operating at 499.8 MHz (for 1H) or 125.7 MHz (for 13C) at ambient temperature. All NMR analyses were performed in DMSO-d6, and chemical shifts are reported in parts per million relative to the internal solvent peak referenced to 2.49 (for 1H) or 39.5 (for 13C). High-resolution mass spectrometry (HRMS) was recorded in positive-ion mode by fast-atom bombardment (FAB+) on a JOEL JMS-600H instrument or by electrospray ionization (ESI+) on a Waters Acquity UPLC-LCT Premiere XE TOF-MS system.
UV Absorption Spectrophotometry
Melting temperature analysis was performed on a Varian Cary 100 spectrophotometer equipped with a thermo-controlled cell holder possessing a cell path length of 1 cm. A degassed aqueous solution of 10 mM sodium cacodylate, 10 mM KCl, 10 mM MgCl2, and 5 mM CaCl2 at pH 7.0 was used as analysis buffer. DNA duplexes and hairpin polyamides were mixed in 1:1 stoichiometry to a final concentration of 2 μM for each experiment. Prior to analysis, samples were heated to 90 °C and cooled to a starting temperature of 23 °C with a heating rate of 5 °C/min for each ramp. Denaturation profiles were recorded at λ = 260 nm from 23 °C to 90 °C with a heating rate of 0.5 °C/min. The reported melting temperatures were defined as the maximum of the first derivative of the denaturation profile.
Measurement of Androgen-Induced PSA mRNA
Experiments were performed as described previously8 with the following modifications: (1) all compounds and controls were prepared in neat DMSO then diluted with media to a final concentration of 0.1% DMSO, and (2) mRNA was isolated with the RNEasy 96 kit (Qiagen, Valencia, CA).
BocHN-(R)β-CbzHNγ-Im-CO2H (8)
A solution of BocHN-(R)β-CbzHNγ-Im-CO2Et 7 (450 mg, 0.894 mmol) dissolved in MeOH (1.0 mL) and aqueous KOH (1N, 2.0 mL, 2.0 mmol) was stirred at 37 °C for 2 h. The reaction mixture was added to a cooled (ice bath) solution of distilled H2O (10 mL) preacidified with aqueous HCl (1N, 2.0 mL, 2.0 mmol), yielding a precipitate that was isolated by centrifugation (~ 4500 rpm). The residual solid was again suspended in distilled H2O (10 mL) and collected by centrifugation. The resultant solid, which contained a small amount of residual H2O, was frozen and lyophilized to dryness, and then suspended in excess anhydrous Et2O, filtered, and the filter cake washed with copious amounts of anhydrous Et2O. Drying of the brown solid in vacuo yielded saponified dimer 8 (404 mg, 95%). 1H NMR: δ 10.46 (s, 1H), 7.47 (s, 1H), 7.31-7.26 (m, 5H), 7.02 (d, J = 8.3 Hz, 1H), 6.79 (t, J = 5.4 Hz, 1H), 4.97 (s, 2H), 3.92 (m, 1H), 3.88 (s, 3H), 3.60 (s, 1H), 3.01 (m, 2H), 2.41 (m, 2H), 1.35 (s, 9H); 13C NMR: δ 167.6, 160.0, 155.8, 155.4, 137.11, 137.09, 131.6, 128.2, 127.66, 127.55, 114.6, 77.7, 65.1, 48.7, 43.5, 38.0, 35.4, 28.2; HRMS (FAB+) calc'd for C22H30N5O7 [M+H]+ 476.2145, found 476.2130.
BocHN-(R)β-CbzHNγ-ImPyPyPy-CO2Me (9)
A solution of BocHN-(R)β-CbzHNγ-Im-CO2H 8 (300 mg, 0.631 mmol) and PyBOP (345 mg, 0.663 mmol) in DMF (3.2 mL) and DIEA (330 μL, 1.9 mmol) was stirred at 23°C for 10 min. The solution was then treated with solid (powdered) HCl•H2N-PyPyPy-CO2Me 6 (288 mg, 0.663 mmol) and stirred at 23°C for 4 h. The solution was then added to distilled H2O (10 mL) pre-acidified with aqueous HCl (1N, 2 mL, 2 mmol), yielding a precipitate that was isolated by centrifugation (~ 4500 rpm). The residual solid was again suspended in distilled H2O (10 mL) and collected by centrifugation (repeated 3X). The resultant solid, which contained a small amount of residual H2O, was frozen and lyophilized to dryness. The solid was triturated with anhydrous Et2O and filtered over a sintered glass funnel. The resultant solid was washed with copious amounts of anhydrous Et2O and dried in vacuo to yield BocHN-(R)β-CbzHNγ-ImPyPyPy-CO2Me 9 as a tan solid (518 mg, 96%). 1H NMR: δ 10.17 (s, 1H), 10.00 (s, 1H), 9.95 (s, 1H), 9.93 (s, 1H), 7.46 (d, J = 1.7 Hz, 1H), 7.44 (s, 1H), 7.31-7.29 (m, 5H), 7.27 (d, J = 1.7 Hz, 1H), 7.23 (d, J = 1.7 Hz, 1H), 7.14 (d, J = 1.7 Hz, 1H), 7.07 (d, J = 1.7 Hz, 1H), 7.04 (d, J = 8.3 Hz, 1H), 6.90 (d, J = 2.0 Hz, 1H), 6.81 (t, J = 5.9 Hz, 1H), 4.98 (s, 2H), 3.96 (m, 1H), 3.95 (s, 3H), 3.85 (s, 3H), 3.84 (s, 3H), 3.83 (s, 3H), 3.73 (s, 3H), 3.03 (m, 2H), 2.46 (m, 2H), 1.36 (s, 9H); 13C NMR: δ 167.8, 160.8, 158.5, 158.4, 155.84, 155.81, 155.5, 137.1, 136.0, 134.0, 128.3, 127.7, 127.6, 123.06, 123.00, 122.5, 122.2, 121.2, 120.7, 118.7, 118.6, 118.5, 114.0, 108.4, 104.9, 77.8, 65.2, 50.9, 48.8, 43.6, 38.2, 36.20, 36.18, 36.09, 34.9, 28.2; HRMS (FAB+) calc'd for C41H49N11O10[M•]+ 855.3663, found 855.3688.
HCl•H2N-(R)β-CbzHNγ-ImPyPyPy-CO2Me (10)
A solution of BocHN-(R)β-CbzHNγ-ImPyPyPy-CO2Me 9 (125 g, 0.146 mmol) in anhydrous HCl in 1,4-dioxane (4.0 M, 10 mL) was stirred at 23°C for 2 h. The mixture was then diluted with 100 mL of anhydrous Et2O and filtered over a sintered glass funnel. The resultant solid was washed with copious amounts of anhydrous Et2O and dried in vacuo to yield HCl•H2N-(R)β-CbzHNγ-ImPyPyPy-CO2Me 10 as a brown solid (114 mg, 99%). 1H NMR: δ 10.38 (s, 1H), 9.98 (s, 1H), 9.96 (s, 1H), 9.94 (s, 1H), 8.10 (m, 3H), 7.46 (d, J = 1.7 Hz, 1H), 7.45 (s, 1H), 7.42 (d, J = 8.3 Hz, 1H), 7.34-7.28 (m, 5H), 7.28 (d, J = 1.7 Hz, 1H), 7.24 (d, J = 1.7 Hz, 1H), 7.16 (d, J = 1.7 Hz, 1H), 7.08 (d, J = 1.7 Hz, 1H), 6.90 (d, J = 1.7 Hz, 1H), 5.02 (m, 2H), 4.14 (m, 1H), 3.95 (s, 3H), 3.85 (s, 3H), 3.84 (s, 3H), 3.83 (s, 3H), 3.73 (s, 3H), 3.02 (m, 2H), 2.63 (m, 2H); 13C NMR: δ 167.0, 160.8, 158.5, 158.4, 155.7, 136.8, 135.7, 134.0, 128.3, 127.8, 127.7, 123.05, 122.97, 122.5, 122.2, 121.1, 120.7, 118.64, 118.60, 118.5, 108.4, 104.9, 65.6, 50.9, 46.7, 42.0, 38.2, 36.2, 36.1, 36.0, 34.9; HRMS (FAB+) calc'd for C36H42N11O8 [M+H]+ 756.3218, found 756.3211.
BocHN-(R)β-CbzHNγ-ImPyPyPy-CO2H (11)
A solution of BocHN-(R)β-CbzHNγ-ImPyPyPy-CO2Me 10 (200 mg, 0.234 mmol) dissolved in 1,4-dioxane (2.3 mL) and aqueous NaOH (1N, 2.3 mL, 2.3 mmol) was stirred at 42 °C for 3 h. The solution was then added to distilled H2O (5 mL) pre-acidified with aqueous HCl (1N, 2.3 mL, 2.3 mmol), yielding a precipitate that was isolated by centrifugation (~ 4500 rpm). The residual solid was again suspended in distilled H2O (10 mL) and collected by centrifugation (repeated 2X). The resultant solid, which contained a small amount of residual H2O, was frozen and lyophilized to dryness, and then suspended in excess anhydrous Et2O, filtered, and the filter cake washed with copious amounts of anhydrous Et2O. Drying of the tan solid in vacuo yielded BocHN-(R)β-CbzHNγ-ImPyPyPy-CO2H 11 (187 mg, 95%). 1H NMR: δ 12.15 (s, 1H), 10.21 (s, 1H), 10.00 (s, 1H), 9.96 (s, 1H), 9.92 (s, 1H), 7.44 (s, 1H), 7.42 (d, J = 1.7 Hz, 1H), 7.31-7.29 (m, 5H), 7.28 (d, J = 1.5 Hz, 1H), 7.24 (d, J = 1.5 Hz, 1H), 7.16 (d, J = 1.5 Hz, 1H), 7.08 (m, 2H), 6.85 (d, J = 1.7 Hz, 1H), 6.82 (t, J = 5.7 Hz, 1H), 4.98 (s, 2H), 3.95 (m, 4H), 3.85 (s, 3H), 3.84 (s, 3H), 3.81 (s, 3H), 3.03 (m, 2H), 2.46 (m, 2H), 1.36 (s, 9H); 13C NMR: δ 167.8, 162.0, 158.44, 158.38, 155.80, 155.77, 155.4, 137.1, 136.0, 133.9, 128.3, 127.7, 127.6, 123.0, 122.7, 122.6, 122.2, 121.2, 120.2, 119.5, 118.6, 118.5, 114.0, 108.4, 104.87, 104.83, 77.7, 65.1, 48.8, 43.5, 38.2, 36.2, 36.13, 36.06, 34.9, 28.2; HRMS (FAB+) calc'd for C40H47N11O10 [M•]+ 841.3507, found 841.3498.
BocHN-(R)β-CbzHNγ-ImPyPyPy-(R)β-CbzHNγ-ImPyPyPy-CO2Me (12)
A solution of BocHN-(R)β-CbzHNγ-ImPyPyPy-CO2H 11 (25 mg, 0.029 mmol) and PyBOP (17 mg, 0.031 mmol) in DMF (150 μL) and DIEA (16 μL, 0.089 mmol) was stirred at 23°C for 20 min. The solution was then treated with solid (powdered) HCl•H2N-(R)β-CbzHNγ-ImPyPyPy-CO2Me 10 (25 mg, 0.031 mmol) and stirred at 23°C for 2 h. The solution was then added to distilled H2O (10 mL) pre-acidified with aqueous HCl (1N, 1 mL, 1 mmol), yielding a precipitate that was isolated by centrifugation (~ 4500 rpm). The residual solid was again suspended in distilled H2O (10 mL) and collected by centrifugation (repeated 3X). The resultant solid, which contained a small amount of residual H2O, was frozen and lyophilized to dryness. The solid was triturated with anhydrous Et2O and filtered over a sintered glass funnel. The resultant solid was washed with copious amounts of anhydrous Et2O and dried in vacuo to yield BocHN-(R)β-CbzHNγ-ImPyPyPy-(R)β-CbzHNγ-ImPyPyPy-CO2Me 12 as a tan solid (44 mg, 94%). 1H NMR: δ 10.20 (s, 1H), 10.16 (s, 1H), 9.98 (s, 2H), 9.94-9.91 (m, 4H), 7.99 (m, 1H), 7.46 (d, J = 1.7 Hz, 1H), 7.45 (s, 1H), 7.44 (s, 1H), 7.32-7.14 (m, 18H), 7.07 (m, 2H), 7.03 (d, J = 8.3 Hz, 1H), 6.92 (s, 1H), 6.90 (d, J = 2.0 Hz, 1H), 6.80 (t, J = 5.6 Hz, 1H), 4.99 (m, 4H), 4.10 (m, 1H), 3.95 (m, 7H), 3.85-3.83 (m, 15H), 3.79 (s, 3H), 3.73 (s, 3H), ~3.30 (m, 2H, obstructed by H2O peak), 3.04 (m, 2H), 2.53-2.44 (m, 4H, partially obstructed by NMR solvent), 1.36 (s, 9H); 13C NMR: δ 167.9, 167.8, 161.6, 160.8, 158.5, 158.44, 158.42, 155.8, 155.6, 155.5, 137.1, 136.0, 134.00, 133.98, 128.3, 127.7, 127.63, 127.60, 123.11, 123.07, 123.00, 122.80, 122.77, 122.5, 122.3, 122.2, 122.1, 121.3, 120.8, 118.69, 118.66, 118.62, 118.52, 118.0, 114.1, 108.4, 104.9, 104.8, 104.5, 77.8, 65.21, 65.16, 50.9, 48.83, 48.78, 43.6, 42.2, 38.4, 38.2, 36.2, 36.10, 36.07, 36.0, 34.9, 28.2; HRMS (TOF-ESI+) calc'd for C76H87N22O17 [M+H]+ 1579.6620, found 1579.6580.
BocHN-(R)β-CbzHNγ-ImPyPyPy-(R)β-CbzHNγ-ImPyPyPy-CO2H (13)
A solution of BocHN-(R)βCbzHNγ-ImPyPyPy-(R)β-CbzHNγ-ImPyPyPy-CO2Me 12 (25 mg, 0.0158 mmol) dissolved in 1,4-dioxane (376 μL) and aqueous NaOH (1N, 253 μL, 0.253 mmol) was stirred at 40 °C for 4 h. The solution was then added to distilled H2O (5 mL) pre-acidified with aqueous HCl (1N, 253 μL, 0.253 mmol), yielding a precipitate which was diluted with another 15 mL H2O and was then isolated by centrifugation (~ 4500 rpm). The resultant solid, which contained a small amount of residual H2O, was frozen and lyophilized to dryness, and then suspended in excess anhydrous Et2O, triturated, filtered, and the filter cake washed with copious amounts of anhydrous Et2O. Drying of the tan solid in vacuo yielded BocHN-(R)β-CbzHNγ-ImPyPyPy-(R)β-CbzHNγ-ImPyPyPy-CO2H 13 (23 mg, 93%). 1H NMR: δ 12.13 (br s, 1H), 10.23 (s, 1H), 10.20 (s, 1H), 9.98-9.90 (m, 6H), 8.01 (m, 1H), 7.443 (s, 1H), 7.439 (s, 1H), 7.42 (d, J = 1.7 Hz, 1H), 7.30-7.15 (m, 18H), 7.07 (m, 3H), 6.92 (m, 1H), 6.84 (d, J = 2.0 Hz, 1H), 6.81 (t, J = 5.6 Hz, 1H), 5.00 (m, 2H), 4.98 (s, 2H), 4.10 (m, 1H), 3.95 (m, 7H), 3.85-3.83 (m, 12H), 3.81 (s, 3H), 3.78 (s, 3H), ~3.30 (m, 2H, obstructed by H2O peak), 3.03 (m, 2H), 2.53 (m, 2H), 2.46 (m, 2H), 1.36 (s, 9H); 13C NMR: δ 167.94, 167.87, 162.4, 162.0, 161.6, 158.54, 158.50, 158.43, 155.8, 155.75, 155.74, 155.6, 155.5, 137.1, 136.0, 133.91, 133.90, 128.3, 127.7, 127.60, 127.57, 123.10, 123.07, 122.8, 122.7, 122.6, 122.3, 122.24, 122.17, 121.16, 121.15, 120.3, 119.5, 118.64, 118.61, 118.5, 118.0, 114.10, 114.06, 108.5, 105.0, 104.5, 77.8, 65.2, 65.1, 48.8, 43.6, 42.2, 38.4, 38.2, 36.2, 36.14, 36.10, 36.08, 36.0, 35.8, 34.9, 28.2; HRMS (TOF-ESI+) calc'd for C75H86N22O17 [M+2H]2+/2 783.3271, found 783.3237.
BocHN-(R)β-CbzHNγ-ImPyPyPy-(R)β-CbzHNγ-ImPyPyPy-CO2Pfp (14)
A solution of BocHN-(R)β-CbzHNγ-ImPyPyPy-(R)β-CbzHNγ-ImPyPyPy-CO2H 13 (250 mg, 0.160 mmol) and DCC (66 mg, 0.320 mmol) in CH2Cl2 (8.8 mL) was stirred at 23°C for 45 min. The solution was then treated with DMAP (2 mg, 0.016 mmol) followed by pentafluorophenol (175 mg, 0.950 mmol) and stirred at 23°C for 12 h. The reaction mixture was then loaded onto a silica gel column with CH2Cl2 and eluted with step gradients of 100% CH2Cl2 to 100% acetone with incremental steps of 5% acetone. The product was concentrated in vacuo to yield BocHN-(R)β-CbzHNγ-ImPyPyPy-(R)β-CbzHNγ-ImPyPyPy-CO2Pfp 14 as a brown solid (221 mg, 80%). 1H NMR: δ 10.20 (s, 1H), 10.16 (s, 1H), 10.08 (s, 1H), 9.99-9.91 (m, 5H), 7.99 (m, 1H), 7.73 (d, J = 1.7 Hz, 1H), 7.444 (s, 1H), 7.440 (s, 1H), 7.30-7.12 (m, 20 H), 7.06 (d, J = 1.2 Hz, 1H), 7.03 (d, J = 8.5 Hz, 1H), 6.92 (s, 1H), 6.80 (t, J = 5.4 Hz, 1H), 5.00 (m, 2H), 4.98 (m, 2H), 4.11 (m, 1H), 3.95 (m, 7H), 3.88 (s, 3H), 3.86-3.84 (m, 12H), 3.78 (s, 3H), ~3.30 (m, 2H, obstructed by H2O peak), 3.04 (m, 2H), 2.52 (m, 2H), 2.46 (m, 2H), 1.36 (s, 9H); HRMS (TOF-ESI+) calc'd for C81H85F5N17[M+2H]2+/2 866.3192, found 866.3236.
cyclo-(-ImPyPyPy-(R)β-H2Nγ-ImPyPyPy-(R)β-H2Nγ-) (1)
A solution of BocHN-(R)β-CbzHNγ-ImPyPyPy-(R)β-CbzHNγ-ImPyPyPy-CO2Pfp 14 (84 mg, 0.049 mmol) in anhydrous CF3CO2H:CH2Cl2 (1:1, 4 mL) was stirred at 23 °C for 10 min prior to being concentrated to dryness in a 500 mL round bottom flask. The residue was then dissolved in cold (0 °C) DMF (10 mL), followed by immediate dilution with MeCN (300 mL) and DIEA (1.6 mL). The reaction mixture was Left at 23 °C for 3 days without stirring. (Note: The solution turns cloudy as the macrocycliztion proceedes.) The reaction mixture was concentrated to a volume of 11 mL and added to a solution of H2O (30 mL) and aqueous HCl (1N, 9.2 mL) at 0 °C. The protected intermediate cyclo-(-ImPyPyPy-(R)β-CbzHNγ-ImPyPyPy-(R)β-CbzHNγ-) 15 was isolated by centrifugation (~ 4500 rpm), lyophilized to dryness, and then suspended in excess anhydrous Et2O, triturated, filtered, and the filter cake washed with copious amounts of anhydrous Et2O. Drying of the tan solid in vacuo yielded the protected intermediate cyclo-(-ImPyPyPy-(R)β-CbzHN γ-ImPyPyPy-(R)βCbzHNγ-) 15, HRMS (TOF-ESI+) calc'd for C70H76N22O14 [M+2H]2+/2 724.2956, found 724.2925. This material was immediately deprotected by dissolving in CF3CO2H (2 mL) followed by addition of CF3SO3H (100 μL) at 23 °C for 5 min. The solution was then frozen and DMF (2 mL) was layered over the frozen solution. The thawed solution was diluted with H2O (6 mL) and purified by reverse-phase HPLC to give a white solid after lyophilization. The solid was suspended in excess anhydrous Et2O, triturated, filtered, and the filter cake washed with copious amounts of anhydrous Et2O. Drying of the white solid in vacuo yielded cyclo-(-ImPyPyPy-(R)β-H2N γ-ImPyPyPy-(R)β-H2Nγ-) 1 (46 mg, 68%). 1H NMR: δ 10.56 (s, 2H), 9.91 (s, 4H), 9.88 (s, 2H), 8.17 (t, J = 5.6 Hz, 2H), 7.96 (m, 6H), 7.40 (s, 2H), 7.31 (d, J = 1.6 Hz, 2H), 7.27 (d, J = 1.6 Hz, 2H), 7.19 (d, J = 1.6 Hz, 2H), 7.00 (d, J = 1.7 Hz, 2H), 6.96 (d, J = 1.6 Hz, 2H), 6.94 (d, J = 1.7 Hz, 2H), 3.94 (s, 6H), 3.83 (s, 12H), 3.80 (s, 6H), 3.71 - 3.66 (m, 2H), 3.49 - 3.27 (m, 4H, partially obstructed by H2O peak), 2.79 (dd, J = 16.1 Hz, 6.0 Hz, 2H), 2.60 (dd, J = 15.2 Hz, 5.2 Hz, 2H). HRMS (TOF-ESI+) calc'd for C54H63N22O10 [M+H]+ 1179.5098, found 1179.5087.
cyclo-(-ImPyPyPy-(R)β-AcHNγ-ImPyPyPy-(R)β-H2Nγ-) (2) and cyclo-(-ImPyPyPy-(R)β-AcHNγ-ImPyPyPy-(R)β-AcHNγ-) (3)
A solution of cyclo-(-ImPyPyPy-(R)β-H2Nγ-ImPyPyPy-(R β-H2Nγ-) 1 (2.81 mg, 2.0 μmol) in anhydrous NMP (200 μL) and DIEA (20 μL) at 23 °C was treated with a solution of Ac2O in NMP (0.122 M, 6.8 μL). After 10 min the reaction mixture was treated with another 6.8 μL of Ac2O in NMP (0.122 M) and allowed to stand for 5 hr. The reaction mixture was then diluted to a volume of 10 mL by addition of a 4:1 solution of aqueous CF3CO2H (0.1% v/v):MeCN (5 mL), followed by additional aqueous CF3CO2H (0.1% v/v, 5 mL), and then purified by reverse-phase HPLC to yield cyclo-(-ImPyPyPy-(R)β-H2Nγ-ImPyPyPy-(R)β-H2Nγ-) 1 (363 nmol, 18%), cyclo-(-ImPyPyPy-(R)β-AcHNγ-ImPyPyPy-(R)β-H2N2g-) 2 (800 nmol, 40%), and cyclo-(-ImPyPyPy-(R)β-AcHNγ-ImPyPyPy-(R)β-AcHNγ-) 3 (432 nmol, 22%). Cyclo-(-ImPyPyPy-(R)β-AcHNγ-ImPyPyPy-(R)β-H2N2g-) 2 HRMS (TOF-ESI+)calc'd for C56H65N22O11 [M+H]+ 1221.5203, found 1221.5204. Cyclo-(-ImPyPyPy-(R)β-AcHNγ-ImPyPyPy-(R)βAcHNγ-) 3 HRMS (TOF-ESI+) calc'd for C58H68N22O12 [M+2H]2+/2 633.2646, found 633.2631.
ImPyPyPy-(R)β-H2Nγ-ImPyPyPy-(+)-IPA (4)
Prepared as described in the preceding manuscript.11
ImPyPyPy-(R)β-AcHNγ-ImPyPyPy-(+)-IPA (5)
A solution of polyamide 411 (7.4 mg, 5.03 μmoles, assumes 4 as the mono-CF3CO2H salt) in DMF (1.76 mL) was treated with a solution of Ac2O in pyridine (10% v/v, 240 μL, 0.254 mmoles Ac2O). The solution was allowed to stand at 23 °C for 30 min, and then acidified with aqueous CF3CO2H (15% v/v, 2 mL). After 5 min the solution was further diluted with distilled H2O (5 mL), purified by preparative RP-HPLC, and lyophilized to dryness. Suspension of the residual solid in anhydrous Et2O, following by filtration and drying under high vacuum yielded 5 (6.7 mg, 95%). HRMS (FAB+) calc'd for C67H79N22O13 [M+H]+ 1399.6191, found 1399.6181.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (GM27681). D.M.C. is grateful for a Caltech Kanel predoctoral fellowship. D.A.H. thanks the California Tobacco-Related Disease Research Program (16FT-0055) for a postdoctoral fellowship. J.W.P. is grateful to the California Breast Cancer Research Program for a predoctoral fellowship. C.D. thanks the Alexander von Humboldt foundation for a postdoctoral research fellowship. The National Science Foundation Chemistry Research Instrumentation and Facilities Program (CHE-0541745) is acknowledged for providing the UPLC-MS instrument.
Footnotes
Supporting Information Available: A summary of ADMET results, 1H and 13C NMR spectra, and analytical HPLC purity analyses of selected compounds. This material is available free of charge via the Internet at http://pubs.acs.org. The full ADMET report including experimental conditions can be accessed via the Dervan laboratory homepage at http://dervan.caltech.edu.
REFERENCES
- (1).Dervan PB. Bioorg. Med. Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- (2).Dervan PB, Edelson JA. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- (3).(a) Trauger JW, Baird EE, Dervan PB. Nature. 1996;382:559–561. doi: 10.1038/382559a0. [DOI] [PubMed] [Google Scholar]; (b) White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Nature. 1998;391:468–470. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]; (c) Kielkopf CL, Baird EE, Dervan PB, Rees DC. Nat. Struct. Biol. 1998;5:104–109. doi: 10.1038/nsb0298-104. [DOI] [PubMed] [Google Scholar]; (d) Kielkopf CL, White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB, Rees DC. Science. 1998;282:111–115. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]
- (4).Hsu CF, Phillips JW, Trauger JW, Farkas ME, Belitsky JM, Heckel A, Olenyuk BZ, Puckett JW, Wang CC, Dervan PB. Tetrahedron. 2007;63:6146–6151. doi: 10.1016/j.tet.2007.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).(a) Belitsky JM, Leslie SJ, Arora PS, Beerman TA, Dervan PB. Bioorg. Med. Chem. 2002;10:3313–3318. doi: 10.1016/s0968-0896(02)00204-3. [DOI] [PubMed] [Google Scholar]; (b) Crowley KS, Phillion DP, Woodard SS, Scheitzer BA, Singh M, Shabany H, Burnette B, Hippenmeyer P, Heitmeier M, Bashkin JK. Bioorg. Med. Chem. Lett. 2003;13:1565–1570. doi: 10.1016/s0960-894x(03)00152-5. [DOI] [PubMed] [Google Scholar]; (c) Best TP, Edelson BS, Nickols NG, Dervan PB. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Edelson BS, Best TP, Olenyuk B, Nickols NG, Doss RM, Foister S, Heckel A, Dervan PB. Nucleic Acids. Res. 2004;32:2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Xiao X, Yu P, Lim HS, Sikder D, Kodadek T. Angew. Chem. Int. Ed. Engl. 2007;46:2865–2868. doi: 10.1002/anie.200604485. [DOI] [PubMed] [Google Scholar]; (f) Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Nucleic Acids. Res. 2007;35:363–370. doi: 10.1093/nar/gkl1042. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Dose C, Farkas ME, Chenoweth DM, Dervan PB. J. Am. Chem. Soc. 2008;130:6859–6866. doi: 10.1021/ja800888d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Hsu CF, Dervan PB. Bioorg. Med. Chem. Lett. 2008;18:5851–5855. doi: 10.1016/j.bmcl.2008.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).(a) Gottesfeld JM, Melander C, Suto RK, Raviol H, Luger K, Dervan PB. Sequencespecific recognition of DNA in the nucleosome by pyrrole-imidazole polyamides. J. Mol. Biol. 2001;309:615–629. doi: 10.1006/jmbi.2001.4694. [DOI] [PubMed] [Google Scholar]; (b) Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, Luger K. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J. Mol. Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]; (c) Edayathumangalam RS, Weyermann P, Gottesfeld JM, Dervan PB, Luger K. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6864–6869. doi: 10.1073/pnas.0401743101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Dudouet B, Burnett R, Dickinson LA, Wood MR, Melander C, Belitsky JM, Edelson B, Wurtz N, Briehn C, Dervan PB, Gottesfeld JM. Chem. Biol. 2003;10:859–867. doi: 10.1016/j.chembiol.2003.09.001. [DOI] [PubMed] [Google Scholar]
- (7).(a) Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin WG, Dervan PB. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kageyama Y, Sugiyama H, Ayame H, Iwai A, Fujii Y, Huang LE, Kizaka-Kondoh S, Hiraoka M, Kihara K. Acta. Oncol. 2006;45:317–324. doi: 10.1080/02841860500486648. [DOI] [PubMed] [Google Scholar]; (c) Nickols NG, Jacobs CS, Farkas ME, Dervan PB. ACS Chem. Biol. 2007;2:561–571. doi: 10.1021/cb700110z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Nickols NG, Dervan PB. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10418–10423. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).(a) Matsuda H, Fukuda N, Ueno T, Tahira Y, Ayame H, Zhang W, Bando T, Sugiyama H, Saito S, Matsumoto K, et al. O. J. Am. Soc. Neph. 2006;17:422–432. doi: 10.1681/ASN.2005060650. [DOI] [PubMed] [Google Scholar]; (b) Yao EH, Fukuda N, Ueno T, Matsuda H, Matsumoto K, Nagase H, Matsumoto Y, Takasaka A, Serie K, Sugiyama H, Sawamura T. Hypertension. 2008;52:86–92. doi: 10.1161/HYPERTENSIONAHA.108.112797. [DOI] [PubMed] [Google Scholar]
- (10).(a) Cho J, Parks ME, Dervan PB. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10389–10392. doi: 10.1073/pnas.92.22.10389. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang Q, Dwyer TJ, Tsui V, Case DA, Cho J, Dervan PB, Wemmer DE. J. Am. Chem. Soc. 2004;126:7958–7966. doi: 10.1021/ja0373622. [DOI] [PubMed] [Google Scholar]; (c) Herman DM, Turner JM, Baird EE, Dervan PB. J. Am. Chem. Soc. 1999;121:1121–1129. [Google Scholar]; (d) Melander C, Herman DM, Dervan PB. Chem. Eur. J. 2000;6:4487–4497. doi: 10.1002/1521-3765(20001215)6:24<4487::aid-chem4487>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- (11).Chenoweth DM, Harki DA, Dervan PB. Solution-phase synthesis of pyrrole-imidazole polyamides. Submitted. doi: 10.1021/ja901307m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).(a) Baird EE, Dervan PB. J. Am. Chem. Soc. 1996;118:6141–6146. [Google Scholar]; (b) Wurtz NR, Turner JM, Baird EE, Dervan PB. Org. Lett. 2001;3:1201–1203. doi: 10.1021/ol0156796. [DOI] [PubMed] [Google Scholar]; (c) Jaramillo D, Liu Q, Aldrich-Wright J, Tor Y. J. Org. Chem. 2004;69:8151–8153. doi: 10.1021/jo048686r. [DOI] [PubMed] [Google Scholar]
- (13).Trauger JW, Dervan PB. Methods Enzymol. 2001;340:450–466. doi: 10.1016/s0076-6879(01)40436-8. [DOI] [PubMed] [Google Scholar]
- (14).For examples of polyamides with Ka values > 2 × 1010 M-1 and a discussion of the limitations of quantitative DNase I footprint titrations, please refer to reference 5g. An analogous polyamide to 4, ImPyPyPy-(R)β-H2Nγ-ImPyPyPy-β-Dp, was found to have Ka > 2 × 1010 M-1.5g Additionally, previous studies with cyclic polyamide cyclo-(-ImPyPyPy-(R)α-H2Nγ-ImPyPyPy-γ-) found Ka values far exceeding 2 × 1010 M-1 by DNase I footprint titrations.10c,10d Cyclic polyamide 1 possesses dual β-amino functionalities; a modification that yields even greater DNA binding affinities compared with α-amino and unsubstituted γ-turns for hairpin polyamides of sequence ImPyPyPy-γ-ImPyPyPy.5g The DNA binding affinity of 1 most likely supersedes that of predecessor cyclo-(-ImPyPyPy-(R)α-H2Nγ-ImPyPyPy-γ-).
- (15).(a) Pilch DS, Poklar N, Gelfand CA, Law SM, Breslauer KJ, Baird EE, Dervan PB. Proc. Natl. Acad. Sci. USA. 1996;93:8306–8311. doi: 10.1073/pnas.93.16.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pilch DS, Poklar N, Baird EE, Dervan PB, Breslauer KJ. Biochemistry. 1999;38:2143–2151. doi: 10.1021/bi982628g. [DOI] [PubMed] [Google Scholar]
- (16).Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4758–4763. doi: 10.1073/pnas.0401123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).For a review of pharmacokinetics in drug discovery see: Ruiz-Garcia A, Bermejo M, Moss A, Casabo VG. J. Pharm. Sci. 2008;97:654–690. doi: 10.1002/jps.21009..
- (18). Apredica, 313 Pleasant St., Watertown, MA 02472 ( http://www.apredica.com/)
- (19).Harki DA, Satyamurthy N, Stout DB, Phelps ME, Dervan PB. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13039–13044. doi: 10.1073/pnas.0806308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.